Abstract
In a wide range of organisms, double-stranded RNA triggers posttranscriptional gene silencing or RNA interference (RNAi). Small interfering RNAs, the 21-nt double-stranded RNA intermediates of this natural pathway, have became a powerful tool to knock down specific gene expression in mammalian cell lines and potentially will be useful for the analysis of loss-of-function phenotypes. In mammalian primary neuronal cultures, where genetic manipulations are especially difficult, RNAi might be developed into a highly efficacious tool to study the roles of specific genes in neuron development and functioning. Neurons, however, have been considered the most resistant to RNAi. We report here an application of RNAi to postmitotic primary neuronal cultures. Synthetic siRNA can be readily introduced into neurons and effectively inhibit the expression of endogenous and transfected genes.
Double-stranded RNAs (dsRNAs) are remarkably effective at suppressing specific gene expression in Caenorhabditis elegans, Drosophila melanogaster, Trypanosoma brucei, and plants by a pathway involving RNA interference (RNAi) or sequence-specific posttranscriptional gene silencing (1–3). dsRNAs are cleaved by ribonuclease III into 21–22 nucleotide RNA duplexes or small interfering RNAs (siRNAs). These molecules trigger the degradation of the cognate mRNA (4, 5). Although dsRNA induces the global shut-down of protein synthesis in mammalian cells, directly introducing siRNAs of less then 30 nt can suppress expression of specific endogenous and heterologous genes in mammalian cell lines (6, 7). From these studies, the investigators concluded that RNA interference could occur in mammalian cells. The mechanism of the siRNA-triggered RNAi in those cells remains to be elucidated. Until recently, existing approaches to suppress specific gene expression in mammalian cells, mainly antisense and dominant–negative, have proven inefficient and inconsistent. Therefore, RNAi, with its characteristic efficacy at very low concentrations of siRNA, in the low nanomolar range, holds great promise for exploring gene function in mammalian cell cultures. A number of genes were successfully knocked-down in mammalian somatic and embryonic cell lines, including HeLa, HEK293, and P19 (8, 9). However, for reasons that are unclear, neurons seemed more resistant to RNAi than other cell types, perhaps because of differences related to the RNA transport across the cell membrane or the RNAi pathway in these cells. Although RNAi is systemic in C. elegans when fed or injected to whole animals, and even silences genes into the next generation, worm neurons seemed resistant to RNAi (10). However, high concentrations of dsRNA (15 μg/ml) can induce a knockdown of targeted gene expression in proliferating and differentiating nematode neuronal culture (11). In D. melanogaster, genomic cDNA hybrids predicted to produce dsRNA are able to target genes expressed in neurons (12). Here, we demonstrate that the introduction of very low concentrations of siRNAs into dissociated postmitotic cultures prepared from the rat hippocampus and forebrain can be effective in suppressing endogenous and heterologous genes.
Materials and Methods
Cortical and Hippocampal Cell Culture.
Pregnant embryonic day 18 (E18) Sprague–Dawley rats were killed by inhalation of CO2, and the embryos were removed immediately by Cesarean section. Cerebral cortices and hippocampi were removed and digested in 0.25% trypsin in Hepes-buffered Hanks' balanced salt solution (HBSS) without calcium or magnesium at 37°C for 15 min. The tissue was washed 3× with HBSS and manually dissociated with a fire-bored Pasteur pipette. Cells were plated at a concentration 12,000–20,000/cm2 on polyl-lysine-coated coverslips in a plating medium containing DMEM and 5% (vol/vol) FBS. After 3 h of incubation, the medium was changed to Neurobasal, containing B27 supplement and 0.5 mM glutamine. All experiments were initiated 5–8 days after plating. Very few glial cells were observed in these cultures.
siRNA Preparation and Transfections.
siRNAs corresponding to pEGFP or dsRed2 reporter genes and to MAP2 or YB-1 mRNAs were designed as recommended (13), with 5′ phosphate, 3′ hydroxyl, and two base overhangs on each strand; they were chemically synthesized by Xeragon. The following gene-specific sequences were used successfully: Si-GFP sense 5′-CAAGCUGACCCUGAAGUUCUU-3′ and antisense 5′-GAACUUCAGGGUCAGCUUGUG-3′; Si-DsRed sense 5′-AGUUCCAGUACGGCUCCAAUU-3′ and antisense 5′-UUGGAGCCGUACUGGAACUUG-3′, cognate for MAP2, siRNA1 sense 5′-CGAGAGGAAAGACGAAGGAUU-3′ and antisense 5′-UCCUUCGUCUUUCCUCUCGUG-3′; siRNA2 sense 5′-CAGGGCACCUAUUCAGAUAUU-3′ and antisense 5′-UAUCUGAAUAGGUGCCCUGUG-3′. Annealing for duplex siRNA formation was performed as described (6).
Cotransfections of reporter plasmids and siRNA were performed with Lipofectamine 2000 (Life Technologies) in 6-well plates. Briefly, Lipofectamine diluted in Opti-MEM was applied to the plasmids/dsRNA mixture, and the formulation was continued for 25 min. Per well, 1 μg of pEGFP-C2, 1 μg of pDsRed2, and 0.01–0.2 μg of 21-bp dsRNA (siRNA) formulated with 2 μl of Lipofectamine were applied in the final volume of 1.7 ml. The medium was changed to Neurobasal 2 h after transfection, and cells were incubated for 24–48 h after fixation and analysis. Transfection efficiencies as determined by 4′,6-diamidino-2-phenylindole staining were from 1% to 8%.
Transfections of siRNA for endogenous gene targeting were carried out with TransMessenger transfection reagent (Qiagen, Chatsworth, CA). siRNA (1 μg per well) was condensed with Enhanser R and formulated with 4 μl of TransMessenger reagent, according to the manufacturer's instructions. The transfection complex was diluted in 900 μl of Neurobasal and was added directly to the cells; it was replaced with Neurobasal after 2 h. Cells were stained and analyzed 48–68 h after transfections. The results were confirmed in three independent experiments.
Immunocytochemistry.
Double immunofluorescence labeling of MAP2 and several proteins was performed. Neurons grown on glass coverslips were washed with PBS, fixed in 4% (wt/vol) paraformaldehyde, permeabilized with 0.25% Triton X-100, washed two more times with PBS, and blocked for 30 min with 10% (vol/vol) neural goat serum. Cells were incubated with primary antibodies [1:200 in 1% (vol/vol) neural goat serum in PBS] overnight at 4°C and washed three times with PBS. Mouse monoclonal anti-MAP2 antibody was produced in our laboratory (14). Rabbit polyclonal anti-YB1 antibody was generously provided by V. Evdokimova (McGill Univ., Montreal, QC, Canada); rabbit polyclonal anti-cortactin was purchased from Santa Cruz Biotechnology. MAP2 was visualized with Alexa 488- or Alexa 594-conjugated goat anti-mouse secondary antibody [1:500 in 1% (vol/vol) goat serum in PBS; Molecular Probes]. Other antibodies were detected with Alexa 488- or Alexa 594-conjugated goat anti-rabbit secondary antibody. Actin was detected with Alexa Fluor 594 phalloidin (1:100; Molecular Probes). Finally, cells were washed in PBS and mounted on the microscope with Antifade mounting medium.
Microscopy and Image Analysis.
Fluorescent microscopy was performed with a confocal laser scanning unit coupled to a Zeiss Axiovert S100. Images were recorded with equal exposure times for specific reporter plasmids or for specific antibodies (in nonsaturating conditions). In each cotransfection experiment, at least 50 randomly chosen transfected neurons were analyzed. In each MAP2-targeting experiment, at least 70 random neurons per experimental condition were analyzed, and gene expression was quantified in both control and targeted cells. Quantification of pixel intensity was performed with Adobe PHOTOSHOP software and normalized to a background.
Results
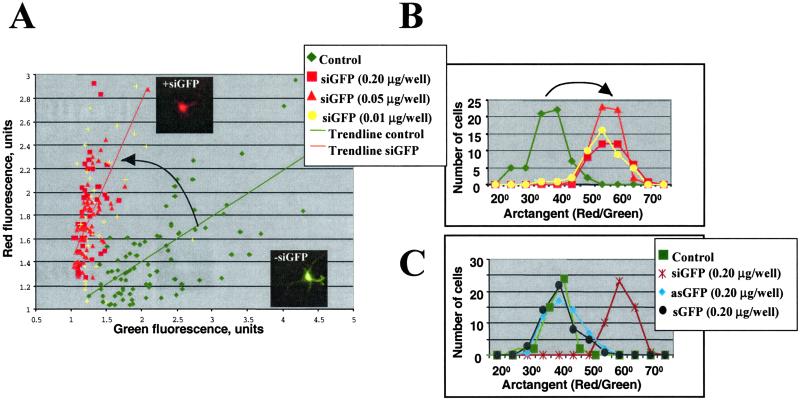
First, to assess the effectiveness of RNAi on cultured neurons, the 21-nt sense and antisense ssRNAs targeted against EGFP-C2 and DsRed2 reporter vectors were selected from the coding regions and designed as described in Materials and Methods. Each strand was synthesized separately, and pairs were annealed to create the duplex dsRNAs with the characteristics of siRNAs (13). Primary neurons were transfected with these two plasmids and one cognate dsRNA 5–8 days after plating. Two hours after transfection, the medium was changed to Neurobasal; cells were fixed, and EGFP and DsRed2 expressions were monitored 24–48 h later. Fluorescent microscopy showed that EGFP or DsRed2 expressions were significantly and specifically inhibited by their corresponding dsRNAs, but not by unrelated dsRNAs in nearly all transfected neurons (Fig. 1 A and B show the data for GFP targeting). At 24 h after cotransfection with two plasmids, most transfected neurons (1–8% efficiency) expressed a high level of GFP and a lower level of DsRed and, therefore, appeared yellow-green when merged (the typical cell is shown in Inset in Fig. 1A). siRNA cognate to green fluorescent protein (siGFP) significantly reduced enhanced green fluorescent protein (EGFP) expression without affecting dsRed expression and, therefore, the cells appeared red. To quantify the effect of siRNA on a GFP expression, images were recorded with equal exposure times for specific reporter plasmids under nonsaturating conditions, with further monitoring of green and red fluorescence for randomly chosen transfected cells. For each cell, green and red fluorescence were normalized to a background and plotted (Fig. 1A). In each experiment, at least 50 randomly chosen transfected control and targeted neurons were compared. This analysis demonstrated that GFP expression was specifically inhibited by the cognate dsRNA duplex as detected by a 42% reduction in the green fluorescence intensity, whereas the red fluorescence remained unchanged. For each analyzed cell, the ratio between red and green fluorescence was calculated as an arctangent function; the results are presented as a distribution function in Fig. 1B. The shift in this distribution represents specific suppression of GFP expression by siGFP. Moreover, almost nonoverlapping distribution functions of control vs. targeted cells clearly indicate that nearly all transfected neurons were affected by siGFP even in its lowest concentration (6 ng/ml). The suppression was specific for dsRNA duplex: both sense and antisense ssRNAs did not inhibit expression of the reporter gene (Fig. 1C).
Figure 1.
Effect of 21nt-siRNA targeting GFP expression (siGFP) in primary cortical neurons. (A) Primary neurons were cotransfected with pEGFP and DsRed2 plasmids and with siRNA cognate to EGFP (siGFP). For each transfected cell, green and red fluorescence were normalized to a background and plotted. In each experiment, at least 50 randomly chosen transfected control (−siGFP) and targeted (+siGFP) neurons were analyzed. Typical control and targeted cells are shown in Insets. (B) For each cell in A, an arctangent function (that represents the ratio between red and green fluorescence) was calculated, and the results are presented as a distribution function. The shift in this distribution represents specific suppression of GFP expression by siGFP. Similar effects were observed with siGFP concentrations from 0.006–0.12 μg/ml. (C) Single strands of siGFP (sense-sGFP and antisense-asGFP 21nt-RNAs) do not suppress GFP expression.
Similar analysis of DsRed2 targeting by its cognate dsRNA demonstrated some overlap between control and targeted cells when plotted as above, with DsRed2 expression inhibited specifically at ≈30% (data not shown). This was due to the later onset of DsRed2 expression and the high variability of its expression level between different control cells.
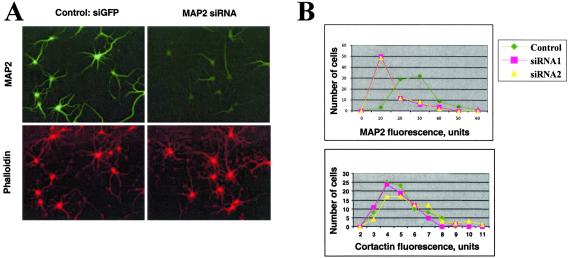
Next, we tested the effect of siRNAs on microtubule-associated protein 2 (MAP2) and RNA-binding protein YB-1 (also called p50) endogenous mRNAs. In the course of these experiments, we noticed that siRNA could be readily introduced into neurons when compared with plasmid DNA. Therefore, there was no need to introduce the reporter vector as a marker of cells transfected with siRNA. The endogenous mRNA for MAP2 was targeted by transfections of cognate 21-nucleotide dsRNAs with TransMessenger transfection reagent. Double immunofluorescence labeling of MAP2 and three unrelated proteins (cortactin, actin, YB-1) was performed 48–68 h after transfections. The expression of MAP2 was specifically reduced by the cognate dsRNA duplex in 70–80% of cells, whereas expression of unrelated proteins was unaffected (Fig. 2A). Among these cells, the decrease in MAP2 fluorescent intensity was ≈4-fold. The fluorescent intensity over the total cell population varied greatly because of variation in transfection efficiency and possibly differential sensitivity of the neurons to siRNA. Nevertheless, even when the fluorescent intensity was measured over the total population of cells, a decrease in MAP2 fluorescence was readily detectable (Fig. 2B). Remarkably, this statistical analysis revealed that two different siRNA cognate to MAP2 (siRNA1 and siRNA2) demonstrated the identical inhibitory effect on MAP2 expression.
Figure 2.
MAP2 suppression by cognate 21nt-siRNAs. (A) Double fluorescence staining of neurons transfected with nonspecific siRNA or with MAP2-siRNA. (Upper) Staining with MAP2 monoclonal antibody (green). (Lower) Staining with actin-bound toxin phalloidin (red). In each experiment, at least 70 random neurons per experimental condition were analyzed, and gene expression was quantified in both control and targeted cells. (B) Distribution of MAP2 and cortactin expression levels in control and targeted cells, two different siRNA (siRNA1 and siRNA2) show a very similar effect.
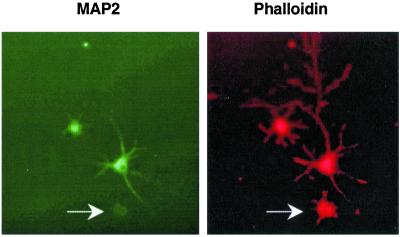
Phenotypic effects of MAP2 suppression on cultured neurons can be observed early on after plating as neurons consolidate their lamellae and elaborate filopodia. It has been shown (15, 16) that MAP2 is required to overcome early transitions of neuritic development and to begin processes elongation. We targeted primary cultures with the cognate siRNAs 3 h after plating and analyzed the effect over 28–40 h. Within this shorter time-frame, fewer neurons suppressed MAP2 expression. However, the degree of MAP2 suppression correlated with the recognized defect in filopodial elaboration (Fig. 3).
Figure 3.
MAP2 suppression correlates with the defect in early staged neuronal development. Double fluorescence staining of young neurons transfected with MAP2-siRNA. (Left) Staining with MAP2 monoclonal antibody (green). (Right) Staining with actin-bound toxin phalloidin (red). The cell with silenced MAP2 expression (labeled by arrow) exhibits severe defect in filopodia elaboration.
Transfection with two different dsRNAs cognate to YB-1 RNA-binding protein did not suppress the target protein even with a 68-h exposure. The reasons for the ineffectiveness of RNAi in this case may arise from the high stability of the extremely abundant YB-1 protein or from intrinsic features of YB-1 mRNA.
Discussion
The use of small interfering RNAs has become a powerful tool to knock down specific gene expression in a wide range of cells. In Drosophila lysate, dsRNA triggers the recognition and targeted cleavage of homologous cellular mRNA (4). In mammalian cells, the mechanism underlying siRNA-mediated gene targeting is unknown; it might interfere with RNA stability and/or translation or, alternatively, transcription.
In cultured mammalian cell lines, chemically synthesized siRNAs can by introduced into cells when formulated with lipophilic reagents (Lipofectamine 2000 or Oligofectamine; refs. 7 and 8). Very recently, several groups developed an alternative approach to suppress genes by intracellular expression of siRNAs from transfected plasmide DNA (17–20). Notably, transfections of postmitotic primary neurons are very inefficient, often toxic, and, therefore, none of the techniques mentioned above would be useful for siRNA-mediated gene targeting in neurons. In principle, RNA can be introduced into a single neuron with cationic lipids (21). We have demonstrated that synthetic 21-nt dsRNA (siRNAs) is readily delivered into primary cortical and hippocampal neurons by cationic TransMessenger transfection reagent and can be used to suppress gene expression. Therefore, the RNAi pathway seems operative in primary mammalian neurons. Even a brief (2-h) exposure to a low concentration of siRNAs (0.006–0.6 μg/ml) caused suppression of endogenous and heterologous gene expression. Antisense ssRNAs in that concentration range did not inhibit gene expression. Higher concentrations of siRNA/transfection reagents seemed to be toxic for neuronal cultures. The effect of siRNA on MAP2 expression was slow and usually became visible 2 days after transfections. We also assume that longer incubation of cells is required for better suppression of transfected GFP and DsRed genes; however, we failed to keep neurons transfected with Lipofectamine 2000 healthy more than 48 h. In nematode, the only other neuronal culture system in which RNAi was shown to be effective, the RNAi effect was also quite slow (visible after 3–4 days with dsRNA) and required much higher amounts of dsRNA (15 μg/ml instead of 0.006–0.6 μg/ml). Currently, we do not know what factors are responsible for the rate and the strength of RNAi–induced posttranscriptional gene suppression. Clearly, particular characteristics of a targeted mRNA (“defensive” secondary structures and bound transfactors, causing ineffectiveness of some siRNA) and protein (different life times) affect RNAi efficacy. In addition, cell-specific factors might be involved in differential siRNA delivery and regulation of RNA interference in various cell types. The recent discovery of predominantly brain-specific microRNAs, a previously uncharacterized class of noncoding RNAs sharing processing pathways with siRNAs (22), together with our data, suggest a role of RNAi-related mechanisms of gene regulation in neuronal development and functioning. The ability to knock down expression of a specific gene is likely to have broad application in neurons, a setting where genetic manipulations have proven difficult.
Abbreviations
- dsRNA
double-stranded RNA
- RNAi
RNA interference
- siRNA
small interfering RNA
- En
embryonic day n
- siGFP
siRNA cognate to green fluorescent protein
- MAP2
microtubule-associated protein 2
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Hutvagner G, Zamore P D. Curr Opin Genet Dev. 2002;12:225–232. doi: 10.1016/s0959-437x(02)00290-3. [DOI] [PubMed] [Google Scholar]
- 2.Sharp P A. Genes Dev. 2001;15:485–490. doi: 10.1101/gad.880001. [DOI] [PubMed] [Google Scholar]
- 3.Hammond S M, Caudy A A, Hannon G J. Nat Rev Genet. 2001;2:110–119. doi: 10.1038/35052556. [DOI] [PubMed] [Google Scholar]
- 4.Zamore P D, Tuschl T, Sharp P A, Bartel D P. Cell. 2000;101:25–33. doi: 10.1016/S0092-8674(00)80620-0. [DOI] [PubMed] [Google Scholar]
- 5.Bernstein E, Caudy A A, Hammond S M, Hannon G J. Nature (London) 2001;409:363–366. doi: 10.1038/35053110. [DOI] [PubMed] [Google Scholar]
- 6.Elbashir S M, Harborth J, Lendeckel W, Yalcin A, Weber K, Tuschl T. Nature (London) 2001;411:494–498. doi: 10.1038/35078107. [DOI] [PubMed] [Google Scholar]
- 7.Caplen N J, Parrish S, Imani F, Fire A, Morgan R A. Proc Natl Acad Sci USA. 2001;98:9742–9747. doi: 10.1073/pnas.171251798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Harborth J, Elbashir S M, Bechert K, Tuschl T, Weber K. J Cell Sci. 2001;114:4557–4565. doi: 10.1242/jcs.114.24.4557. [DOI] [PubMed] [Google Scholar]
- 9.Paddison P J, Caudy A A, Hannon G J. Proc Natl Acad Sci USA. 2002;99:1443–1448. doi: 10.1073/pnas.032652399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Timmons L, Court D L, Fire A. Gene. 2001;263:103–112. doi: 10.1016/s0378-1119(00)00579-5. [DOI] [PubMed] [Google Scholar]
- 11.Christensen M, Estevez A, Yin X, Fox R, Morrison R, McDonnell M, Gleason C, Miller D M, III, Strange K. Neuron. 2002;33:503–514. doi: 10.1016/s0896-6273(02)00591-3. [DOI] [PubMed] [Google Scholar]
- 12.Kalidas S, Smith D P. Neuron. 2002;33:177–184. doi: 10.1016/s0896-6273(02)00560-3. [DOI] [PubMed] [Google Scholar]
- 13.Elbashir S M, Martinez J, Patkaniowska A, Lendeckel W, Tuschl T. EMBO J. 2001;20:6877–6888. doi: 10.1093/emboj/20.23.6877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Escobar M I, Pimienta H, Caviness V S, Jacobson M, Crandall J E, Kosik K S. Neuroscience. 1986;17:975–989. doi: 10.1016/0306-4522(86)90074-6. [DOI] [PubMed] [Google Scholar]
- 15.Gonzalez-Billault C, Engelke M, Jimenez-Mateos E M, Wandosell F, Caceres A, Avila J. J Neurosci Res. 2002;67:713–719. doi: 10.1002/jnr.10161. [DOI] [PubMed] [Google Scholar]
- 16.Caceres A, Mautino J, Kosik K S. Neuron. 1992;9:607–618. doi: 10.1016/0896-6273(92)90025-9. [DOI] [PubMed] [Google Scholar]
- 17.Paul C P, Good P D, Winer I, Engelke D R. Nat Biotechnol. 2002;20:505–508. doi: 10.1038/nbt0502-505. [DOI] [PubMed] [Google Scholar]
- 18.Lee N S, Dohjima T, Bauer G, Li H, Li M J, Ehsani A, Salvaterra P, Rossi J. Nat Biotechnol. 2002;20:500–505. doi: 10.1038/nbt0502-500. [DOI] [PubMed] [Google Scholar]
- 19.Miyagishi M, Taira K. Nat Biotechnol. 2002;20:497–500. doi: 10.1038/nbt0502-497. [DOI] [PubMed] [Google Scholar]
- 20.Yu J Y, DeRuiter S L, Turner D L. Proc Natl Acad Sci USA. 2002;99:6047–6052. doi: 10.1073/pnas.092143499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Crino P B, Eberwine J. Neuron. 1996;17:1173–1186. doi: 10.1016/s0896-6273(00)80248-2. [DOI] [PubMed] [Google Scholar]
- 22.Lagos-Quintana M R R, Yalcin A, Meyer J, Lendeckel W, Tuschl T. Curr Biol. 2002;12:735–739. doi: 10.1016/s0960-9822(02)00809-6. [DOI] [PubMed] [Google Scholar]