Abstract
The genome of the Mastreviruses encodes a replication-associated protein (RepA) that interacts with members of the plant retinoblastoma-related protein family, which are putative cell cycle regulators. Expression of ZmRb1, a maize retinoblastoma-related gene, and RepA inhibited and stimulated, respectively, cell division in tobacco cell cultures. The effect of RepA was mitigated by over-expression of ZmRb1. RepA increased transformation frequency and callus growth rate of high type II maize germplasm. RepA-containing transgenic maize calli remained embryogenic, were readily regenerable, and produced fertile plants that transmitted transgene expression in a Mendelian fashion. In high type II, transformation frequency increased with the strength of the promoter driving RepA expression. When a construct in which RepA was expressed behind its native LIR promoter was used, primary transformation frequencies did not improve for two elite Pioneer maize inbreds. However, when LIR:RepA-containing transgenic embryos were used in subsequent rounds of transformation, frequencies were higher in the RepA+ embryos. These data demonstrate that RepA can stimulate cell division and callus growth in culture, and improve maize transformation.
Like many mammalian DNA viruses, plant geminiviruses have efficient methods to subvert host cell cycle machinery and facilitate their replication (1, 2). This occurs through interactions of viral replicase gene products and host cell components. In the Mastreviral subgroup of geminiviruses, which includes maize streak virus and wheat dwarf virus (WDV), two ORFs are differentially spliced, resulting in a mixture of the full-length replicase protein (Rep) and a truncated protein, RepA (3). Although Rep is required for viral replication, RepA is not (4). Rep and RepA were demonstrated to participate in many overlapping and nonoverlapping interactions with host functions. Examples include interactions of WDV RepA with developmental genes (5), transactivation of genes by Rep and RepA proteins (3, 6–10), and direct interaction of RepA with host cell retinoblastoma-related (Rb) proteins (11–13). Of these, binding to Rb is the most thoroughly studied. Both Rep and RepA proteins have a Rb-binding motif, but it appears that RepA binds Rb more efficiently (9, 14).
The Rb gene family contains critical regulators of the G1/S transition in animal systems. Rb binds to S-phase transcriptional transactivators, such as members of the E2F-family, and masks their activation domain without disrupting DNA binding at cell cycle-regulated promoters. Rb simultaneously recruits transcriptional repressors, such as histone deacetylases and methylases, and DNA helicases, to promote transcriptional quiescence. Animal DNA viruses encode Rb-binding proteins that relieve this repression, stimulate the cell cycle, and create a permissive environment for viral DNA replication (15–17). Similar to Rb-binding proteins in nonplant systems, it is speculated that RepA activates the expression of numerous genes that function in replication and S-phase progression.
Although parallels between plant geminiviral and mammalian oncogenic viruses are striking in terms of viral replicase interactions with cell cycle proteins, there is one incongruous aspect to this comparison. The most obvious phenotypic impact of mammalian oncogenic viruses and the characteristic that led to intense efforts to unravel their biology is their stimulation of host cell proliferation (16, 17). Immunochemical analysis of geminivirus-infected plant cells has identified a stimulation of replication-associated machinery. In terminally differentiated tobacco leaf cells, Rep expression or infection with tomato golden mosaic virus are associated with increases in proliferating cell nuclear antigen (18), an integral component of DNA polymerase regulated by E2F-binding sites in both animals and plants (19). Surprisingly, despite their interactions with Rb and activation of replication machinery, cell proliferation as a result of Rep or RepA expression has not been reported for plant cells. Thus, a consensus has developed that Rep and RepA do not induce dedifferentiation and reentry into the cell cycle, but rather up-regulate S-phase functions to facilitate viral genome replication (2).
Plant transformation is impacted by cell cycle progression. In tobacco Bright-Yellow 2 (BY-2) cell cultures, transient β-glucoronidase (GUS) expression increases when tobacco cells are transformed during G2 or M phase (20, 21). In addition, transformation of synchronized tobacco protoplasts during S–M phase results in increased recovery of selection-resistant colonies (20, 22) and in higher copy, more complex transgene integrations (23) when compared with nonsynchronized cells. More recently, Agrobacterium-mediated delivery of DNA was found to require transition through S phase (24). Although cell cycle dynamics can influence transformation, no practical methods have been reported to positively influence transformation by directly manipulating the cell cycle.
Here we show that expression of the WDV RepA gene stimulates, and the maize ZmRb1341-866 gene inhibits, cell division in tobacco BY2 cells. RepA stimulated cell division is abrogated by the simultaneous expression of ZmRb1341-866. Furthermore, RepA expression stimulates maize embryogenic callus growth, increases transformation frequency, and eliminates the need for chemical selection of transformed calli. Rapidly growing RepA-expressing maize callus maintains its morphogenetic competency and elite maize inbred lines harboring a RepA transgene display an enhanced transformation phenotype.
Materials and Methods
Constructs.
The plasmids in Table 11, which is published as supporting information on the PNAS web site, www.pnas.org, were used for either stable or transient transformation of plant materials. For brevity, only the gene components for individual constructs are described here. Visible marker genes, uidA (GUS; ref. 25) and green fluorescent protein (GFP; ref. 26), and a maize codon-optimized version of GFP (moGFP) were used to identify transformed cells. A plasmid containing the firefly luciferase gene (27) was used to balance the DNA content of particle bombardments in some experiments. For maize cobombardment experiments, a fusion between a maize-optimized phosphinothricin acetyl transferase (moPAT) gene and moGFP was generated (moPAT∼moGFP). In retransformation experiments, a fusion between domains of R and C1 (CRC) encoding the protein functions necessary to activate anthocyanin accumulation (28, 29) was used as a marker to score transgenic calli, and the bar gene (30) was used for bialaphos selection. Promoters driving expression of visible marker genes included a double-enhanced cauliflower mosaic virus (CaMV) 35S promoter (31), the nopaline synthase (Nos) promoter (32), and the maize ubiquitin (Ubi) promoter (33). Downstream 3′ regions used in expression cassettes included those from a proteinase inhibitor (pinII; ref. 34) and CaMV 35S (31).
The coding sequences for the wild-type ZmRb1341-866 and ZmRb1C706G mutant sequences were derived from constructs described by Grafi et al. (11) and are 5′ truncations of ZmRb1, consisting of amino acids 341–866 of the RRB1 sequence (GenBank accession no. AAB69649). pWI-11 was the parental plasmid for all WDV Rep and RepA sequences (35). Two Asp700 restriction sites within the WDV Rep sequence were used to make an internal deletion that removed the intron splice junction sequences. The resulting nucleic acid sequence encodes a RepA protein (RepAAsp700) in which the last five carboxy-terminal amino acids are altered from −PGNGK to −RRGSA. To more completely eliminate intron-splicing and reduce the A/T-content of the intron sequence, additional constructs were made by amplification of the RepA or Rep coding sequences by mutagenic PCR. This did not alter amino acids in Rep, but resulted in a valine to lysine substitution in the RepA coding sequence (at the site of the 5′ splice junction). The “intronless” sequences, are referred to as Repm and RepAm.
Four constructs were used for Agrobacterium-mediated transformation experiments. In the first (designated p108), a DNA fragment containing the Nos promoter, a gene encoding the CRC fusion protein, and the pinII 3′ end was inserted between the T-DNA borders in pSB11. The second, p109, contained two separate T-DNA-border-flanked cassettes, LIR:RepAAsp700 and 35S:bar/Ubi:moGFP:pinII. The third (p111) contained LIR:RepAAsp700, Ubi:moGFP and 35S:bar within a single set of T-borders, and the fourth (p110) contained 35S:bar + Ubi:FLP:pinII within a single set of T-borders. Agrobacterium tumefaciens LBA4404 and vectors pSB1 and pSB11 (36, 37) were obtained from Japan Tobacco (700 Higashibara, Iwata, Japan).
Tobacco Cultures and Transformation.
Nicotiana tabacum L. cv. Bright Yellow 2 (BY-2) suspension cultures were used for evaluation of RepA and ZmRb1 expression on cell division. Suspension cells were subcultured every 7–10 days into fresh Nt1 medium (see Table 12, which is published as supporting information on the PNAS web site, for media formulations) and grown on a gyratory shaker at 150 rpm, 24°C in the dark. Three, six, or nine days after subculturing, cells were pipetted onto solidified agar medium (Nt1) and left in the dark for 24 h before bombardment. Plasmid DNA was precipitated on 1-μm gold particles with polyethylene glycol (PEG) (38). Bombardment was performed with a Bio-Rad PDS-1000 helium gun, 650 PSI rupture discs, a 60-cm Hg vacuum and 8 cm between the stopping plate and Petri dish. Cells were shot once with 500 ng gold and 0.5 μg DNA. All tobacco bombardments used the 35S:GFP expression cassette. Treatments consisted of 35S:RepA, 35S:ZmRb1341–866, 35S:ZmRb1706G, or combinations thereof. Twenty-four hours after bombardment, cells were monitored for GFP expression and cell division by epifluorescence microscopy. The total number of GFP-expressing foci and whether they comprised single or multiple cells was recorded. Data from the controls and treatments were tested for significant differences by Student's t test (39).
Maize Transformation.
A publicly available maize (Zea mays L.), hybrid high type II (Hi-II) (40), and Pioneer proprietary inbreds P38 and N46 were grown in the greenhouse at 16-h day length. Immature embryos (1.0–1.5 mm) were excised from fresh developing kernels and used for transformation.
Particle-mediated transformation followed a standard protocol (41). After 4 or 5 days incubating Hi-II immature embryos on 560P medium (see Table 12 for media formulations) in the dark at 28°C, embryos were transferred onto 560Y and cultured scutellum-side-up for 3 h before transformation. The scutellar surface was targeted with the PDS-1000 Helium Gun from BioRad at one shot per sample using 650 PSI rupture disks. Approximately 67 ng of DNA was delivered per shot. A similar number of embryos per ear were bombarded for each treatment in an experiment, in aliquots of 25 embryos per plate. After bombardment, embryos were maintained on 560L medium. Transformants were transferred 2–7 days after bombardment onto 560R for selection. Plates were maintained at 28°C in the dark, and transferred to fresh medium every 2 weeks. GFP+ and/or bialaphos-resistant (BAR) calli were scored at 6–8 weeks, with each plate of embryos as a replicate.
Calli were examined by epifluorescence with a dissecting microscope using a filter set (Chroma no. 41020) for GFP excitation and emission. When colony size was recorded, two perpendicular measurements were taken for each independent, GFP-expressing multicellular colony. The two measurements were averaged, providing an estimate of the diameter for each colony. These values were used to calculate the colony's presumed spherical volume (V = 4/3πr3).
After approximately 10 weeks of selection, BAR, GFP-positive calli were scored. Positive lines were transferred to a Murashige and Skoog (MS)-based medium with reduced sucrose and hormone levels to initiate plant regeneration (40). After somatic embryo maturation (2–4 weeks), well-developed embryos were transferred to germination medium for 7–10 days and placed in light. Developing plantlets were transferred to medium in tubes for 7–10 days until well established. Plants were then transferred to flats (equivalent to a 6.4 cm pot) containing potting soil, grown for 1 week in a growth chamber and an additional 1–2 weeks in the greenhouse. The plants were finally transferred to 6-liter pots (catalog no. 14-9674-9; Hummert International, Earth City, MO) and grown to maturity. Mature plants were crossed to untransformed plants of the same genotype for analysis of inheritance and retransformation.
Methods for Agrobacterium-mediated transformation of maize followed the general protocol described (42) with the following modifications. Agrobacteria were grown to log phase in liquid minimal A medium containing 100 μM spectinomycin. Cells were transformed by culturing P38 or N46 immature embryos in liquid 700 medium. Embryos were immersed in a log phase suspension of Agrobacterium [5 × 108 colony forming units (cfu) per ml]. Embryos were infected for 5 min by using gentle rotation of the Agrobacterium suspension and then cocultured, embryo axis down, for 7 days in the dark at 20°C on 710 medium. Embryos were then transferred to 720E selection medium. Plates were maintained at 28°C in the dark and observed for colony recovery with transfer to fresh medium every 2 weeks. After 6–8 weeks, selection-resistant and/or GFP expressing colonies were transferred to 288J maturation medium to begin plant recovery. Colonies were maintained for 7 days in the dark at 28°C on this medium, followed by transfer to medium for plant recovery in the light. Recovered plantlets were transferred to culture tubes containing 272 medium for root development before transfer to the greenhouse. Plants were scored based on continued GFP expression, leaf sensitivity to painting with 1% phosphinothricin (Aventis CropScience, Research Triangle Park, NC), and molecular characterization via PCR and Southern blot analyses.
Retransformation.
Primary transformations were done as described above by using the Agrobacterium vectors p111 or p109, and transgenic inbred plants were regenerated and pollinated from nontransformed inbreds. When the “two T-DNA” vector (p109) was used, a high percentage of events was recovered in which the two T-DNA's segregated independently (61). Pollen from wild-type inbred individuals was carried onto inbred transformants hemizygous for either the LIR:RepAAsp700/Ubi:GFP/35S:bar or the LIR:RepAAsp700 locus, respectively, and the resultant embryos, segregating 1:1 for the presence of RepA, were subjected to a subsequent round of transformation (termed “retransformation”). For retransformation, Agrobacterium was used to deliver either CRC (into LIR:RepAAsp700/Ubi:GFP/35S:bar containing embryos) or 35S:bar/Ubi:FLP (into LIR:RepAAsp-700 containing embryos). Stable anthocyanin-accumulating or BAR events were scored 6–8 weeks after Agrobacterium-mediated delivery of the retransformation expression cassette. For 35S:bar retransformation experiments, data were collected as the number of embryos regenerating BAR, PCR-positive plants, relative to the total number of treated embryos. Presence of RepA in segregating material from which transformants were recovered was confirmed by quantitative PCR (62).
Results
RepA Stimulates and ZmRb1 Inhibits Cell Division in BY-2 Cells.
To test whether RepA influences the plant cell cycle, a transfection-based assay of cell cycle regulatory function was done with BY-2 cell cultures. Bombardment with gold particles was used to deliver 35S:GFP and 35S:RepA, and after 24 h GFP-expressing cells were scored as single or divided (Fig. 1a). If mitotically active cells (4 days after subculture) were bombarded with 35S:GFP alone, approximately 38% of fluorescent foci divided within 24 h (Table 1). Codelivery of 35S:RepA and 35S:GFP increased the proportion of divided cells to approximately 60%. These proportions are significantly different when compared by Student's t test (P < 0.01), suggesting that RepA stimulates the cell division cycle in actively growing BY-2 cell suspensions.
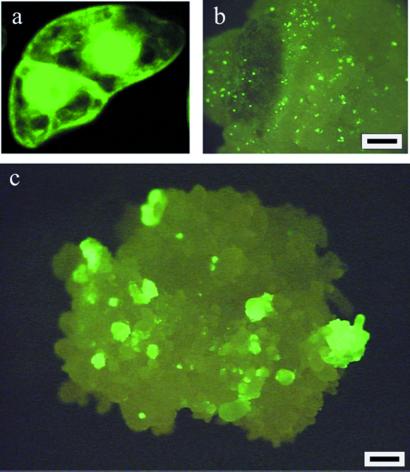
Figure 1.
Effect of RepA expression on plant cell division and callus growth. (a) Recent cell division in a GFP-expressing BY-2 cell culture clearly showing two daughter cells separated by a newly formed transverse wall. (b) GFP expression 2 weeks after particle delivery of Ubi:moPAT∼moGFP:pinII DNA into Hi-II immature embryos; only single cells expressing GFP were observed. (c) GFP expression 2 weeks after particle delivery of Ubi:moPAT∼moGFP:pinII and Ubi:RepA; multiple GFP-expressing multicellular colonies were observed in addition to single cells expressing GFP. (Scale markers in b and c = 500 μm.)
Table 1.
RepA-stimulates cell division in BY-2 cells
| Culture age, days | Treatment | % Divided (±SD) | n | Sig. |
|---|---|---|---|---|
| 4 | Control | 42.0 (5.8) | 307 | |
| 35S:RepA | 59.8 (12.1) | 326 | ** | |
| 7 | Control | 25.3 (5.8) | 529 | |
| 35S:RepA | 50.4 (6.8) | 569 | ** | |
| 14 | Control | 19.9 (0.5) | 450 | |
| 35S:RepA | 35.2 (5.3) | 553 | ** |
Significant difference (Sig.) determined by t test, with
denoting P values < 0.01.
BY-2 cells enter a stationary phase 7 days after subculturing, when cell density stabilizes, the mitotic index decreases, and cells arrest in G1/G0 (43, 44). In 7- and 14-day-old cell cultures transfected with 35S:GFP, the proportion of divided foci were significantly lower (20–25%) than that of 4-day-old cell cultures (t test, P < 0.01; Table 1). Treatment of these cultures with 35S:RepA significantly increased the proportion of divided cells (Table 1). In fact, 35S:RepA induced a greater fold-increase in cell division in the stationary cultures, suggesting that RepA is able to overcome a G0/G1-block in BY-2 cultures.
To test the hypothesis that RepA acts by means of a plant Rb gene family member, the 35S:GFP, 35S:RepA, and 35S:ZmRb1341-866 cassettes were codelivered to 4-day-old BY-2 cell cultures. Bombardment with 35S:GFP produced fluorescent foci with a similar proportion of multicellular events as observed in the previously described experiments (compare Tables 1 and 2). The codelivery of 35S:ZmRb1341-866, but not a mutant 35S:ZmRb1C706G, significantly decreased the proportion of divided cells, demonstrating that the ZmRb1341-866 gene product, which includes the pocket domain of ZmRb1 (11, 13, 45), is sufficient for cell cycle arrest. The codelivery of 35S:RepA and 35S:GFP increased the number of fluorescent foci with multiple cells to approximately 54.3%, similar to proportions seen previously (Tables 1 and 2). When 35S:RepA and the mutant 35S:ZmRb1C706G were codelivered, the number of fluorescent foci was indistinguishable from 35S:RepA treatments (t test, P < 0.01). If GFP, 35S:RepA and 35S:ZmRb1341-866 cassettes were simultaneously delivered, fluorescent foci were found to contain proportions of single and multiple cell clusters indistinguishable from control treatments (t test, P > 0.05) and significantly different from either construct alone (P < 0.01). Thus, RepA and ZmRb1341-866 appear to influence the cell cycle in opposition.
Table 2.
ZmRb1 opposes RepA in BY-2 cells
| Treatment | % Divided (±SD) | n | Sig. |
|---|---|---|---|
| Control | 37.6 (2.8) | 1009 | |
| 35S:ZmRb1341–866 | 15.9 (3.7) | 252 | ** |
| 35S:ZmRb1C706G | 37.7 (4.8) | 372 | NS |
| 35S:RepA | 54.3 (9.9) | 408 | ** |
| 35S:ZmRb1341–866, 35S:RepA | 37.5 (0.2) | 304 | NS |
| 35S:ZmRb1C706G, 35S:RepA | 59.6 (5.6) | 339 | ** |
Significant difference (Sig.) determined by t test, with
and NS denoting P values < 0.01 and > 0.05, respectively.
RepA Stimulates Early Growth of Maize Transformants.
To investigate whether RepA can increase cell division in maize embryos, they were transformed with moPAT∼GFP and Ubi:RepA or Nos:RepA. When compared by fluorometric quantitation of GUS activity, the Ubi promoter is 5- to 10-fold more active than Nos in immature embryos (data not shown). By 16 days after bombardment, GFP-expressing cells were observed on the surface of scutellar-derived tissue. Only single GFP-expressing cells were observed (Fig. 1b) in the majority of control embryos. At this time, single GFP-expressing cells and macroscopic GFP-expressing multicellular clusters were apparent in RepA treatments (Fig. 1c). The proportion of embryos with GFP-containing colonies and the colonies observed per embryo in the Nos:RepA and Ubi:RepA treatments were greater than controls (Table 3). If each embryo producing a transformant was counted, the transformation frequencies for the control, Nos:RepA or Ubi:RepA treatments were ≈3, 16% and 20%, respectively. However, if GFP colonies were scored as independent events, the control, Nos:RepA and Ubi:RepA treatments contained 6, 59, and 80 transformants with transformation frequencies of 3%, 29%, and 46%, respectively. Macroscopic colony size was also sensitive to promoter strength as Ubi:RepA colonies were significantly larger than Nos:RepA colonies (Table 4; Ranked t test, P < 0.01).
Table 3.
RepA increases GFP colonies per embryo in maize
| Treatment (n) | No. of GFP colonies per embryo
|
|||||
|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | |
| Control (200) | 6 | 0 | 0 | 0 | 0 | 0 |
| Nos:RepA (202) | 20 | 8 | 2 | 3 | 1 | 0 |
| Ubi:RepA (175) | 8 | 15 | 9 | 1 | 1 | 1 |
Data collected 16 days after bombardment.
Table 4.
RepA stimulates multicellular cluster growth in maize
| Size class* | Control | Nos:RepA | Ubi:RepA |
|---|---|---|---|
| 0–1.0 | 2 | 17 | 2 |
| 1.1–3.0 | 1 | 11 | 7 |
| 3.1–10 | 0 | 16 | 12 |
| 11–30 | 2 | 7 | 15 |
| 31–100 | 0 | 5 | 21 |
| 101–300 | 1 | 1 | 13 |
| 301–1000 | 1 | 1 | 11 |
| 1000–1300 | 0 | 0 | 1 |
| >1301 | 0 | 0 | 1 |
Volumes of fluorescent cell clusters at 16 days after bombardment represented as mm3 × 103. Scored as numbers of colonies in different size classes.
RepA Increases Maize Transformation Frequency.
As Tables 3 and 4 show, RepA stimulated early growth in GFP-expressing cell clusters and the recovery of transformed calli. To determine whether the Rep protein had a similar impact on maize transformation, Nos:RepAm or Nos:Repm were cobombarded with moPAT-GFP and compared with control treatments. Nos:RepAm significantly increased the recovery of BAR transformants relative to the moPAT∼GFP control treatment, whereas Nos:Repm treatments were indistinguishable from moPAT∼GFP controls (Table 5). To extend this observation and determine whether transformation frequency, like callus growth, is sensitive to promoter strength, embryos were cobombarded with Nos:RepAm or Ubi:RepAm and moPAT∼GFP. When compared with moPAT∼GFP controls, both Nos:RepAm and Ubi:RepAm significantly increased transformation frequencies (Table 6). Furthermore, Ubi:RepAm treatments displayed significantly higher transformation frequencies relative to Nos:RepAm (t test, P < 0.05).
Table 5.
Nos:RepA, but not Nos:Rep, increases transformation efficiency in maize
| Treatment | % BAR (±SD)* | n | Sig. |
|---|---|---|---|
| Nos:RepA | 34.3 (14.7) | 134 | ** |
| Nos:Rep | 10.5 (8.8) | 133 | NS |
| Luciferase (control) | 11.5 (9.3) | 131 |
Scored as percentage of embryos regenerating at least one herbicide resistant plant. Significant difference (Sig.) determined by t test with
and NS denoting P values < 0.01 and > 0.05, respectively.
Table 6.
Maize transformation is sensitive to RepA promoter strength
| Treatment | % BAR (SD)* | n | Sig. |
|---|---|---|---|
| Control | 4.0 (3.3) | 100 | |
| Nos:RepA | 15.0 (12.8) | 100 | ** |
| Ubi:RepA | 40.8 (23.8) | 125 | ** |
Scored as percentage of embryos regenerating at least one herbicide resistant plant. Significant difference (Sig.) determined by t test, with
denoting P values < 0.01.
As RepA promoted growth, we investigated its utility for identification of transgenic calli in the absence of chemical selection. Visual screening for GFP fluorescence recovered one-fourth the number of transformants obtained with bialaphos selection (Table 7). Cobombardment of embryos with Ubi:RepA and moPAT∼GFP substantially increased the frequency of recovering BAR events. In the absence of selection, the inclusion of RepA significantly increased the efficiency of transformant recovery by visual screening for GFP (Table 7), such that it was indistinguishable from that obtained by chemical selection in the presence of RepA (t test, P > 0.05).
Table 7.
Ubi:RepAm increases maize transformation efficiency without chemical selection
| Ubi:RepAm | Selection | % GFP+* | n | RepAm | Selection |
|---|---|---|---|---|---|
| − | + | 11.9 | 126 | ||
| + | + | 68.0 | 150 | ** | |
| − | − | 2.9 | 102 | ** | |
| + | − | 62.0 | 121 | ** | NS |
Scored as percentage of embryos regenerating at least one herbicide resistant plant. Significant differences determined by t test, with
and NS denoting P values < 0.01 and > 0.05, respectively.
RepA+ Inbred Germplasm Exhibits Enhanced Transformation.
All experiments described thus far used embryos from the transformation-competent Hi-II maize hybrid. Transformation of agronomically elite maize inbreds would represent a substantial advancement in maize biotechnology. Our initial attempts to extend these results to elite maize inbreds used LIR:RepAAsp700 introduced, along with 35S:bar and Ubi:moGFP (p111), into maize embryos via Agrobacterium, followed by phosphinothricin selection. As seen in Table 8, although we recovered transformants in both the control and RepA treatments for inbreds N46 and P38, LIR:RepAAsp700 did not significantly enhance the transformation frequencies. However, the transformation frequency for Hi-II embryos was increased by LIR:RepAAsp-700 (t test, P < 0.05). The transgenic plants were regenerated, grown to maturity in the greenhouse and crossed to their respective parents. In progeny, LIR:RepAAsp-700, Ubi:moGFP, and 35S:bar segregated as a single locus that was followed by fluorescence and PCR (data not shown). Agrobacterium-mediated gene transfer was used to deliver Nos:CRC, a cell autonomous stimulator of anthocyanin production (28, 29), to T3 embryos harvested from segregating ears. Calli were screened for multicellular pigmented clusters in the absence of chemical selection. As shown in Table 9, GFP-expressing and nonfluorescent embryos from GFP/bar hemizygotes had similar frequencies of anthocyanin-expressing calli. However, for both inbreds, GFP-positive embryos from ears hemizygous for RepA/GFP/bar exhibited a significantly higher transformation frequency (t test, P > 0.01).
Table 8.
LIR:RepA does not improve primary transformation of maize inbreds
| Genotype | DNA | % BAR* | n | Sig. |
|---|---|---|---|---|
| PHP38 | Control | 4.7 | 1698 | |
| LIR:RepAAsp700 | 3.8 | 1676 | NS | |
| PHN46 | Control | 4.9 | 2215 | |
| LIR:RepAAsp700 | 4.8 | 2231 | NS | |
| Hi-II | Control | 1.1 | 200 | |
| LIR:RepAAsp700 | 6.6 | 200 | * |
Scored as percentage of embryos regenerating at least one herbicide resistant plant. Significant difference (Sig.) determined by t test, with * and NS denoting P values < 0.05 and > 0.05, respectively.
Table 9.
Stable integration of LIR:RepA improves subsequent inbred transformation
| Genotype/transgene | Embryo | % CRC (SD)* | n | Sig. |
|---|---|---|---|---|
| P38/GFP | GFP+ | 0.6 (0.5) | 177 | NS |
| GFP− | 0.5 (0.8) | 210 | ||
| P38/GFP + LIR:RepAΔAsp700 | GFP+ | 27.3 (19.1) | 55 | ** |
| GFP− | 0 | 41 | ||
| N46/GFP + LIR:RepAΔAsp700 | GFP+ | 25.0 (39.3) | 24 | ** |
| GFP− | 0 | 33 |
Calli were screened for anthocyanin accumulation at 16 days. Significant difference (Sig.) determined by t test, with
and NS denoting P values < 0.01 and > 0.05, respectively.
The presence of RepA in the genetic background also improved the efficiency of transformation with chemical selection. Inbreds N46 and P38 were transformed with LIR:RepAAsp-700 and Ubi:moGFP/35S:bar using a “two T-DNA” vector (p109) and backcrossed. After segregating the RepAAsp700 and GFP/bar loci, Southern analyses, fluorescence imaging, and herbicide resistance tests verified that the RepAAsp700 cassette was intact and the GFP/bar locus was no longer present. Hemizygous RepAAsp700 N46-inbred plants (T2 generation) were crossed to nontransformed N46, and the segregating T3 embryos were retransformed by cocultivation with Agrobacterium containing 35S:bar/Ubi:FLP (p110), followed by selection on bialaphos. χ2 tests demonstrated that BAR transformants were more likely to have arisen on RepA transgenic embryos in all transgenic lines tested (Table 10), indicating that stable RepA expression enhanced transformation. Surprisingly, higher transformation frequencies were also seen for nontransgenic embryos harvested from hemizygous ears than for wild-type embryos on nontransgenic ears (Table 10). This result demonstrates that the LIR:RepAAsp700 transgene has a maternal effect on embryo transformation competence.
Table 10.
Stable transformation with LIR:RepAAsp700 improves maize inbred N46 transformation with chemical selection
| Transgene/line | n | Events
|
Sig.
|
||
|---|---|---|---|---|---|
| RepA+ (%) | RepA− (%) | RepA+/−a | Wt/RepA−b | ||
| LIR:RepAASP700/0005 | 1,346 | 28 (4.16%) | 13 (3.04%) | * | ** |
| LIR:RepAASP700/0006 | 2,799 | 44 (3.14%) | 20 (2.29%) | ** | ** |
| LIR:RepAASP700/0008 | 711 | 23 (6.47%) | 8 (4.36%) | ** | ** |
| n.a. (N46 wild-type) | 3,956 | NA | 38 (0.96%) | ||
Data scored as number of embryos regenerating phosphinothricin-resistant plants. % transformation frequency calculated assuming 1:1 segregation. Significant difference (Sig.) was determined by χ2 (
) and t test (
) with
and
denoting P values < 0.01 and < 0.05, respectively.
Discussion
Similarities between interactions of the RepA protein with plant cell proteins (5, 9, 11, 13) and the resultant subversion of host cell cycle machinery led various researchers to suggest a similarity between plant geminiviruses and mammalian oncoviruses (8, 9, 12, 13, 18, 35). We can now expand these similarities to RepA-mediated stimulation of the cell cycle in BY-2 cells and callus growth in maize, observations consistent with the stimulation of cell proliferation associated with oncoviral infection (15–17). The inhibition of cell division by 35S:ZmRb1341–866 also provides direct evidence in favor of the plant Rb gene family encoding cell cycle regulators.
In mammalian cells, oncovirus-induced cell proliferation is typically associated with an undifferentiated cell phenotype. In contrast, despite rapid growth, RepA-treated transgenic calli generally maintain morphogenic competence. However, some of the most rapidly growing Ubi:RepA transformants were less embryogenic and more difficult to regenerate (data not shown). Delivery of RepA to Hi-II derived calli increased transformation efficiency, and increasing RepA expression enhanced this effect. Additionally, RepA transgenes conferred a high-competence transformation phenotype to both Hi-II (data not shown) and elite inbred genetic backgrounds, overcoming genotype-dependence. Furthermore, this effect was manifested by expression of RepA in the embryo and the ear, suggestive of maternal conditioning of transformation competence.
Stimulation of plant cell division and growth has not been reported previously in studies on geminiviral Rep (or RepA). This may be caused by alternative splicing in WDV or maize streak virus, the two Mastreviruses whose Rep have been most thoroughly studied. Inefficient splicing of the single intron in plant cells results in a mixture of Rep and the shorter RepA proteins (2–4, 46). These two proteins have different properties, and likely have different impacts on cell physiology. Consistent with the two proteins having different effects on plant growth, Nos:RepA expression, but not Nos:Rep, resulted in increased maize transformation. As others have not observed cell cycle stimulation (1, 2, 10, 18, 47–50), there may be restricted cell- or tissue-type responsiveness to RepA. Other cell-type-specific responses to geminiviral Rep expression and replication were reported for wheat suspension cultures and scutellar cells (47). Embryonic and/or meristematic cells may be more readily stimulated to divide given an appropriate stimulus, i.e., the presence of RepA, and subsequent patterns of morphogenesis, differentiation, and position-sensitive controls on cell division might simply be more difficult to reverse. This may be particularly true in cereals, where cellular plasticity appears to be strongly suppressed (51, 52). Protein interaction between RepA and Rb's (9, 11–13, 45), supported by our data demonstrating ZmRb1341-866-mediated suppression of RepA-stimulated cell division, suggest that increased transformation is a consequence of the relief of Rb repression of the cell cycle.
The increase in maize transformation could be caused by enhanced transgene integration. If this were occurring, an increase in transgene copy number might be expected. We examined copy number in RepA transgenic material by Southern analyses. Despite an increased transformation frequency, the range of transgene copy number and complexity in RepA T0 transformants was similar to control treatments with no RepA (data not shown) and to typical particle bombardment-generated transformants in the literature (53–57). In a study of Agrobacterium-mediated gene transfer, failure of transformed cells to proliferate and T-DNA silencing were more significant barriers to transformation than T-DNA integration (58). Therefore, we believe the increased transformation frequencies we observed are more likely to reflect increased plant recovery caused by stimulated cell division, rather than enhanced DNA integration.
If RepA acts to stimulate cell division in maize, it should function as a positive “selection” marker. Indeed, in the absence of chemical selection, RepA expression increased the efficacy of visual screening for transformants. The inclusion of bialaphos selection did not further increase transformant recovery, supporting this interpretation. Although visual screening without chemical selection was previously reported for both maize (59) and oats (57, 60), our results with RepA and GFP represent a many fold improvement over previous methods.
Our results demonstrate that RepA improves primary transformation in Hi-II maize embryos. However, Hi-II is a model germplasm for maize tissue culture (40). Although Hi-II transformation has continued to improve, most elite maize inbreds are recalcitrant to transformation. Results from primary transformation of two Pioneer elite inbreds with LIR:RepAAsp700 confirmed this. Nonetheless, in subsequent generations LIR:RepAAsp700 transgenic germplasm was more transformable than siblings lacking RepA. Experiments using CRC as a marker gene demonstrated that RepA integration improved transformation efficiency from 0.5% to ≈25%. Subsequent experiments using herbicide selection showed that LIR:RepAAsp700 conditions both higher and more consistent transformation frequencies in the T3 generation. Thus, the positive impact on transformation is a heritable trait. Furthermore, increased transformation was observed for nontransgenic embryos harvested from LIR:RepAAsp700 hemizygous ears, strongly suggesting that RepA expression also exerts a maternal influence on the transformability of embryos.
Our observation that RepA-enhanced transformation is heritable has an important implication. First, an initial transformation with RepA can be used to create transformable inbred germplasm. On subsequent retransformation, for example with an agronomically important gene, the resultant plants can easily be crossed and the RepA transgene (which exists as a separate locus) segregated away from the agronomically important transgenes. When this approach is used, the RepA-gene becomes a laboratory and greenhouse tool that is readily left behind before the newly generated transgenic inbreds are moved into the field.
Supplementary Material
Acknowledgments
This research was supported by grants from the Department of Energy (DE-FG03-96ER20242) and Pioneer Hi-Bred (to B.A.L.).
Abbreviations
- BAR bialaphos-resistant
Hi-II, high type II, BY-2 bright yellow 2
- WDV wheat dwarf virus
GUS, β-glucuronidase
- GFP
green fluorescent protein
- moGFP
maize codon-optimized GFP
- Rep
replicase protein
- RepA
replication associated protein A
- Rb
retinoblastoma-related
- pinII
proteinase inhibitor
- Ubi
ubiquitin
- Nos
nopaline synthase
- moPAT
maize-optimized phosphinothricin acetyl transferase
Footnotes
References
- 1.Gutierrez C. EMBO J. 2000;19:792–799. doi: 10.1093/emboj/19.5.792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gutierrez C. Plant Mol Biol. 2000;43:763–772. doi: 10.1023/a:1006462028363. [DOI] [PubMed] [Google Scholar]
- 3.Palmer K E, Rybicki E P. Adv Virus Res. 1998;50:183–234. doi: 10.1016/s0065-3527(08)60809-x. [DOI] [PubMed] [Google Scholar]
- 4.Schalk H J, Matzeit V, Schiller B, Schell J, Gronenborn B. EMBO J. 1989;8:359–364. doi: 10.1002/j.1460-2075.1989.tb03385.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Xie Q, Sanz-Burgos A P, Guo H, Garcia J A, Gutierrez C. Plant Mol Biol. 1999;39:647–656. doi: 10.1023/a:1006138221874. [DOI] [PubMed] [Google Scholar]
- 6.Lazarowitz S G, Wu L C, Rogers S G, Elmer J S. Plant Cell. 1992;4:799–809. doi: 10.1105/tpc.4.7.799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bisaro D M. In: DNA Replication in Eukaryotic Cells. de Pamphelis M, editor. Plainview, NY: Cold Spring Harbor Lab. Press; 1996. pp. 833–854. [Google Scholar]
- 8.Collin S, Fernandez-Lobato M, Gooding P S, Mullineaux P M, Fenoll C. Virology. 1996;219:324–329. doi: 10.1006/viro.1996.0256. [DOI] [PubMed] [Google Scholar]
- 9.Horvath G V, Pettko-Szandtner A, Nikovics K, Bilgin M, Boulton M, Davies J W, Gutierrez C, Dudits D. Plant Mol Biol. 1998;38:699–712. doi: 10.1023/a:1006076316887. [DOI] [PubMed] [Google Scholar]
- 10.Hanley-Bowdoin L, Settlage S B, Orozco B M, Nagar S, Robertson D. Crit Rev Biochem Mol Biol. 2000;35:105–140. [PubMed] [Google Scholar]
- 11.Grafi G, Burnett R J, Helentjaris T, Larkins B A, DeCaprio J A, Sellers W R, Kaelin W G., Jr Proc Natl Acad Sci USA. 1996;93:8962–8967. doi: 10.1073/pnas.93.17.8962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xie Q, Suarez-Lopez P, Gutierrez C. EMBO J. 1995;14:4073–4082. doi: 10.1002/j.1460-2075.1995.tb00079.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xie Q, Sanz-Burgos A P, Hannon G J, Gutierrez C. EMBO J. 1996;15:4900–4908. [PMC free article] [PubMed] [Google Scholar]
- 14.Liu L, Saunders K, Thomas C L, Davies J W, Stanley J. Virology. 1999;256:270–279. doi: 10.1006/viro.1999.9616. [DOI] [PubMed] [Google Scholar]
- 15.Chellappan S, Kraus V B, Kroger B, Munger K, Howley P M, Phelps W C, Nevins J R. Proc Natl Acad Sci USA. 1992;89:4549–4553. doi: 10.1073/pnas.89.10.4549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Moran E. FASEB J. 1993;7:880–885. doi: 10.1096/fasebj.7.10.8344487. [DOI] [PubMed] [Google Scholar]
- 17.Moran E. Nature (London) 1988;334:168–170. doi: 10.1038/334168a0. [DOI] [PubMed] [Google Scholar]
- 18.Nagar S, Pedersen T J, Carrick K M, Hanley-Bowdoin L, Robertson D. Plant Cell. 1995;7:705–719. doi: 10.1105/tpc.7.6.705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Egelkrout E M, Robertson D, Hanley-Bowdoin L. Plant Cell. 2001;13:1437–1452. doi: 10.1105/tpc.13.6.1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Okada K, Takebe I, Nagata T. Mol Gen Genet. 1986;205:398–403. [Google Scholar]
- 21.Iida A, Yamashita T, Yamada Y, Morikawa H. Plant Physiol. 1991;97:1585–1587. doi: 10.1104/pp.97.4.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Meyer P, Walgenbach E, Bussmann K, Hombrecher G, Saedler H. Mol Gen Genet. 1985;210:513–518. [Google Scholar]
- 23.Kartzke S, Saedler H, Meyer P. Plant Sci. 1990;67:63–72. [Google Scholar]
- 24.Villemont E, Dubois F, Sangwan R S, Vasseur G, Bourgeois Y, SangwanNorreel B S. Planta. 1997;201:160–172. [Google Scholar]
- 25.Jefferson R A, Kavanagh T A, Bevan M W. EMBO J. 1987;6:3901–3907. doi: 10.1002/j.1460-2075.1987.tb02730.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chalfie M, Tu Y, Euskirchen G, Ward W W, Prasher D C. Science. 1994;263:802–805. doi: 10.1126/science.8303295. [DOI] [PubMed] [Google Scholar]
- 27.de Wet J R, Wood K V, Helinski D R, DeLuca M. Proc Natl Acad Sci USA. 1985;82:7870–7873. doi: 10.1073/pnas.82.23.7870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bowen B. In: Transgenic Plants. Kung S-D, Wu R, editors. San Diego: Academic; 1993. pp. 89–123. [Google Scholar]
- 29.Bruce W, Folkerts O, Garnaat C, Crasta O, Roth B, Bowen B. Plant Cell. 2000;12:65–79. doi: 10.1105/tpc.12.1.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.White J, Chang S Y P, Bibb M J, Bibb M J. Nucleic Acids Res. 1990;18:1062–1062. doi: 10.1093/nar/18.4.1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Odell J T, Nagy F, Chua N H. Nature (London) 1985;313:810–812. doi: 10.1038/313810a0. [DOI] [PubMed] [Google Scholar]
- 32.An G, Ebert P R, Yi B Y, Choi C H. Mol Gen Genet. 1986;203:245–250. [Google Scholar]
- 33.Christensen A H, Sharrock R A, Quail P H. Plant Mol Biol. 1992;18:675–689. doi: 10.1007/BF00020010. [DOI] [PubMed] [Google Scholar]
- 34.An G H, Mitra A, Choi H K, Costa M A, An K S, Thornburg R W, Ryan C A. Plant Cell. 1989;1:115–122. doi: 10.1105/tpc.1.1.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Timmermans M C, Das O P, Messing J. Nucleic Acids Res. 1992;20:4047–4054. doi: 10.1093/nar/20.15.4047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Komari T. Plant Cell Rep. 1990;9:303–306. doi: 10.1007/BF00232856. [DOI] [PubMed] [Google Scholar]
- 37.Komari T, Hiei Y, Saito Y, Murai N, Kumashiro T. Plant J. 1996;10:165–174. doi: 10.1046/j.1365-313x.1996.10010165.x. [DOI] [PubMed] [Google Scholar]
- 38.Lowe K, Bowen B, Hoerster G, Ross M, Bond D, Pierce D, Gordonkamm B. Bio/Technology. 1995;13:677–682. [Google Scholar]
- 39.Student W S. Biometrika. 1907;5:351–360. [Google Scholar]
- 40.Armstrong C L, Romeroseverson J, Hodges T K. Theor Appl Genet. 1992;84:755–762. doi: 10.1007/BF00224181. [DOI] [PubMed] [Google Scholar]
- 41.Songstad D D, Armstrong C L, Petersen W L, Hairston B, Hinchee M A W. In Vitro Cell Dev Biol Plant. 1996;32:179–183. [Google Scholar]
- 42.Ishida Y, Saito H, Ohta S, Hiei Y, Komari T, Kumashiro T. Nat Biotechnol. 1996;14:745–750. doi: 10.1038/nbt0696-745. [DOI] [PubMed] [Google Scholar]
- 43.Nagata T, Nemoto Y, S. H. Int Rev Cytol. 1992;132:1–30. [Google Scholar]
- 44.Planchais S, Glab N, Trehin C, Perennes C, Bureau J M, Meijer L, Bergounioux C. Plant J. 1997;12:191–202. doi: 10.1046/j.1365-313x.1997.12010191.x. [DOI] [PubMed] [Google Scholar]
- 45.Ach R A, Durfee T, Miller A B, Taranto P, Hanley-Bowdoin L, Zambryski P C, Gruissem W. Mol Cell Biol. 1997;17:5077–5086. doi: 10.1128/mcb.17.9.5077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wright E A, Heckel T, Groenendijk J, Davies J W, Boulton M I. Plant J. 1997;12:1285–1297. doi: 10.1046/j.1365-313x.1997.12061285.x. [DOI] [PubMed] [Google Scholar]
- 47.Gooding P S, Batty N P, Goldsbrough A P, Mullineaux P M. Nucleic Acids Res. 1999;27:1709–1718. doi: 10.1093/nar/27.7.1709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gutierrez C. Cell Mol Life Sci. 1999;56:313–329. doi: 10.1007/s000180050433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hanley-Bowdoin L, Elmer J S, Rogers S G. Plant Cell. 1989;1:1057–1067. doi: 10.1105/tpc.1.11.1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hanley-Bowdoin L, Elmer J S, Rogers S G. Proc Natl Acad Sci USA. 1990;87:1446–1450. doi: 10.1073/pnas.87.4.1446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Potrykus I. Physiol Plant. 1990;79:125–134. [Google Scholar]
- 52.Potrykus I. Trends Biotechnol. 1989;7:269–273. [Google Scholar]
- 53.Gordon-Kamm W J, Spencer T M, Mangano M L, Adams T R, Daines R J, Start W G, Obrien J V, Chambers S A, Adams W R, Willetts N G, et al. Plant Cell. 1990;2:603–618. doi: 10.1105/tpc.2.7.603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Register J C, III, Peterson D J, Bell P J, Bullock W P, Evans I J, Frame B, Greenland A J, Higgs N S, Jepson I, Jiao S, et al. Plant Mol Biol. 1994;25:951–961. doi: 10.1007/BF00014669. [DOI] [PubMed] [Google Scholar]
- 55.Cho M J, Jiang W, Lemaux P G. Plant Sci. 1998;138:229–244. [Google Scholar]
- 56.Bano M S, Christou P. Mol Breed. 1999;5:471–480. [Google Scholar]
- 57.Kaeppler H F, Carlson A R, Menon G K. In Vitro Cell Dev Biol Plant. 2001;37:120–126. [Google Scholar]
- 58.Weld R, Heinemann J, Eady C. Plant Mol Biol. 2001;45:377–385. doi: 10.1023/a:1010798625203. [DOI] [PubMed] [Google Scholar]
- 59.Gordon-Kamm B, Spencer T M, Baszczynski C L, Bruce W, Tomes D T. In: Molecular Improvements of Cereal Crops. Vasil I K, editor. London: Kluwer Academic; 1999. [Google Scholar]
- 60.Kaeppler H F, Menon G K, Skadsen R W, Nuutila A M, Carlson A R. Plant Cell Rep. 2000;19:661–666. doi: 10.1007/s002999900167. [DOI] [PubMed] [Google Scholar]
- 61.Miller M, Tagliani L, Wang N, Berka B, Bidney D, Zhao Z-Y. Transgenic Res. 2002;11:381–396. doi: 10.1023/a:1016390621482. [DOI] [PubMed] [Google Scholar]
- 62.Ferre F. Gene Quantification (Advanced Biomedical Technologies) Boston: Birkhäuser; 1998. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.