Abstract
A variety of posttranscriptional mechanisms affects the processing, subcellular localization, and translation of messenger RNAs (mRNAs). Translational control appears to occur primarily at the initiation rather than the elongation stage. It has been suggested that translation is mediated largely by means of a cap-binding/scanning mechanism. On the basis of recent findings, we propose here that differential binding of particular mRNAs to eukaryotic 40S ribosomal subunits before translation may also selectively affect rates of polypeptide chain production. In this view, ribosomal subunits themselves are considered to be regulatory elements or filters that mediate interactions between particular mRNAs and components of the translation machinery. Differences in these interactions affect how efficiently individual mRNAs compete for ribosomal subunits. These competitive interactions would depend in part on the complementarity between sequences in mRNA and rRNA, as well as on structural differences among ribosomes in different cell types. By these means, translation may either be enhanced through increased recruitment of ribosomes or inhibited through strong interactions that sequester mRNAs. We propose that ribosomal filters may be important in cell differentiation and describe experimental tests for the filter hypothesis.
The repertoire of proteins expressed by eukaryotic cells at different developmental stages is restricted by their mRNA levels, which are affected to some extent by mechanisms that control transcription. In addition, mRNA levels are regulated by the rates at which various mRNAs are degraded (1), by alternative splicing of some mRNAs (2, 3), and by localization of certain mRNAs to specific regions of the cell (4, 5). Protein synthesis rates are also controlled by a number of posttranscriptional mechanisms that affect the efficiency with which individual mRNAs or groups of mRNAs are translated.
Translation of the messenger population is affected by both global mechanisms that influence the mRNA population as a whole and selective mechanisms that influence individual mRNAs or small groups of mRNAs. Although a variety of regulatory mechanisms are known to affect translation (9), the ribosome itself is not generally considered to be a regulatory element. Results of our studies and the work of others suggest that specific sequences within some mRNAs are sites of direct binding to ribosomes and that these interactions affect translation efficiency. We postulate here a hypothesis suggesting that ribosomes are not simply translation machines, but are regulatory elements that can selectively influence or filter the translation of various mRNAs.
This filter hypothesis proposes that specific mRNA⋅rRNA and mRNA⋅ribosomal protein interactions at sites on the ribosomal subunits are important in controlling translation. The binding interactions depend on sequences within different mRNAs that compete for sites on the ribosomal subunits. The hypothesis further predicts that these competitive interactions between mRNAs and ribosomal subunits may be modulated by ribosome heterogeneity, manifested as differences in the affinity for mRNAs of various sites on the ribosomal subunits. This heterogeneity has been shown to arise either as a result of variation in rRNA or ribosomal protein composition, or as a result of interactions with various associated factors (e.g., refs. 6–8). According to the filter hypothesis, ribosome heterogeneity is expected to lead to differential rates of mRNA translation in different cell types or even within the same cell. As background for the hypothesis, we first consider the known mechanisms of translational control.
Mechanisms of Translational Control
Control of the rate of translation of the messenger population appears to be restricted mainly to initiation rather than elongation (reviewed in ref. 9). Although the molecular details of translation initiation are not fully understood, they provide examples of biological degeneracy and complexity (10).
One mechanism by which translation is initiated involves the cap, a 7-methylguanosine that is linked to the 5′ ends of RNA polymerase II transcripts by a 5′-5′-triphosphate bond (11). In addition to initiation of translation, the cap has been implicated in mRNA stability, splicing, and transport (12). During initiation, the cap facilitates the recruitment of 40S ribosomal subunits through a complex of factors (13), including the eukaryotic initiation factor eIF4E, which binds to the cap, the poly(A)-binding protein (PABP), which binds to the 3′ end of the mRNA, and eIF4G, which links both ends of the mRNA by binding to both eIF4E and PABP. Initiation factor eIF4G also binds to the 43S preinitiation complex, which contains the 40S ribosomal subunit, eIF3, eIF1A, eIF2, the initiator methionine tRNA, and GTP. After recruitment, the 40S subunit is thought to interact with the 5′ leader of the mRNA and proceed (or “scan”) to the AUG initiation codon. The complex then attaches to the 60S subunit, and peptide elongation begins (reviewed in ref. 13). The translation of some mRNAs, such as β-globin mRNA, appears to be strictly cap-dependent (e.g., refs. 14 and 15).
Not all mRNAs initiate translation in a cap-dependent manner. For example, picornaviral RNAs are uncapped, but are still able to initiate translation. These RNAs recruit ribosomes at sequences contained within their 5′ leaders that are commonly referred to as an internal ribosome entry site (IRES) (16, 17). Internal initiation of different viral IRESes appears to occur by more than one mechanism, and different IRESes vary dramatically in their initiation factor requirements (18). The initiation of translation by IRESes is not limited to viral mRNAs. Certain cellular mRNAs also contain IRESes, some of which appear to facilitate translation when cap-dependent initiation is compromised, as occurs during the G2/M phase of the cell cycle and under different types of cellular stress (e.g., refs. 19–26). The mechanisms by which cellular IRESes facilitate translation are poorly understood but, as we discuss below, certain cellular IRESes appear to be composed of short regulatory sequences, or modules, some of which are complementary to rRNA. The translation efficiency of some mRNAs is affected by such regulatory cis sequences, which are often found within the 5′ leader and 3′ noncoding regions. If these sequences are introduced into unrelated mRNAs, they can affect the translation of those mRNAs in a fashion similar to that of their native messages (e.g., ref. 27).
The effects of some mRNA cis sequences are related to their ability to form stem-loop and pseudoknot structures, whereas other sequences serve as binding sites for trans factors. For example, the iron-responsive element (IRE) is a hairpin structure contained within the ferritin mRNA and a few other mRNAs whose translation is controlled by iron (28). In the absence of iron, the iron-regulatory protein (IRP) binds to the IRE, and blocks translation. In the presence of iron, the IRP no longer binds to the IRE and translation proceeds. Other sequences that affect translation include the terminal oligopyrimidine tract (TOP), the binding sites for the pyrimidine-tract binding protein (PTB), and sites for the upstream of n-ras (unr) binding protein (29–31). Trans factors that interact with cis sequences also include RNAs. For example, in Caenorhabditis elegans, two small noncoding RNAs, lin-4 and let-7, block the translation of target mRNAs by base pairing to complementary segments contained within the 3′ untranslated regions of those mRNAs (32).
The Ribosome Filter Hypothesis
As suggested by our studies (27, 33–36) and by the observations of others (see below), some mRNA cis-regulatory sequences may affect translation by binding directly to 40S ribosomal subunits. These interactions may be further influenced by the fact that eukaryotic ribosomes differ structurally in different cell types or during different stages of development (e.g., ref. 6). The hypothesis that cis sequences at various locations in mRNAs interact directly with ribosomes was prompted by our earlier observation that large numbers of mRNAs contain segments that are similar or complementary to either 18S or 28S rRNAs (33). We proposed that mRNA sequences similar to those of rRNA might mimic it and bind ribosomal proteins, whereas sequences complementary to those of rRNA might base pair to the rRNA itself. The rRNA-like sequences range in size from ≈10 to several hundred nucleotides and match sequences located in both the conserved regions and in the expansion segments of the rRNAs. It is striking that 4 segments of the 18S rRNA each match several hundred rRNA-like sequences in mRNAs. In an independent study, shorter (7–14 nucleotides) GC-rich segments that are complementary to 13 regions of the 18S rRNA have been postulated to occur in all mRNAs (37). It has been suggested that these so-called “clinger” fragments function by attaching mRNAs to 18S rRNA, thus increasing their chances of being translated. As will be reviewed later, some complementary mRNA sequences bind ribosomes through base pairing, affecting translation efficiency and functioning as IRES modules.
A key question raised by these observations is whether ribosomal subunits themselves can selectively affect translation in a manner that is different from the effects of initiation or elongation factors, from those of mRNA secondary structures, or from those of binding sites for trans factors. Inasmuch as a variety of different individual messages in mRNA populations may compete for ribosomal subunits and because ribosomal subunits may be structurally different in different cell types, we suggest that the specificity of filter interactions may be modified during development and differentiation. We will first outline the filter hypothesis, then propose some mechanisms for its action, review a body of supporting evidence, and finally suggest some experimental tests.
In basic outline, the hypothesis postulates that:
(i) The ribosome is a regulatory structure that embodies mechanisms for preferentially translating particular members of the message population over others. The proposed filter mechanisms depend on specific interactions between ribosomal subunits and segments of mRNAs. Direct interactions include base pairing between complementary segments of mRNAs and rRNA, between segments that are noncomplementary, or by the binding of mRNA sequences to ribosomal proteins. Most of the interactions between mRNAs and ribosomes are proposed to occur with components of the smaller 40S ribosomal subunit. However, interactions with components of the larger 60S ribosomal subunit or with 80S ribosomal complexes are not excluded.
(ii) As filters for translation, ribosomes may display a continuum of effects ranging from interactions that act to recruit particular mRNAs and enhance translation to interactions that sequester certain mRNAs and diminish their translation rates.
(iii) The hypothesis emphasizes competitive interactions among various mRNA sequences for binding to rRNA or ribosomal proteins. Because the filter interactions are binary, the presence of similar or identical cis sequences in different mRNAs could lead to competition for binding sites on the ribosomal subunits. Segments of mRNAs may therefore act as competitive modules or domains. It follows that mRNAs that bind to particular complementary sites on ribosomal subunits might affect the translation of other mRNAs or groups of mRNAs that bind to these same sites, thereby modulating the specificity of the filter itself.
(iv) The filter might also be modulated by altering or masking particular binding sites on ribosomes. This might occur as a consequence of ribosome heterogeneity or by interaction with extraribosomal factors, including ribosome-associated proteins and noncoding RNAs such as microRNAs (32, 38, 39). Some of these short (21- to 24-nucleotide) RNAs appear to be riboregulators that affect the translation of other RNAs by base pairing to them (32).
A central mechanism for filtering translation during development may result from the evolution of a series of similar cis sequences in particular subsets of structurally different mRNAs. These sequences might occur in 5′ leaders, coding regions, or 3′ untranslated regions. The presence of such sequences would enable the coordinate regulation of translation of these subsets of otherwise unrelated mRNAs.
Modulation of Translation Initiation by the Filter.
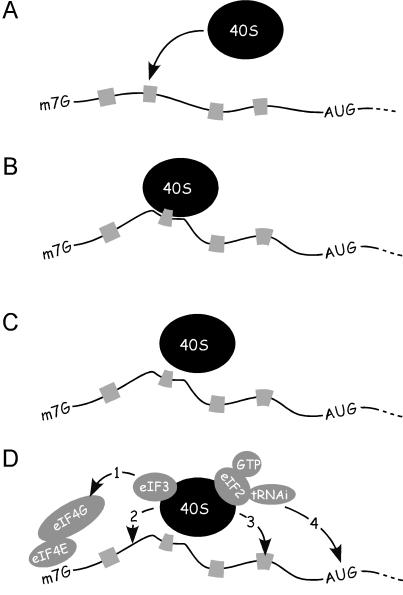
How might the filter operate? Strong interactions between ribosomal subunits and mRNA cis sequences might prevent translation, whereas weaker interactions might allow detachment and clustering, thereby increasing the local concentration of 40S subunits (Fig. 1). Binding to mRNA cis sequences might be followed by movement of the ribosomal subunit on the mRNA to initiate translation. This movement could be toward the cap by means of initiation factors, or the subunit could begin scanning toward the initiation codon directly. As suggested in Fig. 1, the clustering of 40S subunits as a result of mRNA–ribosome interactions would provide an advantage for nonlinear movement or shunting of the 40S subunit along the 5′ leader. Such interactions between segments of mRNA and 18S rRNA have been proposed to be the basis for shunting in adenovirus mRNA (40). A subset of mRNA binding sites may act as IRES modules, whereas the same or perhaps other binding sites might facilitate shunting. Alternatively, as shown in the figure, after its initial binding interaction, the 40S ribosomal subunit may move directly to the initiation codon by means of the initiator tRNA of the ternary complex.
Fig 1.
mRNA–40S ribosomal subunit interactions and their effects on translation initiation as predicted by the ribosome filter hypothesis. (A) A 40S ribosomal subunit binds to the mRNA at binding sites, which are indicated schematically as gray bars. A subset of such binding sites may function as IRES modules. (B) In some cases, strong interactions between 40S subunits and mRNA binding sites may slow or prevent subsequent movement of the ribosomal subunit and block translation. (C) Weaker interactions between 40S subunits and mRNA binding sites would allow detachment and local clustering, which may favor an initiation event by increasing the local concentration of the 40S subunits. For clarity, only one ribosomal subunit is indicated; local clustering would actually occur as a result of multiple binary interactions. (D) After the initial binding of the 40S subunit to a binding site on the mRNA, the subunit might interact with another region of the mRNA, which may or may not require prior detachment from the initial binding site. The 40S subunit might: 1, move to the cap by means of initiation factors as indicated; 2, reorient on the mRNA and begin scanning; 3, shunt to another binding site; or 4, move to the initiation codon by means of the Met-tRNAi of the ternary complex (eIF2, GTP, and Met-tRNAi). Note that the initiation factors indicated in D are not indicated in A–C, but they may also be present during the events depicted. m7G, methylguanosine cap.
The interactions described in Fig. 1 could also occur after recruitment of the 40S subunit to the 5′ end or 3′ end of the mRNA. The 40S subunit can be linked to the 5′ cap, to the 3′ poly(A) tail, or to a circularized mRNA by using various combinations of eIF4E, eIF4G, eIF3, and poly(A)-binding protein. In any of these scenarios, the potential interactions are predicted to be the same as those depicted in Fig. 1.
Evidence for Binary Interactions of Ribosomal Subunits with Various Sites of mRNA
Interactions with Ribosomal Subunits.
It has been postulated that interactions between the mRNA and the components of the 43S complex, including the 40S subunit itself, might be important for translation initiation (13). One of the best-characterized examples of a direct interaction between an mRNA sequence and a 40S ribosomal subunit comes from the hepatitis C virus (HCV) IRES, which was shown to bind directly to the 40S subunit (41). This binding has been visualized by cryoelectron microscopy, which showed interactions between the HCV IRES and the head and platform of the 40S subunit (42).
Interactions with rRNA.
Numerous studies now indicate that direct physical interactions between segments of mRNA and rRNA occur and affect translation efficiency. Data from our own and other laboratories suggest that many segments of 18S rRNA within 40S ribosomal subunits are accessible and these are able to base pair to complementary sequences contained within some mRNAs (e.g., refs. 34, 35, 37, 43). In addition, the accessibility of the α-sarcin domain within 28S rRNA of the large ribosomal subunit has been shown by using oligonucleotides complementary to this segment to stop protein synthesis (44).
These results are consistent with the high-resolution structures of the 30S subunit of Thermus thermophilus and of the 50S ribosomal subunit of Haloarcula marismortui, which indicate that the functional portion of these subunits consists of rRNA. Much of the prokaryotic rRNA is accessible, and although the structures of eukaryotic ribosomal subunits are not yet as well resolved, it is expected that the structures will be fundamentally similar (45). Direct evidence that prokaryotic rRNA is accessible for base pairing comes from studies of the small and large subunit rRNAs in Escherichia coli (46, 47). These studies used several hundred fluorescently labeled oligonucleotide probes to target all regions of these rRNAs and showed that most regions of both rRNAs were accessible to complementary probes. Indeed, only 7% and 13% of the probes were targeted to inaccessible regions of the large and small ribosomal subunits, respectively.
We have analyzed in greater detail the rRNA binding and the effects on translation of the mRNAs that encode mouse ribosomal protein S15 and the mouse Gtx homeodomain protein. Both mRNAs contain complementary sequence matches to 18S rRNA, which were found to inhibit translation when introduced into unrelated mRNAs (34, 35). A strong correlation between the degree of complementarity and the extent to which translation was inhibited suggested that complementary matches might base pair and sequester the mRNA, thus preventing further translation.
We also identified a 9-nucleotide sequence within the 5′ leader of Gtx homeodomain mRNA that was perfectly complementary to 18S rRNA. This sequence, which is not predicted to contain much secondary structure, nevertheless could function as an IRES (27). When multiple copies of this small module were linked together, IRES activity increased greatly and, in some cases, the activity of the linked modules was much greater than the sum of their individual activities. We proposed that a certain degree of complementarity (but not too much) was important for 40S ribosomal subunit recruitment and subsequent translation. This conclusion was supported by more recent studies in which we created mutations in the modules that increased or decreased their degree of complementarity. Constructs with increased complementarity were translated poorly and a polysome analysis showed that these highly complementary mRNAs became strongly associated with 40S ribosomal subunits (unpublished results). Similar results were obtained in another study (48), which showed that an mRNA that was engineered to contain an extensive complementary match to a segment of the 18S rRNA became stably associated with 40S ribosomal subunits and was translated poorly.
On the assumption that the results obtained with Gtx were unlikely to be an isolated case, we used a selection methodology to search for other short sequences with similar properties (36). We found several such cis sequences within the 5′ leader of the mRNA for the Rbm3 protein induced by cold stress (27). A 22-nucleotide segment functioned independently as a potent IRES, and several relatively short sequences adjoining the IRES enhanced or inhibited translation in a cell-type-specific manner (unpublished observations). The results prompt an analogy to enhancers and silencers in transcriptional control.
Consistent with the filter hypothesis, many of the short cis sequences identified in our studies contained significant complementary matches to 18S rRNA. These studies also showed that each of three different IRES modules containing short complementary sequence matches to 18S rRNA had effects on translation that varied in different cell lines. The results raise the possibility that the accessibility of the complementary sequences in ribosomal subunits may differ in such cell lines. An alternative possibility is that different cell-type-specific proteins bind either to the IRES sequences or to the rRNA sequences with differential effects.
The ability of ribosomes to base pair to mRNAs and affect their translation is well illustrated in a study that introduced sequences with complementarity to particular cellular mRNAs into the Tetrahymena thermophila 28S rRNA (49). Three different complementary sequences were tested in the 28S rRNA, and in each case, expression of the corresponding mRNA was specifically inhibited, suggesting that the complementary sequence within the 28S rRNA base paired to its complement in the mRNA and blocked its expression. Base pairing of mRNAs to 18S rRNA has also been postulated to be important in the recruitment of ribosomes at the poliovirus and other viral IRESes (50).
Interactions with Ribosomal Proteins.
In contrast to the analysis of complementary interactions between mRNAs and rRNA, the interaction with ribosomal subunits of segments of mRNAs that resemble or mimic rRNA sequences has not yet been investigated extensively. We will not deal with this possibility at length here, except to point out that the investigation of these interactions may yield additional support for the filter hypothesis. It has been shown that, in prokaryotes, the translation of a number of ribosomal protein mRNAs is controlled by their interactions with ribosomal proteins, some of which appear to mimic the rRNA targets of these proteins (51). Also, the expression in eukaryotes of particular ribosomal proteins is controlled by specific interactions with the mRNAs that encode them (52–56). However, it remains to be determined whether binding can occur in the context of intact ribosomal subunits.
Potential Effects of Ribosome Heterogeneity on the Filter
The filter hypothesis predicts that ribosomes will initiate translation of various individual mRNAs in a population with different relative efficiencies. It follows that these efficiencies may be altered when the accessibility or structure of the binding sites on the ribosomal subunits has been altered. Observations indicating that ribosomal subunits are heterogeneous led to the suggestion, as early as 1970 (57), that this heterogeneity is important for the expression of specific proteins. Although the methodology used to show ribosomal protein heterogeneity in some of the older studies was questioned (58), heterogeneity was also noted by using improved methodologies (reviewed in ref. 6). It may be significant that studies of differential gene expression under numerous experimental paradigms have revealed dynamic regulation of ribosomal protein mRNAs (e.g., ref. 59), in some cases resulting in changes in ribosomal protein production (60). Although the finding that various ribosomal proteins have extraribosomal functions adds to the complexity in interpretation of the data (61), ribosome heterogeneity in differentiated cells may well alter the interactions that form the basis for the ribosome filter.
Heterogeneity in Ribosomal Proteins.
In maize and yeast, a set of very acidic ribosomal proteins (P-proteins) that are associated with 60S ribosomal subunits are heterogeneous (62, 63). In yeast, ribosomes from stationary phase are deficient in P-proteins when compared with those from exponentially growing cells (63). By using cell-free lysates, it was demonstrated that yeast ribosomes lacking P-proteins translated particular mRNAs differentially as compared with ribosomes containing P-proteins (64). The addition of exogenous P-proteins to lysates lacking them abrogated the specific translational differences, suggesting that differences in translation efficiency are attributable to heterogeneity in the P-proteins.
Ribosome heterogeneity has been well documented in the cellular slime mold Dictyostelium discoideum, in which ribosomes of vegetative amoebae differ from those of spores and cells at different stages of development (reviewed in ref. 6). These differences included qualitative and quantitative differences in 6 and 29 ribosomal proteins, respectively, methylation differences in 14 ribosomal proteins, and phosphorylation differences in 2 ribosomal proteins.
A source of ribosome heterogeneity that has received much attention is the phosphorylation status of ribosomal protein S6, a protein that is phosphorylated when cells divide or are stimulated to grow (65). There is some evidence that the phosphorylation of this ribosomal protein affects the translation of a particular subset of mRNAs (TOP mRNAs), but this is based on correlations, and definitive experiments are lacking (65).
Another possible source of heterogeneity is genetic. Most eukaryotes have one functional gene and several pseudogenes for each ribosomal protein, and there are genetic variants of some ribosomal proteins in higher eukaryotes. These include ribosomal protein S4, which is present on the X and Y chromosomes (66), and S3, S6, S17, and L17, each of which appear to have multiple homologous variants.
A remarkable example of ribosome heterogeneity has been seen in Drosophila embryos. Mitochondria-type ribosomes, comprised of large and small mitochondrial rRNAs and at least two mitochondrial ribosomal proteins, were found to be present in the germ plasm on polysomes, together with cytoplasmic ribosomes (67). These results, together with those from earlier studies, suggest that the activity of mitochondrial-type ribosomes in the germ plasm is essential for the production of proteins required for formation of germ cells (68).
Heterogeneity in Ribosomal RNA.
In addition to heterogeneity in ribosomal protein composition, there is evidence that variations in rRNA can contribute to ribosome heterogeneity. rRNAs are transcribed from multigene families and heterogeneity arises from nucleotide substitutions and deletions among the individual rRNA genes. An analysis of human 28S rRNA genes revealed that 28S rRNA comprises a heterogeneous group of RNAs (7, 69). These rRNAs were shown to be transcribed and to be present both in monosomes and in polysomes (70). Most of the sequence differences in the 28S rRNAs are contained within expansion segments. These regions account for most of the increase in size between prokaryotic and eukaryotic ribosomes and they show great variability, even between closely related species (71). In one study, 35 different variants of human 28S rRNA were obtained in an analysis of expansion segment V8 (72). It was also noted that different human cell lines expressed different 28S rRNA genes. Similarly, an analysis of variants of expansion segment V5 within a single individual identified four sites of variability (73). In addition to the sequence variation that occurs in expansion segments, heterogeneity in the 28S rRNA genes has also been noted in regions that are conserved between species (70). Although 18S rRNA appears to be much less variable than 28S rRNA, some heterogeneity in 18S rRNA genes has also been described (69, 74).
In addition to the 28S and 18S rRNAs that comprise the bulk of the large and small ribosomal subunits, respectively, the sequence of the shorter 5S rRNA can also be heterogeneous. Heterogeneity in 5S rRNA has been noted in a variety of eukaryotes. In the fungus Neurospora crassa, there are at least seven types of 5S rRNA genes, and it has been suggested that this heterogeneity may lead to the production of functionally heterogeneous ribosomes (75). In Xenopus laevis, there are two variants of 5S rRNA that differ at 6 of the 120 nucleotides; one variant is expressed in oocytes, and the other is expressed in somatic cells (76).
Ribosome heterogeneity might also result from posttranscriptional modification of rRNA at specific nucleotides. One such modification involves methylation, a process directed by the base pairing of small nucleolar RNAs (snoRNAs) to complementary segments of rRNA (77).
Other Sources of Heterogeneity.
Ribosome heterogeneity can occur as a result of interactions with cellular molecules. Interactions with ribosomes of extraribosomal proteins such as the laminin-binding protein precursor p40 (LBP/p40) (8), an Hsp70 cytosolic molecular chaperone SSb (78), and the fragile X mental retardation protein (FMRP) (79), are likely to affect the structure of the ribosomal subunits and alter the accessibility of different sites on these subunits.
Finally, another potential source of heterogeneity that must be considered is the degradation of ribosomes. It is known that rabbit reticulocyte ribosomes can translate efficiently in cell-free lysates, even when these lysates are nuclease-treated and the rRNAs are fragmented (e.g., ref. 80). Inasmuch as these ribosomes are still active despite the presence of partially degraded rRNA, it may be fruitful to consider the possibility that aging ribosomes can contribute to ribosome heterogeneity within cells. It is not yet known whether ribosomes continue to translate while they are being degraded, or whether changes that occur during degradation, such as rRNA fragmentation, have any specific effects on translation.
In summary, numerous examples from the literature indicate that ribosomes within a species are heterogeneous. It now remains to be determined whether any of the reported heterogeneities affect the ribosome filter by altering the interactions of ribosomal subunits with particular mRNAs and thus the efficiency with which those mRNAs are translated.
Consequences and Tests
The filter hypothesis posits that ribosomes are regulatory elements that affect the translation of particular mRNAs by binding differentially to them. If this occurs generally, then we would expect to find a subset of mRNAs that remain tightly associated with highly purified 40S or 60S ribosomal subunits. The filter hypothesis also predicts that structural differences in ribosome populations and in mRNA populations may affect the filter. If this assumption is valid, one would expect that different ribosomes (for example, those that differ in their rRNA or ribosomal protein composition) might bind to particular mRNAs to different extents, thus altering the relative efficiencies of translation.
The notion that ribosome heterogeneity can lead to differences in the relative efficiencies of translation of various messages can be tested by examining ribosomes from different sources. For example, ribosomes that differ in their rRNA or ribosomal protein composition can be tested for their abilities to translate members of the same population of mRNAs with different relative efficiencies. Although this prediction is difficult to test in vivo, it can be approached in vitro by testing isolated ribosomes from different cell types for evidence of selective translation. This can be done by purifying ribosomal subunits from these cells and testing their function in a common ribosome-free background containing a fixed set of mRNAs and all other necessary translation factors, including initiation factors and tRNAs. In this type of experiment, filter effects would lead to protein expression patterns that differ with ribosomes of different origins. Ribosomes for these studies might come from different tissues, from cells at different developmental stages, or even from distinct subcellular locations within the same cell.
A consistency test of the filter mechanism might come by comparing ribosomes from radically different organisms that vary considerably in rRNA and ribosomal protein compositions. An examination of the relative efficiencies with which particular mRNAs of a given species are translated by using yeast vs. mammalian ribosomal subunits may provide a useful example.
Subcellular heterogeneity affecting filtering might be demonstrable in highly specialized cells such as neurons. Certain neurons have been shown to contain a small number of dendritically localized mRNAs and their dendrites have been found to be sites of protein synthesis (81). Earlier studies have identified IRESes in five dendritically localized mRNAs and indicated that some IRESes might be relatively more efficient in dendrites than in the cell body (19).
In considering tests of the hypothesis, a critical issue concerns the relative number of mRNAs and ribosomes in a particular cell at any one time. Would translation of a given mRNA in a population be affected by the amounts and types of other mRNAs in that population? A variety of evidence suggests that this is the case (e.g., ref. 82). Other issues concern ribosome concentrations during development and in differentiated tissues—are ribosomes in excess, or limiting, for translation? Very little is known about whether these relative concentrations change during development, in different cell types, or under different cellular conditions. Altering the relative concentrations of mRNAs and ribosomes might increase or decrease competition for binding sites on ribosomes and might, therefore, be a way to engage or disengage a ribosomal filter.
If the filter hypothesis is confirmed and the control of gene expression lies in part with the ribosome itself, it may be possible to identify ribosome-binding proteins and RNAs that modulate ribosome structure and accessibility and thereby coordinately up- or down-regulate the translation of particular subsets of mRNAs. The notion of a ribosome filter opens the possibility that pathological events affecting the structure or accessibility of sequences in ribosomes or in mRNAs may alter mRNA–ribosome interactions and thereby affect patterns of protein production. Numerous diseases have been attributed to both specific and global disruptions in translation (e.g., refs. 83 and 84). It is therefore conceivable that some pathologies may arise from events that disrupt the filter. If so, specific ribosome-binding molecules that can reverse the effects of a disrupted filter may have therapeutic applications.
Acknowledgments
We are indebted to Drs. B. A. Cunningham, J. A. Gally, and G. N. Reeke for valuable criticism. Funding was generously provided to V.P.M. by the G. Harold and Leila Y. Mathers Charitable Foundation, National Science Foundation Grant MCB9982574, and National Institutes of Health Grant GM61725, and to G.M.E. by U.S. Public Health Service Grant NS39837.
Abbreviations
IRES, internal ribosome entry site
eIF, eukaryotic initiation factor
References
- 1.Guhaniyogi J. & Brewer, G. (2001) Gene 265, 11-23. [DOI] [PubMed] [Google Scholar]
- 2.Modrek B. & Lee, C. (2002) Nat. Genet. 30, 13-19. [DOI] [PubMed] [Google Scholar]
- 3.Brett D., Pospisil, H., Valcarcel, J., Reich, J. & Bork, P. (2002) Nat. Genet. 30, 29-30. [DOI] [PubMed] [Google Scholar]
- 4.Mohr E. & Richter, D. (2001) Int. J. Biochem. Cell Biol. 33, 669-679. [DOI] [PubMed] [Google Scholar]
- 5.Gao F. B. (1998) BioEssays 20, 70-78. [DOI] [PubMed] [Google Scholar]
- 6.Ramagopal S. (1992) Biochem. Cell Biol. 70, 269-272. [DOI] [PubMed] [Google Scholar]
- 7.Gonzalez I. L., Gorski, J. L., Campen, T. J., Dorney, D. J., Erickson, J. M., Sylvester, J. E. & Schmickel, R. D. (1985) Proc. Natl. Acad. Sci. USA 82, 7666-7670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sato M., Saeki, Y., Tanaka, K. & Kaneda, Y. (1999) Biochem. Biophys. Res. Commun. 256, 385-390. [DOI] [PubMed] [Google Scholar]
- 9.Mathews M. B., Sonenberg, N. & Hershey, J. W. B. (1996) in Translational Control, eds. Hershey, J. W. B., Mathews, M. B. & Sonenberg, N. (Cold Spring Harbor Lab. Press, Plainview, NY), pp. 1–29.
- 10.Edelman G. M. & Gally, J. A. (2001) Proc. Natl. Acad. Sci. USA 98, 13763-13768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shatkin A. J. (1976) Cell 9, 645-653. [DOI] [PubMed] [Google Scholar]
- 12.Varani G. (1997) Structure (London) 5, 855-858. [DOI] [PubMed] [Google Scholar]
- 13.Pestova T. V. & Hellen, C. U. T., (2001) The Ribosome (Cold Spring Harbor Lab. Press, Plainview, NY), pp. 389–396.
- 14.Lockard R. E. & Lane, C. L. (1978) Nucleic Acids Res. 5, 3237-3347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Keiper B. D. & Rhoads, R. E. (1997) Nucleic Acids Res. 25, 395-402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pelletier J. & Sonenberg, N. (1988) Nature (London) 334, 320-325. [DOI] [PubMed] [Google Scholar]
- 17.Jang S. K., Krausslich, H. G., Nicklin, M. J., Duke, G. M., Palmenberg, A. C. & Wimmer, E. (1988) J. Virol. 62, 2636-2643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pestova T. V., Kolupaeva, V. G., Lomakin, I. B., Pilipenko, E. V., Shatsky, I. N., Agol, V. I. & Hellen, C. U. (2001) Proc. Natl. Acad. Sci. USA 98, 7029-7036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pinkstaff J. K., Chappell, S. A., Mauro, V. P., Edelman, G. M. & Krushel, L. A. (2001) Proc. Natl. Acad. Sci. USA 98, 2770-2775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cornelis S., Bruynooghe, Y., Denecker, G., Van Huffel, S., Tinton, S. & Beyaert, R. (2000) Mol. Cell 5, 597-605. [DOI] [PubMed] [Google Scholar]
- 21.Pyronnet S., Pradayrol, L. & Sonenberg, N. (2000) Mol. Cell 5, 607-616. [DOI] [PubMed] [Google Scholar]
- 22.Holcik M. & Korneluk, R. G. (2000) Mol. Cell. Biol. 20, 4648-4657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Johannes G. & Sarnow, P. (1998) RNA 4, 1500-1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chappell S. A., Owens, G. C. & Mauro, V. P. (2001) J. Biol. Chem. 276, 36917-36922. [DOI] [PubMed] [Google Scholar]
- 25.Stein I., Itin, A., Einat, P., Skaliter, R., Grossman, Z. & Keshet, E. (1998) Mol. Cell. Biol. 18, 3112-3119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fernandez J., Yaman, I., Mishra, R., Merrick, W. C., Snider, M. D., Lamers, W. H. & Hatzoglou, M. (2001) J. Biol. Chem. 276, 12285-12291. [DOI] [PubMed] [Google Scholar]
- 27.Chappell S. A., Edelman, G. M. & Mauro, V. P. (2000) Proc. Natl. Acad. Sci. USA 97, 1536-1541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rouault T. A. & Harford, J. B. (2000) in Translational Control of Gene Expression, eds. Sonenberg, N., Hershey, J. W. B. & Mathews, M. B. (Cold Spring Harbor Lab. Press, Plainview, NY), pp. 655–670.
- 29.Meyuhas O. & Hornstein, E. (2000) in Translational Control of Gene Expression, eds. Sonenberg, N., Hershey, J. W. B. & Mathews, M. B. (Cold Spring Harbor Lab. Press, Plainview, NY), pp. 671–693.
- 30.Kaminski A. & Jackson, R. J. (1998) RNA 4, 626-638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hunt S. L., Hsuan, J. J., Totty, N. & Jackson, R. J. (1999) Genes Dev. 13, 437-448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Banerjee D. & Slack, F. (2002) BioEssays 24, 119-129. [DOI] [PubMed] [Google Scholar]
- 33.Mauro V. P. & Edelman, G. M. (1997) Proc. Natl. Acad. Sci. USA 94, 422-427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tranque P., Hu, M. C.-Y., Edelman, G. M. & Mauro, V. P. (1998) Proc. Natl. Acad. Sci. USA 95, 12238-12243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hu M. C.-Y., Tranque, P., Edelman, G. M. & Mauro, V. P. (1999) Proc. Natl. Acad. Sci. USA 96, 1339-1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Owens G. C., Chappell, S. A., Mauro, V. P. & Edelman, G. M. (2001) Proc. Natl. Acad. Sci. USA 98, 1471-1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Matveeva O. V. & Shabalina, S. A. (1993) Nucleic Acids Res. 21, 1007-1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ambros V. (2001) Cell 107, 823-826. [DOI] [PubMed] [Google Scholar]
- 39.Moss E. G. (2002) Curr. Biol. 12, R138-R140. [DOI] [PubMed] [Google Scholar]
- 40.Yueh A. & Schneider, R. J. (2000) Genes Dev. 14, 414-421. [PMC free article] [PubMed] [Google Scholar]
- 41.Pestova T. V., Shatsky, I. N., Fletcher, S. P., Jackson, R. J. & Hellen, C. U. T. (1998) Genes Dev. 12, 67-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Spahn C. M., Kieft, J. S., Grassucci, R. A., Penczek, P. A., Zhou, K., Doudna, J. A. & Frank, J. (2001) Science 291, 1959-1962. [DOI] [PubMed] [Google Scholar]
- 43.Nakashima K., Darzynkiewicz, E. & Shatkin, A. J. (1980) Nature (London) 286, 226-230. [DOI] [PubMed] [Google Scholar]
- 44.Saxena S. K. & Ackerman, E. J. (1990) J. Biol. Chem. 265, 3263-3269. [PubMed] [Google Scholar]
- 45.Lafontaine D. L. & Tollervey, D. (2001) Nat. Rev. Mol. Cell. Biol. 2, 514-520. [DOI] [PubMed] [Google Scholar]
- 46.Fuchs B. M., Wallner, G., Beisker, W., Schwippl, I., Ludwig, W. & Amann, R. (1998) Appl. Environ. Microbiol. 64, 4973-4982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fuchs B. M., Syutsubo, K., Ludwig, W. & Amann, R. (2001) Appl. Environ. Microbiol. 67, 961-968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Verrier S. B. & Jean-Jean, O. (2000) RNA 6, 584-597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sweeney R., Fan, Q. & Yao, M.-C. (1996) Proc. Natl. Acad. Sci. USA 93, 8518-8523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Scheper G. C., Voorma, H. O. & Thomas, A. A. M. (1994) FEBS Lett. 352, 271-275. [DOI] [PubMed] [Google Scholar]
- 51.Nomura M., Gourse, R. & Baughman, G. (1984) Annu. Rev. Biochem. 53, 75-117. [DOI] [PubMed] [Google Scholar]
- 52.Presutti C., Ciafre, S. A. & Bozzoni, I. (1991) EMBO J. 10, 2215-2221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tasheva E. S. & Roufa, D. J. (1995) Genes Dev. 9, 304-316. [DOI] [PubMed] [Google Scholar]
- 54.Fewell S. W. & Woolford, J. L., Jr. (1999) Mol. Cell. Biol. 19, 826-834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Li H., Dalal, S., Kohler, J., Vilardell, J. & White, S. A. (1995) J. Mol. Biol. 250, 447-459. [DOI] [PubMed] [Google Scholar]
- 56.Neumann F., Hemmerich, P., von Mikecz, A., Peter, H. H. & Krawinkel, U. (1995) Nucleic Acids Res. 23, 195-202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sussman M. (1970) Nature (London) 225, 1245-1246. [DOI] [PubMed] [Google Scholar]
- 58.Sherton C. C. & Wool, I. G. (1974) J. Biol. Chem. 249, 2258-2267. [PubMed] [Google Scholar]
- 59.Bortoluzzi S., d'Alessi, F., Romualdi, C. & Danieli, G. A. (2001) Bioinformatics 17, 1152-1157. [DOI] [PubMed] [Google Scholar]
- 60.Boon K., Caron, H. N., van Asperen, R., Valentijn, L., Hermus, M. C., van Sluis, P., Roobeek, I., Weis, I., Voute, P. A., Schwab, M. & Versteeg, R. (2001) EMBO J. 20, 1383-1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wool I. G. (1996) Trends Biochem. Sci. 21, 164-165. [PubMed] [Google Scholar]
- 62.Szick-Miranda K. & Bailey-Serres, J. (2001) J. Biol. Chem. 276, 10921-10928. [DOI] [PubMed] [Google Scholar]
- 63.Saenz-Robles M. T., Remacha, M., Vilella, M. D., Zinker, S. & Ballesta, J. P. (1990) Biochim. Biophys. Acta 1050, 51-55. [DOI] [PubMed] [Google Scholar]
- 64.Remacha M., Jimenez-Diaz, A., Bermejo, B., Rodriquez-Gabriel, M. A., Guarinos, E. & Ballesta, J. P. (1995) Mol. Cell. Biol. 15, 4754-4762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Volarevic S. & Thomas, G. (2001) Prog. Nucleic Acid Res. Mol. Biol. 65, 101-127. [DOI] [PubMed] [Google Scholar]
- 66.Watanabe M., Zinn, A. R., Page, D. C. & Nishimoto, T. (1993) Nat. Genet. 4, 268-271. [DOI] [PubMed] [Google Scholar]
- 67.Amikura R., Kashikawa, M., Nakamura, A. & Kobayashi, S. (2001) Proc. Natl. Acad. Sci. USA 98, 9133-9138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kobayashi S. & Okada, M. (1989) Development (Cambridge, U.K.) 107, 733-742. [DOI] [PubMed] [Google Scholar]
- 69.Maden B. E., Dent, C. L., Farrell, T. E., Garde, J., McCallum, F. S. & Wakeman, J. A. (1987) Biochem. J. 246, 519-527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Gonzalez I. L., Sylvester, J. E. & Schmickel, R. D. (1988) Nucleic Acids Res. 16, 10213-10224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Gerbi S. A. (1996) in Ribosomal RNA Structure, Evolution, Processing, and Function in Protein Biosynthesis, eds. Zimmermann, R. A. & Dahlberg, A. E. (CRC, Boca Raton, FL), pp. 71–87.
- 72.Leffers H. & Andersen, A. H. (1993) Nucleic Acids Res. 21, 1449-1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kuo B. A., Gonzalez, I. L., Gillespie, D. A. & Sylvester, J. E. (1996) Nucleic Acids Res. 24, 4817-4824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Tautz D., Hancock, J. M., Webb, D. A., Tautz, C. & Dover, G. A. (1988) Mol. Biol. Evol. 5, 366-376. [DOI] [PubMed] [Google Scholar]
- 75.Selker E. U., Stevens, J. N. & Metzenberg, R. L. (1985) Science 227, 1340-1343. [DOI] [PubMed] [Google Scholar]
- 76.Wolffe A. P. & Brown, D. D. (1988) Science 241, 1626-1632. [DOI] [PubMed] [Google Scholar]
- 77.Kiss T. (2002) Cell 109, 145-148. [DOI] [PubMed] [Google Scholar]
- 78.Lopez N., Halladay, J., Walter, W. & Craig, E. A. (1999) J. Bacteriol. 181, 3136-3143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Tamanini F., Meijer, N., Verheij, C., Willems, P. J., Galjaard, H., Oostra, B. A. & Hoogeveen, A. T. (1996) Hum. Mol. Genet. 5, 809-813. [DOI] [PubMed] [Google Scholar]
- 80.Kennedy T. D., Hanley-Bowdoin, L. K. & Lane, B. G. (1981) J. Biol. Chem. 256, 5802-5809. [PubMed] [Google Scholar]
- 81.Steward O. & Schuman, E. M. (2001) Annu. Rev. Neurosci. 24, 299-325. [DOI] [PubMed] [Google Scholar]
- 82.Temple G. & Lodish, H. F. (1975) Biochem. Biophys. Res. Commun. 63, 971-979. [DOI] [PubMed] [Google Scholar]
- 83.Mendell J. T. & Dietz, H. C. (2001) Cell 107, 411-414. [DOI] [PubMed] [Google Scholar]
- 84.Delepine M., Nicolino, M., Barrett, T., Golamaully, M., Lathrop, G. M. & Julier, C. (2000) Nat. Genet. 25, 406-409. [DOI] [PubMed] [Google Scholar]