Abstract
The synthesis of the catecholic siderophore bacillibactin is accomplished by the nonribosomal peptide synthetase (NRPS) encoded by the dhb operon. DhbE is responsible for the initial step in bacillibactin synthesis, the activation of the aryl acid 2,3-dihydroxybenzoate (DHB). The stand-alone adenylation (A) domain DhbE, the structure of which is presented here, exhibits greatest homology to other NRPS A-domains, acyl-CoA ligases and luciferases. It's structure is solved in three different states, without the ligands ATP and DHB (native state), with the product DHB-AMP (adenylate state) and with the hydrolyzed product AMP and DHB (hydrolyzed state). The 59.9-kDa protein folds into two domains, with the active site at the interface between them. In contrast to previous proposals of a major reorientation of the large and small domains on substrate binding, we observe only local structural rearrangements. The structure of the phosphate binding loop could be determined, a motif common to many adenylate-forming enzymes, as well as with bound DHB-adenylate and the hydrolyzed product DHB*AMP. Based on the structure and amino acid sequence alignments, an adapted specificity conferring code for aryl acid activating domains is proposed, allowing assignment of substrate specificity to gene products of previously unknown function.
Keywords: adenylation domain, nonribosomal peptide synthesis, X-ray crystal structure, antibiotic biosynthesis, siderophore formation
Iron is an essential trace element for most organisms. As iron concentrations below 1 μM can be growth limiting, many bacteria synthesize low molecular weight iron chelating compounds, termed siderophores (1). Siderophores can be classified into two major groups, catecholic and hydroxamate, recently supplemented by a third main group, the carboxylate siderophores (2). Siderophores successfully compete for iron with other iron chelating agents, such as haem, and are therefore often important pathogenicity factors (3), e.g., mycobactin secreted by Mycobacterium tuberculosis, vibriobactin secreted by Vibrio cholerae, and yersiniabactin secreted by Yersinia pestis. Disruption of their production is an attractive target for the design of compounds to eliminate the growth of such pathogenic germs.
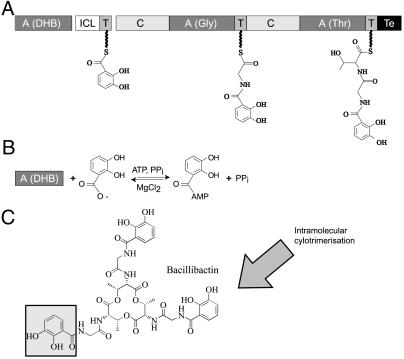
Bacillibactin (DHB–Gly–Thr)3 (4) is a cyclic catecholic siderophore produced by the soil bacterium Bacillus subtilis. Biosynthesis of bacillibactin is achieved by three enzymes encoded by the iron-regulated dhb operon (5) (Fig. 1). The three gene products DhbB, DhbE, and DhbF together form a nonribosomal peptide synthetase (NRPS) exhibiting a three-module architecture. In contrast to the universal nucleic-acid-dependent ribosomal peptide synthesis, NRPS rely solely on proteinaceous enzymatic activity. Individual residues are added to the nascent peptide chain by dedicated modules, granting the assembly of a wide variety of chemically diverse products (6, 7), including many siderophores and peptide antibiotics. DhbE is a stand-alone adenylation (A) domain that recognizes and activates 2,3-dihydroxybenzoate (DHB), whereas the two modules contained in DhbF incorporate Gly and Thr and result in the product release (Fig. 1).
Fig 1.
(A) Bacillibactin NRPS cluster from B. subtilis with the corresponding domain organization of synthetase modules. ICL, isochorismatase; C, condensation domain; T, thiolation domain. (B) Structure of the trilactone Bacillibactin, with one of the catecholic moiety activated by DhbE shaded in gray. (C) The DhbE-dependent aryl acid adenylation in peptide synthesis is an ATP-consuming process leading to a protein-bound adenylate.
Being selective for substrate recognition and activation, the A domains represent the gatekeepers of the NRPS assembly line. They belong to a superfamily of so-called adenylate-forming enzymes, which includes also the 4-coumarate-CoA ligases, acyl-CoA ligases, and the oxidoreductases. These enzymes are defined by an adenylate (AMP) binding motif that is conserved in all members (8). The A domain, either as a separate protein (as in DhbE) or fused into an NRPS module (as in DhbF), catalyzes the same first step of the catalytic reaction as the aminoacyl-tRNA-synthetases, yet share with them neither primary nor tertiary structural homology (9).
The crystal structures of two members of the adenylate-forming superfamily have been solved: the oxidoreductase luciferase from Photinus pyralis (10) and the phenylalanine-activating A domain (PheA) of the first module of the Gramicidin S NRPS of Bacillus brevis (11). Although they possess very low sequence identity (only 16% in 550 residues), luciferase and PheA exhibit an almost identical fold. Each enzyme consists of a large and a small domain, with the active site at the junction between them. Despite the similar secondary structure, however, the two proteins showed different relative orientations of the domains, suggesting that substrate binding and adenylation results in large-scale conformational changes.
The A domains of NRPSs share sequence identities of ≈30–60% (8), making the PheA structure an archetype for all amino acid activating NRPS A domains. Insights were obtained into substrate recognition and activation, and selectivity determinants could be deduced in other adenylate-forming enzymes (12). This resulted in a series of conserved sequence or “core” motifs that served as functional anchors. In the case of the aryl-acid activating domains such as DhbE, however, several of these motifs were not evident, presumably as a result of the more distant evolutionary relationship to the amino acid activating A domains.
Here we report the structure of DhbE, both in the presence and absence of substrates. In contrast to the excised PheA domain structure, DhbE is a distinct protein that activates not an amino acid, but an aryl acid. The enzyme has been cocrystallized to reveal three states: (i) in the absence of substrates (native), (ii) with bound DHB-adenylate (adenylate), and (iii) with hydrolyzed DHB*AMP (hydrolyzed; Table 1, which is published as supporting information on the PNAS web site, www.pnas.org). Incorporation of DHB reveals the substrate binding pocket, which shows an altered specificity conferring code for aryl-acid activating A-domains compared with that reported for amino acid recognition (12). The DhbE structure also confirms the roles of highly conserved residues in the superfamily of adenylate-forming enzymes.
Methods
Cloning, Overproduction, and Purification of DhbE.
DhbE was amplified from chromosomal DNA from B. subtilis ATCC21332, and cloned into the pQE60 vector (Qiagen, Hilden, Germany) by using standard techniques (13) as described (4). After successful cloning, the plasmid was sequenced by using the chain termination method (14). For heterologous expression, plasmids were transformed in Escherichia coli strain M15 [pREP4] (Qiagen; ref. 15). Subsequent expression was carried out in 2× YT medium (16) under aerobic conditions at 30°C with induction at an OD600 of 0.5 by using isopropyl-β-d-thiogalactopyranoside (final concentration of 0.25 mM) for 2–3 h. Cells were harvested by centrifugation and resuspended in Hepes buffer A (50 mM Hepes/100 mM NaCl, pH 7.8) and lysed by three passages through a French pressure cell. Cell debris were removed by centrifugation for 30 min at 35,000 × g. Supernatant was applied to a Ni2+-NTA column and purified by immobilized metal ion absorption chromatography (IMAC) on a FPLC system. Purity was estimated by Coomassie-stained SDS/polyacrylamide gels. Fractions containing the recombinant protein were pooled and dialyzed against buffer A. Afterward, the protein was concentrated by using a 50-ml Amicon cell with YM30 membrane and Centricon concentrators with the same membrane size to a final concentration of 10 mg/ml, determined photometrically at a wavelength of 280 nm. The ATP–PPi exchange reaction was performed as described (4), and reaction mixtures (100 μl final volume) contained 1 mM amino acid, 2 mM ATP, and 500 nM enzyme in assay buffer (50 mM Hepes, pH 7.8/20 mM MgCl2). Reactions were initiated by the addition of 0.15 mCi of sodium [32P]pyrophosphate and 0.1 mM PPi at pH 7.8 (1 Ci = 37 GBq).
Se-methionine (Se-Met)-labeled protein was obtained as described (17). During the purification procedure of Se-Met-DhbE, all buffers contained 5 mM 2-mercaptoethanol. The replacement of methionine by Se-Met was checked by using matrix-assisted laser desorption ionization time-of-flight (MALDI-TOF) analysis. From the 8 methionine residues in DhbE, 7 were replaced by Se-Met. Activity of Se-DhbE, determined by ATP-PPi exchange as described above, was equivalent to that of the wild-type protein.
Crystallization Conditions.
Trigonal crystals of the His-tagged DhbE were grown at 18°C by using the sitting drop vapor diffusion method with an in-house factorial screen. The protein was diluted to 8 mg/ml and mixed with an equal volume of reservoir solution. Crystals for structure determination were obtained within 4 to 5 days by using 30% polyethylene glycol (PEG) 4000/0.1 M Tris⋅HCl/0.2 M LiSO4, pH 8.6, for crystals with the substrates and in 30% PEG 4000/0.1 M Na citrate/0.1 M LiSO4, pH 5.5, for DhbE alone. For complex crystals, ATP (5 mM), DHB (2 mM), and MgCl2 (2 mM) were added to the protein solution and incubated for 5 min at 37°C to carry out the adenylation before crystallization.
Measurements and Data Refinement.
Crystals were mounted in nylon loops by using 20% glycerol as cryo-protectant and shock frozen in a cryostream (X-stream; Rigaku/MSC, Woodlands, TX) at −173°C. A native data set was collected for native (no substrate) crystals of DhbE in-house by using an R-AxisIV image plate system (Rigaku/MSC) installed on a Rigaku rotating anode generator with Yale mirrors. Multiwavelength anomalous dispersion (MAD) data sets with Se-Met labelled protein were collected of the substrate complex on the Max-Planck Gesellschaft/Gesellschaft für Biotechnologische Forschung (MPG/GBF) wiggler beam line BW6/DORIS at Deutches Elektronen-Synchrotron (DESY), Hamburg. Additionally, complex crystals with wild-type enzyme were measured on ID14–4, European Synchrotron Radiation Faculty, Grenoble, France. All data were indexed and scaled by using DENZO/SCALEPACK (18) All crystals exhibit the P31 space group with one molecule per asymmetric unit. In the presence of substrate, the crystallographic c axis increases by 8 Å (Table 1). MAD phasing was carried out by using cns (19), with density modification involving solvent flipping. The wild-type complex crystals were isomorphous to the Se-Met crystals, whereas the native crystals were solved by using Patterson Search techniques with cns. Model building was performed by using the program o (20), and the structures were refined by using cns. Data collection and refinement statistics are given in Table 1.
Results
Description of the Overall Structure.
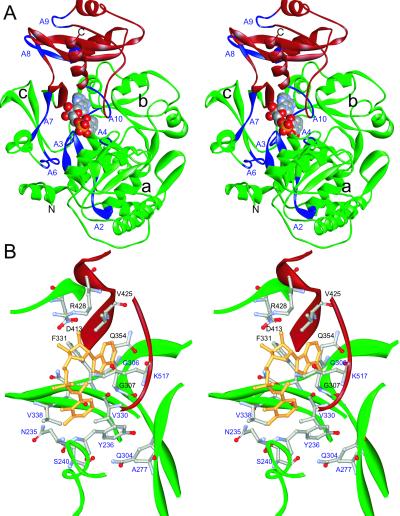
The DhbE enzyme folds into two distinct domains, a large N-terminal domain comprising residues 1–420 and a smaller compact C-terminal domain formed by residues 421–536 (Fig. 2). The fold has been described for the firefly luciferase as a hammer-and-anvil model (10, 21). In the following, the residue numbering corresponds to the DhbE sequence; exceptions referring to the PheA or luciferase sequences will be written in italics.
Fig 2.
(A) Stereo presentation of the DhbE structure (adenylated state) in presence of the product DHB-adenylate (CPK model). The core motifs are shown in blue, the N-terminal domain is shown in green, and the lid domain is shown in red. The N-terminal domain can be further divided into three, defined by a 6-strand/5-helix subdomain (a), an 8-strand/6-helix part (b), and a 5-stranded β-barrel (c). The N-terminal helix nestles between a and c. Core motif locations are also indicated. (B) Focus on the substrate-binding pocket in the adenylated state. The 10 residues determining substrate specificity are labeled in blue, those coordinating AMP in black. The side chains of these residues and the product are shown as ball-and-stick model.
The N-terminal domain can be further divided into three subdomains (Fig. 2). The cores of two of the subdomains exhibit a similar α+β topology that juxtapose one another to build up a five-layered αβαβα sandwich. The third subdomain is a β-barrel that abuts the other two. The relative arrangements of these elements resemble more closely those of luciferase. Peculiar to DhbE is the presence of a pronounced kink at cisPro-241 in the specificity determining helix (see below). The compact C-terminal domain, termed the “lid,” is made up of five β-strands and three α-helices (Fig. 2A). The lid is separated by a large cleft from the N-terminal domain, connected by only a short hinge of no regular distinct secondary structure. In luciferase, this lid is rotated by 94° with respect to the PheA structure (11). In the 3 structures presented here (native, adenylated and hydrolyzed), DhbE displays the same relative orientation as in PheA.
In DhbE, the substrate binding site for DHB and ATP is a deep compartment on the surface of the N-terminal domain at the interface to the small domain. It is partially covered by a loop formed by residues 190–199. We divide this ligand binding site into a DHB site and an adenine site, described in the following sections.
Adenylate Binding.
The adenosine moiety in DhbE is bound at the top of the catalytic cavity at the surface of the N-terminal domain, coordinated by highly conserved residues as seen in PheA (11). The adenine moiety is held in place by means of hydrophobic and hydrogen-bonding interactions, sandwiched between the aromatic side chain of F323 and the main-chain segment 306GGA308 (Fig. 2). The only residues that differ between DhbE and PheA are V425 (Y425) and F331 (Y323), both involved in coordinating the sugar moiety. Owing to the absence of a side-chain hydroxyl group in V425, no hydrogen bond can be made with the 2′-hydroxyl group of the sugar as found for PheA Y425. In PheA, the Oh group of Y323 makes a hydrogen bond with the carboxy group of D413 (D413); in DhbE, this role is substituted by a water molecule at the vertex of the F331 side chain. D413 is conserved among the superfamily of adenylate-forming enzymes (core A7: YxTGD), and is found not only in the structure of PheA but also luciferase (11) (Fig. 3). By coordinating the 2′- and 3′-hydroxyl groups of the sugar, it fixes the central portion of the AMP. D413 in turn contacts the folded side chain of R428 both in the absence of substrate and the presence of adenylate product (Fig. 4 A and C), as observed in PheA (11). In the presence of DHB and AMP, however, the R428 side chain extends away to form a salt bridge with the α-phosphate (Fig. 4B). R428 is strictly conserved in the superfamily of adenylate-forming enzymes, and site-directed mutagenesis of this residue leads to a complete loss of protein activity (22).
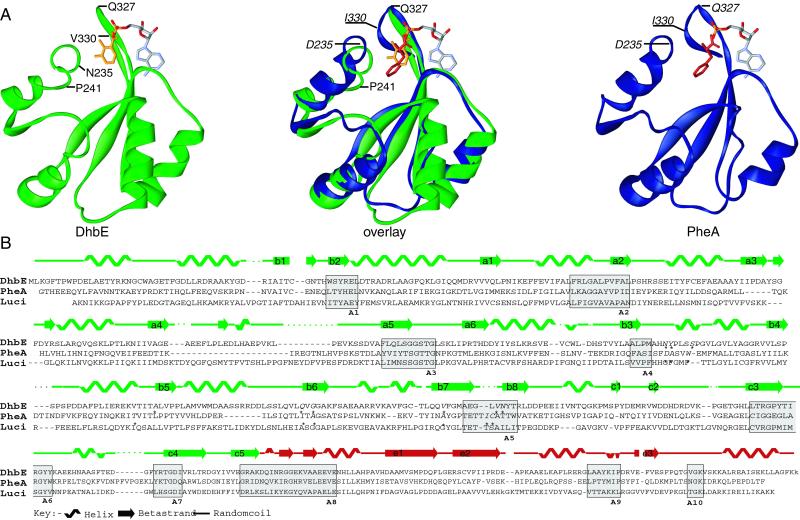
Fig 3.
(A) Close-up of the catalytic site of DhbE and PheA in the region between core A4 and core A5 (see B). The route of the main chain is represented as solid ribbon; the ligands Phe (orange), DHB (red), and AMP are represented as sticks. Residues that influence the main chain route or are involved in substrate binding are labeled. (B) Secondary structure and sequence comparison of DhbE, PheA, and Luciferase. The core motifs of NRPS A domains are boxed and labeled. The asterisks in the primary sequence indicate the 10 residues responsible for substrate specificity of the A domains.
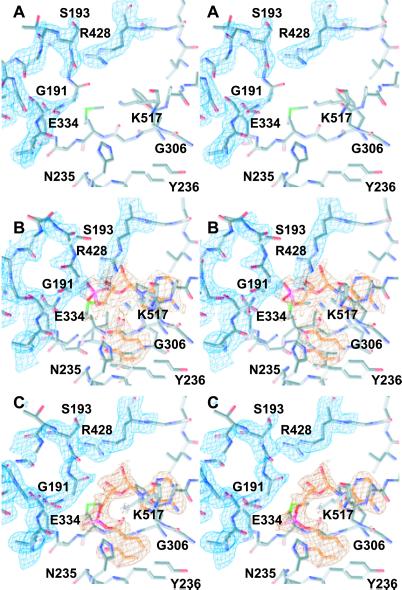
Fig 4.
Experimental (2Fo − Fc) electron density contoured at 0.9 σ, showing the active site region of DhbE. Phases were calculated after a cycle of simulated annealing omitting residues shown; remaining electron density is not shown for the sake of clarity. (A) Electron density of the p-loop in the native state (residues Ser-190–Lys-198); the loop is located above the active site, with a break in the density at Gly-191. (B) In the presence of DHB and AMP (orange, hydrolyzed state), the loop moves away from the position shown in A. Furthermore, the side chain of Arg-428 extends down to interact with the α-phosphate group. (C) On adenylate formation (orange, adenylated state), the loop returns to a conformation similar to that seen in A, as does the side chain of Arg-428, where it is in contact with Asp-413.
P-Loop Motif.
The so called p-loop (phosphate-binding loop or Walker A motif) is a common motif in many ATP-binding proteins (23), including adenylate kinases, elongation factors, Ras proteins, and ATP synthases. It is also found in all NRPS adenylation domains (6, 11) as well as in the luciferases (10, 21). The p-loop signature typically starts with a Ser or Thr residue, followed by a glycine-rich sequence and ends with a highly conserved Lys. In NRPSs, the signature sequence of this motif is S(T)G(S)GS(T)TGXXP(S)KL(G)I(V) (designated as core A3) and connects the β-strands 5 and 6 in the N-terminal domain (Fig. 3). Only few crystallographic data exist for this region, but it is expected to undergo a transition from a nonsubstrate to a substrate-bound state (23). Our data supports this theory. In each of our crystals, this segment forms an open loop (Fig. 4), anchored at its base by a hydrogen bond between the side chain hydroxyl of S190Og to the backbone amide of K198N. The density is partially broken, prohibiting an unambiguous tracing of all atoms of the loop. In the absence of ligands, no interpretable density is observed for G191. In the hydrolyzed complex, the loop moves away from the substrate (Fig. 4B), with T194 becoming disordered. On adenylate formation, segment 190SGGS193 is in a conformation similar to that of the unbound enzyme (Fig. 4C), protecting the bound adenylate, whereas residues 194TGL196 are disordered. In the latter case, an additional hydrogen bond is found between S197Og and the main-chain carbonyl S190O.
Substrate-Binding Pocket.
Like PheA, the substrate-binding site of DhbE has a channel-like entrance situated at the surface of the large N-terminal domain. The adenosine ligand blocks the entrance to the DHB binding pocket (flanked by the side chains of residues F331, V425, and Q354 on one side and the main-chain atoms of 306GGA on the other side) (Fig. 2), suggesting that productive ATP binding can only take place after DHB binding. The major determinants for DHB binding are provided by residues H234–S240. The pocket is much shallower than in PheA, and is restricted at its base by the side chain of Y236. The DHB hydroxyl groups are coordinated by S240Oγ (3′-OH) and N235Nδ2 (bivalent hydrogen bonds to 2′- and 3′-OH). The DHB carboxylate group is coordinated by H234Nɛ2 and K517Nζ of the small domain (Fig. 2). This contrasts to the situation in PheA, where only one ligand (K517) is found for the Phe carboxylate group, a second interaction being provided by a salt bridge between D235 and the Phe amino function.
A Previously Uncharacterized Specificity Code.
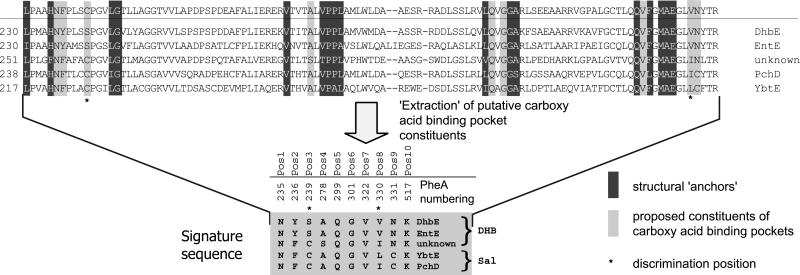
Although the PheA structure provided the means to decode substrate specificity for various members of the amino acid adenylation domain superfamily (12), aryl acid domains appeared to be different. In the light of the present study on DhbE, it is possible (i) to validate the general rules for the deduction of substrate specificity for amino- and aryl acid-activating domains, (ii) to refine the functional anchors for this class of substrates, and (iii) to discriminate between DHB (e.g., EntE) and salicylate (SAL) (e.g., YbtE) activating enzymes on the basis of their primary sequences (Fig. 5).
Fig 5.
Determination of the specificity conferring code of ca-activating domains. The primary sequence between core A4 and A5 (see Fig. 3) of nine ca activating domains are aligned by using the Clustal method. Based on the structural data of DhbE, extraction of the 10 residues conferring the substrate specificity leads to the identification of the signature sequence of ca activating A domain. The asterisks define the residues that allow discrimination between DHB and SAL.
The first anchor in amino acid-activating domains (e.g., in PheA) is usually provided by the so-called core motif A4 (YxFDxS) (Fig. 3), whose invariant Asp moiety (D235) stabilizes the α-amino group of the amino acid substrate. In aryl acid-activating domains like DhbE, however, this core motif is lacking, as the substrate contains no α-amino group. As revealed by the cocrystallization with ATP and DHB, the anchor is represented by the sequence motif “234HNYPLSSPG242” (234FDASVWEMF242), where the neutral Asn replaces the otherwise invariant Asp residue. Previous in silico studies misinterpreted this motif, aligning the conserved Tyr/Phe and first Ser residues (underlined). As revealed by the present structure, H234 aligns with F234 as a result of an altered main-chain route, caused by the presence of cisPro-241 (Fig. 3A), conserved in the aryl acid-activating domains. This results in a two-residue shift compared with the previous alignment, bringing residues N235 and Y236 responsible for DHB specificity into register with D235 and A236.
The conserved N235 hydrogen bonds with the 2′-hydroxy group of DHB, whereas aryl acid-activating domains like rifamycin synthetase A, whose substrate (3-amino-5-hydroxybenzoate) lacks the 2′-OH moiety, exhibit a different sequence at this position (24, 25). The next specificity determining function is provided by the side chain of S240, which results again in an insertion with respect to PheA as a result of the altered main-chain conformation (residue W239). S240Oγ hydrogen bonds to the 3′-hydroxyl moiety of DHB. Although sequence alignments reveal this Ser to be conserved throughout DHB-activating enzymes, it is replaced by a Cys residue in SAL-activating domains (e.g., YbtE). The larger thiol group of C240 would impede access of the 3′-OH group of DHB, whereas allowing SAL to bind, leading to an increased selectivity of the protein.
A similar structural alteration can be seen in the region of core A5 (328QVFGMAEGLVN338), which is absolutely conserved among aryl acid-activating enzymes (Fig. 5), and clearly differs from the corresponding structural anchor observed in amino acid-activating domains (NxYGPTETTxx) (Fig. 3). This alternative core motif sequence therefore acts as a fingerprint to identify as yet unknown aryl acid activating adenylation domains and to discriminate at a glance between aryl and amino acid A domains. The strictly conserved first G331 (underlined) is situated at the same spatial location in all three structures (luciferase, PheA, and DhbE). The main chain of DhbE then takes another shortcut to bring the residue V337 in a position (Fig. 3A) to interact with the carbon-6 of the substrate. Intriguingly, this residue corresponds to the highly variant position 331 in PheA, whereas the only moderately variant position 330 is bypassed. Although this residue plays no direct role in binding the substrate, it remains in the specificity-conferring code as it interacts with the substrate in the amino-acid-activating A domains. V337 is conserved throughout all DHB-activating enzymes, whereas SAL-activating enzymes exhibit a more sterically demanding Leu or Ile residue at the same position.
The two remaining structural anchors (279VPPL282 and 306GGA308) are found at their expected positions, confirming the other six proposed selectivity-conferring amino acid residues. The observed homologies among aryl acid-activating domains allow more reliable predictions about specificity to be made by using the modified anchors shown in Fig. 5. This code also allows differentiation between DHB- and SAL-activating enzymes by considering the two residues corresponding to S239 and V330.
Discussion
In this study, we present the structure of the aryl acid-activating domain DhbE both in the presence and absence of its substrates DHB and ATP, revealing three different catalytic stages of an adenylate-forming enzyme, namely without substrates, with the bound substrate aryl acid DHB, and with the product DHB-adenylate, respectively. In contrast to the former proposed model, in which a relative movement of the two domains was deemed necessary for the catalytic activity (10, 11), our results suggest that only local structural changes take place in the course of substrate binding and product formation. A more recent structure of luciferase with bound bromoform also showed only limited conformational changes, restricted to a loop formed by luciferase residues 314 to 319 (26). According to the structures presented here, however, such a movement probably does not take place in the course of the catalytic reaction of DhbE.
Another barely understood structural feature of adenylate-forming enzymes is the so-called p-loop. Although highly conserved among the super family of adenylate-forming enzymes (8), none of the previously solved structures showed an ordered conformation of this loop, indicating high flexibility. Our crystal structure of DhbE, in contrast, reveals electron density for the p-loop, defining its fold and its position in proximity to the enzyme's catalytic center. The p-loop is situated at the entrance to the catalytic cavity, and moves in the course of substrate binding, presumably to isolate the substrate ATP from surrounding water molecules and to start the reaction.
In our snapshots of three different stages of the DhbE reaction, the movement is visible as a rotation around the dipeptide 191GG192 and Ser S197 at the very beginning and the end of the motif, respectively. This observation is in agreement with mutational studies on NRPS A domains, where the substitution of the equivalent of Gly-192 in TycA (27) to Ala led to a 20% reduction of the enzyme's activity; substitution to the larger and negatively charged Asp led to a complete loss of activity. This can be explained by a loss of structural flexibility because of the fact that the loop is no longer able to rotate around these residues. On the other hand, substitution of the Gly-195 equivalent leads to no loss in activity (27), supporting a rigid body movement of the loop. Substitution of K198 to Ser (22) or to Arg or Thr (27) lead to a drastic reduction of activity, in accordance with its proposed role of coordinating the substrate ATP. In the light of these studies, we conclude that the p-loop interacts with the ATP substrate, shuttling it to the catalytic cavity by the described movement and sealing the binding pocket from surrounding water. It therefore appears to represent an integral motif for the initiation of the adenylation reaction.
Particularly intriguing is the observation of two different substrate-bound states. The electron density from the multiwavelength anomalous dispersion experiment suggests hydrolysis of the adenylate DHB*AMP (Fig. 4B), analogous to that observed for PheA (11), though a DHB-ATP complex with disordered pyrophosphate cannot be ruled out. In the DHB*AMP complex, the α-phosphate interacts with the guanidinium group of R428; before substrate binding and after adenylate formation, this residue adopts a folded conformation, approaching the ribose-anchor D413. The characteristic U-shaped conformation of ATP known for aminoacyl-tRNA-synthetases can be modeled to the AMP conformation observed here (9), such that R428 would mimic R262 in Lysyl–tRNA synthetase (28). The latter residue acts as a mediator between the substrate lysine carboxylate group and the ATP α-phosphate, where it also coordinates the phosphate group of the bound adenylate. It is tempting to speculate that R428 and the p-loop could steer the ATP toward the DHB carboxylate, allowing an in-line nucleophilic attack. After adenylate formation, the R428 side chain flips away (reducing the chances of a back reaction leading to hydrolysis), and the p-loop appears to close up on the adenylate, so that G191 protects the adenylate phosphate from bulk solvent. This would assist sequestration of the aryl acid adenylate, ready for transfer to the phosphopantetheine arm of the subsequent peptidyl-carrier protein.
Despite minor changes, the nonribosomal specificity conferring code has so far proved to be universal for the NRPS adenylate-forming enzymes. Based on sequence comparisons and biochemical studies, the code has recently been applied to the subclass of 4-coumarate ligases (29). Our study on DhbE, however, reveals some significant differences for the subclass of aryl acid-activating enzymes, which may reflect the different binding behavior of aryl acids compared with amino acids. The observed rearrangement of the main-chain of DhbE compared with PheA, caused by the presence of cisPro-241, results in small but significant displacements that force at least four residues out of the positions originally predicted to be involved in substrate recognition. Further analysis of the substrate-binding site has already permitted discrimination between different SAL- and DHB-activating enzymes (Fig. 5).
Adenylate-forming enzymes are characterized by the presence of highly conserved core-motifs, a fact that is frequently exploited for the determination of unknown NRPS gene clusters using degenerated PCR with primers targeted against these sequence motifs (30). We expected to find similar core motifs for the subclasses of aryl acid-activating enzymes represented by DhbE, 4-coumarate ligases, chlorobenzoate dehalogenase, and luciferase. Surprisingly, however, these cores are frequently highly distorted or even completely different (Fig. 3). In the case of core A5 (DhbE residues 330–339) for instance, which is directly involved in substrate coordination, the originally annotated sequence “xNxYGPTExx ” is replaced by the sequence motif “QQVxFMAEGL” in aryl acid-activating enzymes. Performing a blast search with this sequence only, we found the highest homologies among unknown or putative aryl acid activating enzymes. Aligning the sequences of these unknown proteins with other members of the superfamily show that they cluster together with the other aryl acid activating domains and are clearly separate from the amino-acid-activating A domains. Biochemical examination will be required to prove whether the sequence motif “QQVxFMAEGL” could be a tool for the prediction of putative aryl acid-activating domains.
In conclusion, the data presented here give insights into how the adenylate-forming enzymes work and how they create their selectivity. Together with previous studies, it also shows the generality of many aspects of this superfamily, and provides sound evidence for an evolutionary relationship over the borders of the different kingdoms, with members of this superfamily ranging from proteins in bacteria to those in higher organisms. Finally, the structure serves as a basis for the development of inhibitors of siderophore synthesis, and provides ground rules for the incorporation of alternative chemical groups into novel compounds of bioactive potential.
Supplementary Material
Acknowledgments
We thank Gleb Bourenkov and Hans Bartunik (Max Planck Institute beamline, Deutches Elektronen-Synchrotron) and Raimond Ravelli (beamline ID14–4, European Synchrotron Radiation Faculty) for providing assistance with data collection, and Gerhard Klebe for encouragement and support. We also thank Torsten Stachelhaus and Henning Mootz for valuable discussions. This work was supported by Deutsche Forschungsgemeinschaft, Fonds der Chemischen Industrie (to M.A.M.), and Graduiertenkolleg “Proteinfunktion auf atomarer Ebene”, Marburg (to M.T.S. and M.A.M.).
Abbreviations
DHB, 2′-3′-dihydroxybenzoate
NRPS, nonribosomal peptide synthetase
SAL, salicylate
A domain, adenylation domain
PheA, phenylalanine-activating A domain
Se-Met, Se-methionine
This paper was submitted directly (Track II) to the PNAS office.
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accessison no. AY138812). The atomic coordinates and crystal structures reported in this paper have been deposited in the Protein Data Bank, www.rcsb.org [PDB ID codes (DhbE hydrolyzed), (DhbE adenylated), and (DhbE native)].
References
- 1.Ratledge C. & Dover, L. G. (2000) Annu. Rev. Microbiol. 54, 881-941. [DOI] [PubMed] [Google Scholar]
- 2.Winkelmann G. & Carrano, C. J., (1998) Transition Metals in Microbial Metabolism (Harwood Academic, New York).
- 3.Quadri L. E. (2000) Mol. Microbiol. 37, 1-12. [DOI] [PubMed] [Google Scholar]
- 4.May J. J., Wendrich, T. M. & Marahiel, M. A. (2001) J. Biol. Chem. 276, 7209-7217. [DOI] [PubMed] [Google Scholar]
- 5.Rowland B. M., Grossman, T. H., Osburne, M. S. & Taber, H. W. (1996) Gene 178, 119-123. [DOI] [PubMed] [Google Scholar]
- 6.Marahiel M. A., Stachelhaus, T. & Mootz, H. D. (1997) Chem. Rev. 97, 2651-2673. [DOI] [PubMed] [Google Scholar]
- 7.Marahiel M. A. (1997) Chem. Biol. 4, 561-567. [DOI] [PubMed] [Google Scholar]
- 8.Turgay K., Krause, M. & Marahiel, M. A. (1992) Mol. Microbiol. 6, 529-546. [DOI] [PubMed] [Google Scholar]
- 9.Weber T. & Marahiel, M. A. (2001) Structure (London) 9, R3-R9. [DOI] [PubMed] [Google Scholar]
- 10.Conti E., Franks, N. P. & Brick, P. (1996) Structure (London) 4, 287-298. [DOI] [PubMed] [Google Scholar]
- 11.Conti E., Stachelhaus, T., Marahiel, M. A. & Brick, P. (1997) EMBO J. 16, 4174-4183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stachelhaus T., Mootz, H. D. & Marahiel, M. A. (1999) Chem. Biol. 6, 493-505. [DOI] [PubMed] [Google Scholar]
- 13.Sambrook J., Fritsch, E. F. & Maniatis, T., (1989) Molecular Cloning: A Laboratory Manual (Cold Spring Harbor Lab. Press, Plainview, NY).
- 14.Sanger F., Miklen, S. & Coulson, A. R. (1977) Proc. Natl. Acad. Sci. USA 74, 5463-5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zamenhof P. J. & Villarejo, M. (1972) J. Bacteriol. 110, 171-178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Harwood C. R. & Cutting, S. M., (1990) Molecular Biological Methods in Bacillus subtilis (Wiley, Chichester, U.K.).
- 17.Hendrickson W. A., Horton, J. R. & LeMaster, D. M. (1990) EMBO J. 9, 1665-1672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Otwinowski Z. & Minor, W. (1996) Methods Enzymol. 276, 307-326. [DOI] [PubMed] [Google Scholar]
- 19.Bruenger A. T., Adams, P. D., Clore, G. M., DeLano, W. L., Gros, P., Grosse-Kunstleve, R. W., Jiang, J. S., Kuszewski, J., Nilges, M., Pannu, N. S., et al. (1998) Acta Crystallogr. D 54, 905-921. [DOI] [PubMed] [Google Scholar]
- 20.Jones T. A., Zou, J. Y., Cowan, S. W. & Kjeldgaard, M. (1991) Acta Crystallogr. A 47, 110-119. [DOI] [PubMed] [Google Scholar]
- 21.Baldwin T. O. (1996) Structure (London) 4, 223-228. [DOI] [PubMed] [Google Scholar]
- 22.Stuible H., Buttner, D., Ehlting, J., Hahlbrock, K. & Kombrink, E. (2000) FEBS Lett. 467, 117-122. [DOI] [PubMed] [Google Scholar]
- 23.Saraste M., Sibbald, P. R. & Wittinghofer, A. (1990) Trends Biochem. Sci. 15, 430-434. [DOI] [PubMed] [Google Scholar]
- 24.August P. R., Tang, L., Yoon, Y. J., Ning, S., Muller, R., Yu, T. W., Taylor, M., Hoffmann, D., Kim, C. G., Zhang, X., Hutchinson, C. R. & Floss, H. G. (1998) Chem. Biol. 5, 69-79. [DOI] [PubMed] [Google Scholar]
- 25.Kim C. G., Yu, T. W., Fryhle, C. B., Handa, S. & Floss, H. G. (1998) J. Biol. Chem. 273, 6030-6040. [DOI] [PubMed] [Google Scholar]
- 26.Franks N. P., Jenkins, A., Conti, E., Lieb, W. R. & Brick, P. (1998) Biophys. J. 75, 2205-2211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gocht M. & Marahiel, M. A. (1994) J. Bacteriol. 176, 2654-2662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Desogus G., Todone, F., Brick, P. & Onesti, S. (2000) Biochemistry 39, 8418-8425. [DOI] [PubMed] [Google Scholar]
- 29.Stuible H. P. & Kombrink, E. (2001) J. Biol. Chem. 276, 26893-26897. [DOI] [PubMed] [Google Scholar]
- 30.Turgay K. & Marahiel, M. A. (1994) Pept. Res. 7, 238-241. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.