Abstract
The conformational changes in Ras that accompany the hydrolysis of GTP are critical to its function as a molecular switch in signaling pathways. Understanding how GTP is hydrolyzed by revealing the sequence of intermediary structures in the reaction is essential for understanding Ras signaling. Until now, no structure of an intermediate in GTP hydrolysis has been experimentally determined for Ras alone. We have solved the crystal structure of the Ala-59 to Gly mutant of Ras, (RasA59G), bound to guanosine 5′-imidotriphosphate or GDP to 1.7-Å resolution. In the guanosine 5′-imidotriphosphate-bound form, this mutant adopts a conformation that is intermediate between the GTP- and GDP-bound forms of wild-type Ras and that is similar to what has been predicted by molecular dynamics simulation [Ma, J. P. & Karplus, M. (1997) Proc. Natl. Acad. Sci. USA 94, 11905–11910]. This conformation is stabilized by direct and water-mediated interactions between the switch 1 and switch 2 regions and is characterized by an increase in the binding affinity for GTP. We propose that the structural changes promoted by the Ala-59 to Gly mutation exhibit a discrete conformational state assumed by wild-type Ras during GTP hydrolysis.
Ras is an essential component of signal transduction pathways that regulate growth, proliferation, differentiation, and apoptosis (1–3). Ras functions as a molecular switch by cycling between an active GTP-bound form and an inactive GDP-bound form. The conversion from the GTP- to the GDP-bound form is mediated by the intrinsic GTPase activity of Ras, a reaction that is accelerated by the binding of the GTPase-activating-protein (GAP) (4, 5). The impairment of the hydrolytic reaction is the most common biochemical defect associated with oncogenic Ras mutations (6). Although the conformational endpoints of the GTPase reaction are well understood, relatively little is known about the structural characteristics of the reaction intermediates. Comparison of the GTP- and GDP-bound structures of Ras has identified two regions referred to as switch 1 (residues 30–40) and switch 2 (residues 60–76), respectively, that change conformation on γ-phosphate release (7–9). These regions are located in close proximity on the surface of the protein and contain conserved residues important for nucleotide and Mg2+-ion coordination. We have identified a Ras mutant containing an alanine to glycine substitution at position 59 (RasA59G) that adopts a novel conformation in the guanosine 5′-imidotriphosphate (GppNp)-bound form. This conformation bears features remarkably similar to a structural intermediate predicted by molecular dynamics simulations of the GTPase reaction path (10) and thus potentially corresponds to a transient intermediate of Ras-catalyzed GTP hydrolysis.
Methods
Crystallization and Structure Determination.
Wild-type Ras and RasA59G mutant (residues 1–166) were cloned as His-tagged proteins in pProEX HTb vector and expressed in Escherichia coli BL21 strain (11). Proteins were purified on a Ni-NTA column (Qiagen, Chatsworth, CA) followed by a Q-Sepharose (Sigma) and a gel filtration column. The GDP-bound nucleotide was exchanged to GppNp (12), and the exchanged protein was checked on an HPLC C18 reverse-phase column. For diffraction experiments, crystals were grown at room temperature by mixing 4 μl of 25 mg/ml RasA59G (in 20 mM Hepes/100 mM NaCl/10 mM MgCl2, pH 7.5) and 4 μl of the reservoir. For the GDP-bound form of RasA59G, the reservoir consisted of 16–22% (wt/vol) polyethylene glycol (PEG)8000, 100 mM calcium acetate, and 100 mM Tris⋅HCl, pH 7.5. For the GppNp-bound form of RasA59G, the reservoir consisted of 20% (wt/vol) PEG1500, 100 mM CaCl2, and 100 mM Tris⋅HCl, pH 7.5. Both Ras forms crystallized in space group R32 with one copy in the asymmetric unit and slightly different unit cells: (a = 93.1 Å, c = 120.3 Å) for the GDP-bound form and (a = 89.2 Å, c = 134.7 Å) for the GTP analog-bound form. In each case, data from one crystal were collected on beamline X26C at the National Synchrotron Laboratory Source (Brookhaven, NY) on a charge-coupled device detector (ADSC) with a wavelength of 1.1 Å. Data were processed to 1.7 Å with denzo and scaled with scalepack (13). The structures of the RasA59G mutant were solved by molecular replacement (14). For the GDP- and GppNp-bound forms, the deposited coordinates of wild-type Ras (Protein Data Bank accession nos. and 5P21) were used as search models, respectively (8, 15). The GppNp- and GDP-bound structures were refined with the program cns (16) to 1.7-Å resolution to final crystallographic residuals R/Rfree of 18.5/20.8% and 22.2/24.6%, respectively, and the stereochemistry checked with the program procheck (17). Data collection and refinement statistics are summarized in Table 1. The final electron density map of the GDP-bound RasA59G showed two different conformations for residues Arg-41, Val-44, Ile-84, and Arg-149. For the GppNp-bound form, residues Glu-3, Ser-39, Gln-43, Leu-79, Arg-102, Glu-126, and Glu-153 were modeled with two distinct conformations.
Table 1.
Data collection and refinement statistics
| GDP-bound form | GppNp-bound form | |
|---|---|---|
| Resolution range, Å | 20.0–1.7 | 20.0–1.7 |
| Rsym, % overall (1.74–1.70 Å) | 5.4 (19.6) | 4.6 (10.8) |
| Completeness, %, overall (1.74–1.70 Å) | 98.4 (89.4) | 92.7 (86.4) |
| Total no. of observations | 267,511 | 255,064 |
| Multiplicity | 12.2 | 12.0 |
| No. of unique reflections | 21,891 | 21,166 |
| Protein atoms | 1,252 | 1,300 |
| Heterogeneous atoms | 29 | 35 |
| Solvent atoms | 89 | 118 |
| B factor, Å2 overall (from Wilson plot) | 30.0 (23.5) | 19.2 (17.8) |
| Rfree, %, overall (1.81–1.70 Å) | 24.5 (31.9) | 20.8 (23.3) |
| Rcryst, % overall (1.81–1.70 Å) | 22.2 (26.6) | 18.5 (19.5) |
| rmsd in bond length, Å | 0.008 | 0.009 |
| rmsd in bond angle, ° | 1.2 | 1.4 |
| Estimated coordinate error, Å | 0.21/0.14 | 0.17/0.04 |
| Ramachandran plane, % | 93.8/6.2 | 93.3/6.7 |
Rsym = Σi,hkl|〈I(hkl)〉 − Σj,hkl/Ij(hkl).
Rfree = Σ(hkl)ɛT∥Fobs| − |Fcalc∥/Σ(hkl)ɛT|Fobs|, where T is the test set (reference) obtained by randomly selecting 10% of the data.
Rcryst = Σ(hkl)∥Fobs| − |Fcalc∥/Σ(hkl)|Fobs|.
Estimated coordinate error calculated for the data in the (5.0–1.70 Å) resolution range from the Luzzati/sigmaa statistics.
Most favored/additional allowed regions.
Results and Discussion
We have determined the crystal structure of the RasA59G mutant bound to the nonhydrolyzable GTP analogue, GppNp. The final model is refined to 1.7-Å resolution with low crystallographic indicators and excellent stereochemistry (Table 1). The final electron density is clear for all of the atoms and is weak for the side chains of switch 2 residues 61–64. The average temperature factor of this segment is higher than for the rest of the protein.
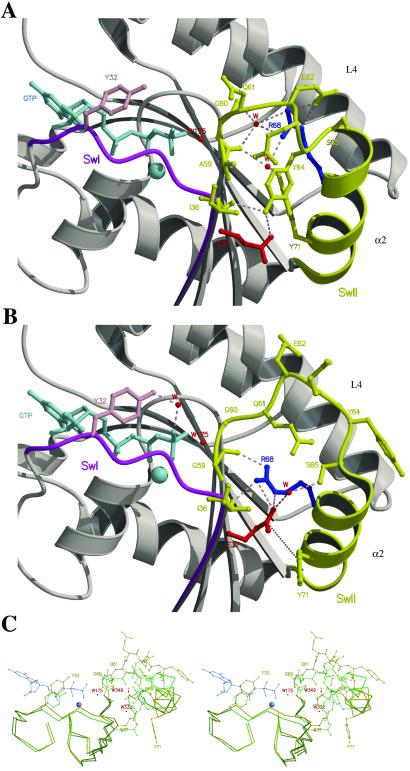
Overall, the structures of the GppNp-bound forms of wild-type Ras and RasA59G superpose well. The calculated rms deviation after superposition of the 166 Cα is 1.0 Å, and the Mg2+ ion and the GppNp have conserved interactions with the protein (15). However, the switch 2 region is extensively restructured as a consequence of introducing a glycine at position 59 (Fig. 1). Gly-60 seems to be the initiator of the switch 2 restructuring. The (φ,ψ) dihedral angles of this residue change from (−80°, −10°) in the wild-type Ras structure to (−57°, 153°) in the RasA59G structure. This change is propagated to the next six residues (Gly-60 to Ser-65), which also undergo large changes in their dihedral angles and are displaced from the nucleotide (Fig. 1C). The Cα of residues Gln-61 and Tyr-64 are shifted, for example, by as much as 3.0 and 5.4 Å from their positions in wild-type Ras, respectively. Another consequence of the A59G mutation is that the N-terminal part (Ser-65 to Met-67) of helix α2 (65–74), which makes a short 310-helix in the wild-type Ras structure (15), is now a regular α-helix with the classical i to i + 4 hydrogen bond pattern. In addition, the last turn (Gln-70 to Met-72) of α2 has lost this hydrogen bonding pattern and thus unwinds. Furthermore, α2 has undergone a 12° rotation toward its position in the GDP-bound conformation such that it now occupies a position intermediate between the GTP- and GDP-bound structures (Fig. 2).
Fig 1.
Structural changes induced in Ras switch regions by the glycine for alanine substitution at position 59. Close-up view of Glu-37 (switch 1) and Arg-68 (switch 2) in the crystal structures of wild-type Ras (A) (15) and RasA59G (B) in the GppNp-bound forms. Switch 1 (SwI) and switch 2 (SwII) are in magenta and yellow, respectively. Loop L4 and helix-α2 of switch 2 are labeled. The GppNp and the Mg2+-ion are in light blue, and water molecules (W) are represented by red spheres. Tyr-32 is in pink, Glu-37 in red, and Arg-68 in blue. Hydrogen bonds are represented by dashed lines and the van der Waals interactions between Glu-37 and Tyr-71 (B) are indicated by a dotted line. In wild-type Ras (A), Arg-68 stabilizes the N terminus of the switch 2 region (60–62) through direct or water-mediated hydrogen bonds. In RasA59G (B), Glu-37 makes hydrogen bonds with Arg-68 and van der Waals interactions with Tyr-71, which protects it partially from the solvent. Tyr-64, which is essential for Sos-binding by Ras, adopts a position that inhibits the docking of the two proteins. The catalytic residue Gln-61 is positioned far from W175. Tyr-32 is making a water-mediated hydrogen bond with the γ-phosphate, and its bulky phenol group is protecting the phosphates from the surrounding solvent. (C) Stereo representation of the superposition of the Cα of switch 1 and 2 regions between the structures of wild-type Ras (green) and RasA59G (gold) in the GppNp-bound form. The residues of L4 are shown, whereas only the side chains of Tyr-32, Glu-37, Gln-61, Arg-68, and Tyr-71 are shown. The two water molecules (W332 and W349) in wild-type Ras that coordinated Arg-68 and that have been exchanged with the solvent on the reorientation of the switch 2 region, are shown. Prepared with MOLSCRIPT (24) and RASTER3D (25).
Fig 2.
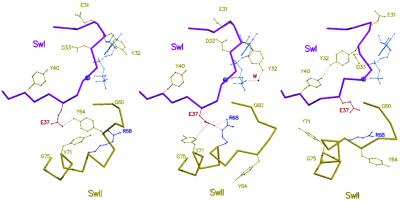
Structural changes that affect the switch 1 and switch 2 regions of Ras along the path for GTP hydrolysis. Structures proceed from the GTP-bound form (Left, PDB coordinates 5P21), to the intermediate (Center) that is represented by the A59G mutant, and finally to the GDP-bound form (Right, PDB coordinates ). Water molecules are shown as red spheres; the nucleotide and the Mg2+-ion are in light blue. Switch 1 is in magenta and switch 2 in gold. Close contacts are represented by dotted lines.
In contrast to the extensive remodeling of switch 2, switch 1 exhibits subtle conformational differences between wild-type Ras and the RasA59G structures. These changes are confined to the side chains of Tyr-32 and Glu-37. In the wild-type Ras structure, Tyr-32 points to the solvent (Fig. 1A), whereas in the RasA59G structure, the bulky phenol group rests above the phosphates in a crevice between switch 1 and the P-loop (Fig. 1B). The hydroxyl group of Tyr-32 is interacting with the γ-phosphate through a water molecule-mediated hydrogen bond. Glu-37 is solvent exposed in wild-type Ras, whereas in the RasA59G structure, it is partially shielded from the surrounding solvent by the bulky phenol group of Tyr-71 (Fig. 1 A and B).
The most dramatic feature in the RasA59G GppNp-bound structure is the interaction between the switch regions (Fig. 1B). The nucleus of this interaction consists of Ile-36 and Glu-37 of switch 1 and Ser-65, Arg-68, and Tyr-71 of switch 2. In the wild-type GTP-bound structure of Ras, Ile-36 is solvent exposed, whereas the side chains of Glu-37, Tyr-64, Arg-68, Tyr-71, Leu-56, Ala-59, and Thr-58 form a pocket that is stabilized by van der Waals interactions and water-mediated hydrogen bonds. The main chain carbonyl (mcCO) of Ile-36 is interacting with the hydroxyl side chain group of Tyr-64, whereas the side chain of Arg-68 is stabilizing the 60–62 loop through direct or water-mediated hydrogen bonds (Fig. 1A). In the RasA59G GppNp-structure, this pocket unfolds and, as a consequence, the side chain of Arg-68 adopts a new conformation in which the guanidinium group is making strong hydrogen bonds with the side chain of Glu-37, the mcCO of Ile-36, and the mcCO of Gly-59 (Fig. 1B). These interactions pull the Ile-36/Glu-37 polypeptide by 1 Å toward the switch 2 region (Fig. 1C). The reorientation of Arg-68 is incompatible with the conformations of Tyr-64 and Tyr-71 observed in wild-type Ras. Consequently, these two tyrosines have changed their conformations in RasA59G and point to the solvent with the phenol group of Tyr-71 at van der Waals contact distances from the carboxylic group of Glu-37 (Fig. 1 B and C).
The conformational changes displayed by the RasA59G-GppNp structure have not been described before. In addition, no experimental structure for an intermediate for GTP hydrolysis of Ras alone has been reported. However, the structure of the RasA59G mutant is remarkably similar to the predicted structure of an intermediate on the reaction path between the GTP- and GDP-bound forms, which has been identified by molecular dynamics (MD) simulations (10). In particular, the reorientation of the side chains of Tyr-32, -64, and -71, the restructuring of loop L4 (59–64), and the coupling between the switch regions through a transient hydrogen bonding complex between Glu-37 and Arg-68 observed are all depicted in the structure predicted by the MD simulations (Fig. 2, ref. 10). This similarity, along with the observation that the positioning of helix-α2 in the RasA59G structure is intermediate to its positions in the GTP- and GDP-bound forms of wild-type Ras, suggests that the structure of RasA59G corresponds to the structure of an intermediate during the conformational transition from Ras-GTP to Ras-GDP.
Alanine 59 is located in the conserved 57DXXGQ61 motif that is important for Mg2+ coordination, GTP hydrolysis, and nucleotide binding (18, 19). This motif is part of a sharp flexible turn (L4) connecting strand β3 and helix α2 of Ras. The flexibility of this loop is essential for the conformational change that accompanies GTP hydrolysis and γ-phosphate release. Specifically, it has been postulated that the conformational changes that accompany GTP hydrolysis are initiated by an increase in the flexibility of the L4 motif because of the uncoupling of Gly-60 from the γ-phosphate. The increase in backbone flexibility that is likely to occur as a result of substituting alanine 59 for glycine could in principle trigger a similar conformational change. Indeed, the mcCO of Gly-60 undergoes a significant rotation in the RasA59G structure (change in the ψ main chain angle of Gly-60 of ≈Δψ = 160°, Fig. 1C). As a result of this rotation, Gly-60 adopts a unique conformation that allows it to form tighter interactions with the γ-phosphate in comparison with wild type (2.8 vs. 3.0 Å, respectively). To test whether this interaction could be responsible for trapping the RasA59G-GppNp in the intermediate structure, we solved the crystal structure of RasA59G in the GDP-bound form to 1.7-Å resolution (Table 1). The electron density of this structure is very clear, with the exception of switch 2 residues 60–64, where it is missing, and switch 1 residues 33–35 and switch 2 residues 65–70, where it is weak. Overall, the structure of the mutant superposes well with the structure of wild-type Ras (8) (rms deviation of 0.61 Å for all of the Cα atoms). The Mg2+ ion and the GDP are making conserved interactions with the protein (8). Significantly, the interactions between the switch regions and the conformational transition of the switch 2 region observed in the GppNp-bound form of RasA59G are absent in the GDP-bound form of the mutant. This suggests that the intermediate conformation adopted by RasA59G in the GppNp-bound form is stabilized by the interaction of Gly-60 with the γ-phosphate. On the basis of this analysis, we propose that the A59G mutation triggers a conformational change that resembles one conformation assumed by wild-type Ras after the cleavage of the γ-phosphate but before its release. However, the mutant protein cannot complete the conformational transition to the GDP-bound form, because the movement of switch 2 is constrained because of the interaction of the γ-phosphate of the GppNp with the main chain amide of Gly-60. Thus, the GppNp-bound form of the RasA59G mutant is locked in a conformation that is intermediate between that of the GTP- and GDP-bound forms.
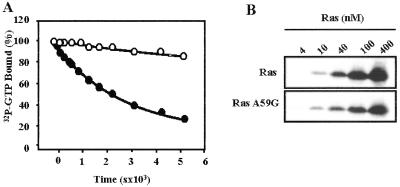
In an effort to understand the functional significance of this intermediate conformation, we characterized the biochemical properties of the RasA59G mutant. The rate of intrinsic GTP hydrolysis for the mutant protein (3.5 × 10−5 s−1) is ≈10 times slower than for the wild-type Ras (Fig. 3A). The impairment in GTPase activity can be attributed to the displacement of Gln-61 from the catalytic site. This residue plays a critical role in orienting and stabilizing the water responsible for the nucleophilic attack on the γ-phosphorus, identified as Wat175 in wild-type Ras (7). The final electron density of the GppNp-bound form of RasA59G clearly shows a water molecule equivalent to Wat175. It is positioned at 3.5 Å from the γ-phosphorus and is hydrogen bonded to the mcCO of Thr-35 and the γ-phosphate. Because Gln-61 is pulled away from the active site and is very mobile in the final electron density, it is unlikely that this catalytic residue will stabilize the water molecule for an attack on the GTP. This observation is consistent with RasA59G mimicking a structure of an intermediate occurring after the nucleophilic attack but before the complete rearrangements of the switch regions.
Fig 3.
(A) Comparison of the GTP-hydrolysis rate of wild-type Ras (closed circles) and RasA59G (open circles). The GTP-hydrolysis rate was measured by using the filter-binding assay. Histidine-tagged Ras proteins (40 pM) were loaded with [γ-32P]GTP (25 Ci/mmol; ICN; 1 Ci = 37 GBq) in 40 mM Hepes/100 mM NaCl/10 mM MgCl2/1 mM DTT, pH 8.0/2 mM EDTA at room temperature. After incubation, MgCl2 was added to a final concentration of 20 mM and samples were placed on ice. GTPase reaction was initiated by incubating at 30°C. Aliquots (20 μl) were taken at appropriate times, and the remaining radioactivity was measured on nitrocellulose filter discs (Millipore HAWPO2500) by using a scintillation counter. Results are shown as percentages of radioactivity bound at time 0. Hydrolysis rates (kcat) were calculated by fitting the experimental points to a single exponential by using SIGMA PLOT (SPSS, Chicago) giving values of 2 × 10−4 s−1 and 3.5 × 10−5 s−1 for wild-type Ras and RasA59G, respectively. (B) Binding of RasA59G to the Ras-binding domain (RBD) of the Raf kinase. Glutathione S-transferase-fusion RafRBD was incubated with increasing amounts of wild type (Upper) or A59G (Lower) histidine-tagged Ras loaded with guanosine 5′-O-(3-thiotriphosphate), as indicated. RafRBD/Ras complexes were precipitated with glutathione Sepharose, and the bound material was analyzed by Western blotting with antihistidine antibodies.
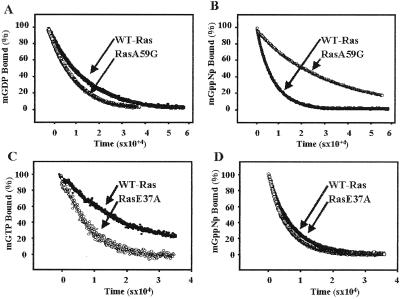
Next, we compared the relative affinities of wild-type Ras and RasA59G mutant for nucleotides by measuring the intrinsic dissociation rates of the fluorescent analogues N-methylanthranoyl-GDP (mant-GDP) and mant-GppNp (20). Fig. 4A shows that for the RasA59G mutant, the dissociation rate of GDP is not significantly altered. In contrast, the RasA59G mutant displays a 4-fold decrease in the rate of GppNp dissociation (koff = 4.2 × 10−5 s−1) (Fig. 4B). Several structural features of the mutant could account for this difference. First, the γ-phosphate is held tighter in the RasA59G structure by virtue of shorter hydrogen bonds with Lys-16 and Gly-60 and water-mediated hydrogen bond with the hydroxyl group of Tyr-32. Second, the bulky phenol group of Tyr-32, which in RasA59G rests in a crevice between switch 1 and the P-loop, shields the phosphates from the surrounding solvent. Third, the new interface created by the direct hydrogen bonding between Arg-68 in switch 2 and Glu-37 in switch 1 could stabilize switch 1 and impose sterical constraints that prevent nucleotide release. We took advantage of this last feature to test the hypothesis that the intermediate conformation adopted by RasA59G might serve to transiently stabilize the nucleotide during the structural transition that accompanies GTP hydrolysis. If this hypothesis were correct, one would then predict that the failure of switch 1 and switch 2 to interact properly would destabilize the nucleotide during GTP hydrolysis. To test this prediction, the switch 1/switch 2 interactions were disrupted by mutating glutamate 37 to alanine, and the effects of this mutation on the rate of nucleotide dissociation and GTP hydrolysis were investigated. Fig. 4C shows that the E37A mutation increased the rate of GTP dissociation from Ras by ≈3-fold (koff = 1.2 × 10−4 s−1). In contrast, the same mutation had no effect on the dissociation rate for the nonhydrolyzable GTP analogue, GppNp (Fig. 4D). Because the E37A mutation did not affect the rate of GTP hydrolysis (data not shown), this observation suggests that the intermediate conformation adopted by RasA59G in the GppNp-bound form, particularly the remodeling of the switch 2 region and the Glu-37/Arg-68 interaction, is important for nucleotide stability during the conformational changes that accompany GTP hydrolysis.
Fig 4.
Intrinsic nucleotide dissociation rates of Ras proteins. Nucleotide dissociation was followed by using the fluorescent nucleotide analogues mant-GDP, mant-GppNp (a nonhydrolyzable GTP-analogue), and mant-GTP, as described in ref. 26. Purified Ras proteins (1 μM) were loaded with mant-GDP, mant-GppNp, or mant-GTP, as indicated, and incubated in the presence of excess GDP or guanosine 5′-O-(3-thiotriphosphate), respectively. The rate for nucleotide dissociation was determined by the decrease in fluorescence emission at 450 nm over time at room temperature. Data shown are from a single experiment and are representative of three independent measurements. Results are plotted as percentages of mant nucleotide bound to Ras at time 0. Off rates were calculated by fitting the experimental plots to a single exponential. (A) GDP dissociation from wild-type Ras (closed circles, koff = 6.4 × 10−5 s−1) and RasA59G (open circles, koff = 9.2 × 10−5 s−1). (B) GppNp-dissociation from wild-type Ras (closed circles, koff = 1.5 × 10−4 s−1) and RasA59G (open circles, koff = 4.2 × 10−5 s−1). (C) GTP dissociation from wild-type Ras (closed circles, koff = 4.3 × 10−5 s−1) and RasE37A (open circles, koff = 1.2 × 10−4 s−1). (D) GppNp dissociation from wild-type Ras (closed circles, koff = 1.4 × 10−4 s−1) and RasE37A (open circles, koff = 1.5 × 10−4 s−1).
The conformational changes in the switch regions triggered by GTP hydrolysis result in the dissociation of Ras from effector molecules. To examine the effects of the intermediate conformation adopted by the RasA59G mutant on the binding of downstream effectors, we have used an in vitro pull-down assay by using the Ras-binding domain of Raf kinase (RafRBD). As illustrated in Fig. 3B, the RasA59G mutant binds RafRBD in a GTP-dependent manner with a comparable affinity to wild-type Ras. This result is not surprising because in the RasA59G-GppNp structure, the switch 1 residues Glu-31, Asp-33, and Asp-38, which are mainly responsible for the RafRBD binding, adopt a conformation suitable for binding similar to the one they have in Rap in the Rap/RafRBD complex (21, 22). This observation suggests that the conformation of the RasA59G mutant represents an intermediate in the reaction path for GTP hydrolysis corresponding to a state that follows the β/γ-phosphate bond cleavage but precedes the dissociation of the terminal phosphate from Ras. Further, we predict that the rotation of Tyr-32 and the surrounding residues (Fig. 2), coupled with the increase in flexibility of loop L4, is required for effector displacement.
We have previously shown that the RasA59G mutant is refractory to Sos-catalyzed GTP exchange in vitro (11). The structure of the RasA59G mutant provides a molecular explanation for this defect. Most of the binding determinants for Sos are located within the switch 2 region of Ras (23). The rearrangement of switch 2 in the GppNp-bound form of RasA59G alters the solvent-exposed interface for recognition and interaction with Sos. In particular, Tyr-64, which is critical for maintaining high-affinity binding of Ras to Sos, is displaced 6 Å in the RasA59G mutant from its position in the Ras/Sos structure (Figs. 1 and 2).
In conclusion, the crystal structure of the RasA59G mutant in the GppNp-bound form reveals a novel conformation distinct from any structure described for the GTP- or GDP-bound form of Ras. Most significantly, the switch regions interact directly through strong hydrogen bonds, and the α2-helix of the switch 2 region adopts a conformation that is intermediate to the positions it assumes in the GTP- and GDP-bound states. Thus, in the presence of GppNp, the alanine 59 to glycine mutation appears to stabilize an otherwise transient intermediate of Ras-catalyzed GTP hydrolysis, thereby uncovering a new conformational state along the reaction path. This conformation impedes intrinsic and guanine nucleotide exchange factor-catalyzed nucleotide dissociation, suggesting that during GTP hydrolysis, Ras increases its affinity toward the nucleotide. This increase in affinity could provide a mechanism to ensure that the nucleotide stays bound to the protein during the structural changes initiated by the water-mediated nucleophilic attack and until the stable endpoint conformation is reached. The higher affinity for the nucleotide is elegantly achieved by a strong interaction between the switch regions. In doing so, Ras is a catalyst that binds its substrate during the chemical reaction and protects it from falling into the solvent until the product is formed.
Acknowledgments
We thank Dieter Schneider for technical assistance at X26C and Suzanne Scarlata for help with fluorescence data measurements. We thank Todd Miller for critical reading of the manuscript. This work was supported by National Institutes of Health Grant CA28146 (to D.B.-S.) and the Carol Baldwin Foundation (to N.N.). The National Synchrotron Light Source is supported by the Department of Energy and National Institutes of Health, and beamline X26C is supported in part by Stony Brook University and its Research Foundation.
Abbreviations
GppNp, guanosine 5′-imidotriphosphate
mcCO, main chain carbonyl
mant, N-methylanthranoyl
Data deposition: The atomic coordinates reported in this paper have been deposited in the Protein Data Bank, www.rcsb.org (PDB ID codes: RasA59G, GDP-bound form, ; RasA59G, GppNp-bound form, ).
References
- 1.Barbacid M. (1987) Annu. Rev. Biochem. 56, 779-827. [DOI] [PubMed] [Google Scholar]
- 2.Lowy D. R. & Willumsen, B. M. (1993) Annu. Rev. Biochem. 62, 851-891. [DOI] [PubMed] [Google Scholar]
- 3.Vojtek A. B. & Der, C. J. (1998) J. Biol. Chem. 273, 19925-19928. [DOI] [PubMed] [Google Scholar]
- 4.McCormick F. (1989) Cell 56, 5-8. [DOI] [PubMed] [Google Scholar]
- 5.Bourne H. R., Sanders, D. A. & McCormick, F. (1990) Nature (London) 348, 125-132. [DOI] [PubMed] [Google Scholar]
- 6.Bos J. L. (1989) Cancer Res. 49, 4682-4689. [PubMed] [Google Scholar]
- 7.Pai E. F., Kabsch, W., Krengel, U., Holmes, K. C., John, J. & Wittinghofer, A. (1989) Nature (London) 341, 209-214. [DOI] [PubMed] [Google Scholar]
- 8.Milburn M. V., Tong, L., Devos, A. M., Brunger, A., Yamaizumi, Z., Nishimura, S. & Kim, S. H. (1990) Science 247, 939-945. [DOI] [PubMed] [Google Scholar]
- 9.Scheidig A. J., Burmester, C. & Goody, R. S. (1999) Structure (London) 7, 1311-1324. [DOI] [PubMed] [Google Scholar]
- 10.Ma J. P. & Karplus, M. (1997) Proc. Natl. Acad. Sci. USA 94, 11905-11910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hall B. E., Yang, S. S., Boriack-Sjodin, P. A., Kuriyan, J. & Bar-Sagi, D. (2001) J. Biol. Chem. 276, 27629-27637. [DOI] [PubMed] [Google Scholar]
- 12.John J., Sohmen, R., Feuerstein, J., Linke, R., Wittinghofer, A. & Goody, R. S. (1990) Biochemistry 29, 6058-6065. [DOI] [PubMed] [Google Scholar]
- 13.Otwinowski Z. & Minor, W. (1997) Methods Enzymol. 276, 307-326. [DOI] [PubMed] [Google Scholar]
- 14.Navaza J. (1994) Acta Crystallogr. A 50, 157-163. [Google Scholar]
- 15.Pai E. F., Krengel, U., Petsko, G. A., Goody, R. S., Kabsch, W. & Wittinghofer, A. (1990) EMBO J. 9, 2351-2359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brünger A. T., Adams, P. D., Clore, G. M., DeLano, W. L., Gros, P., Grosse-Kunstleve, R. W., Jiang, J. S., Kuszewski, J., Nilges, M., Pannu, N. S., et al. (1998) Acta Crystallogr. D 54, 905-921. [DOI] [PubMed] [Google Scholar]
- 17.Laskowski R. A., MacArthur, M. W., Moss, D. S. & Thornton, J. M. (1993) J. Appl. Crystallogr. 26, 283-291. [Google Scholar]
- 18.Mistou M. Y., Jacquet, E., Poullet, P., Rensland, H., Gideon, P., Schlichting, I., Wittinghofer, A. & Parmeggiani, A. (1992) EMBO J. 11, 2391-2397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sung Y. J., Hwang, M. C. C. & Hwang, Y. W. (1996) J. Biol. Chem. 271, 30537-30543. [DOI] [PubMed] [Google Scholar]
- 20.Lenzen C., Cool, R. H., Prinz, H., Kuhlmann, J. & Wittinghofer, A. (1998) Biochemistry 37, 7420-7430. [DOI] [PubMed] [Google Scholar]
- 21.Nassar N., Horn, G., Herrmann, C., Block, C., Janknecht, R. & Wittinghofer, A. (1996) Nat. Struct. Biol. 3, 723-729. [DOI] [PubMed] [Google Scholar]
- 22.Block C., Janknecht, R., Herrmann, C., Nassar, N. & Wittinghofer, A. (1996) Nat. Struct. Biol. 3, 244-251. [DOI] [PubMed] [Google Scholar]
- 23.Boriack-Sjodin P. A., Margarit, S. M., Bar-Sagi, D. & Kuriyan, J. (1998) Nature (London) 394, 337-343. [DOI] [PubMed] [Google Scholar]
- 24.Kraulis P. J. (1991) J. Appl. Crystallogr. 24, 946-950. [Google Scholar]
- 25.Merritt E. A. & Bacon, D. J. (1997) Methods Enzymol. 277, 505-524. [DOI] [PubMed] [Google Scholar]
- 26.Lenzen C., Cool, R. H. & Wittinghoffer, A. (1995) Methods Enzymol. 255, 95-109. [DOI] [PubMed] [Google Scholar]