Abstract
Addition of ubiquitin or ubiquitin chains to target proteins leads to their mono- or polyubiquitination, respectively. Whereas polyubiquitination targets proteins for degradation, monoubiquitination is thought to regulate receptor internalization and endosomal sorting. Cbl proteins are major ubiquitin ligases that promote ligand-dependent polyubiquitination and degradation of receptor tyrosine kinases. They also recruit CIN85-endophilin in the complex with activated receptors, thus controlling receptor endocytosis. Here we show that the adaptor protein CIN85 and its homologue CMS are monoubiquitinated by Cbl/Cbl-b after epidermal growth factor (EGF) stimulation. Monoubiquitination of CIN85 required direct interactions between CIN85 and Cbl, the intact RING finger domain of Cbl and a ubiquitin acceptor site present in the carboxyl terminus of CIN85. Cbl-b and monoubiquitinated CIN85 are found in the complex with polyubiquitinated EGF receptors during prolonged EGF stimulation and are degraded together in the lysosome. Dominant interfering forms of CIN85, which have been shown previously to delay EGF receptor degradation, were also impaired in their monoubiquitination. Thus, our data demonstrate that Cbl/Cbl-b can mediate polyubiquitination of cargo as well as monoubiquitination of CIN85 to control endosomal sorting and degradation of receptor tyrosine kinases.
Regulation of protein tyrosine kinase activity is implicated in the control of almost all cellular functions, whereas its deregulation is often associated with human diseases such as cancer (1, 2). Binding of growth factors to receptor tyrosine kinases (RTKs) promotes receptor dimerization and subsequent activation (3), which enhances autophosphorylation of RTKs, phosphorylation of numerous cellular proteins, and recruitment of adaptor molecules, which initiate signaling cascades (1). Receptors are also recruited to clathrin-coated pits, and within a few minutes they are internalized into endocytic vesicles (4). Subsequently, receptors and associated complexes are transferred into multivesicular bodies (MVBs), with characteristic inner vesicles and membranes (5). In these structures receptors undergo sorting and are either recycled back to the plasma membrane or directed to lysosomes for destruction (6). Lysosomal degradation is the ultimate step in negative regulation of RTKs that controls the strength and duration of RTK-induced signals and is critical to prevent RTK hyperactivation, commonly associated with tumorigenesis (2, 7, 8).
The mechanisms underlying epidermal growth factor (EGF) receptor (EGFR) trafficking are probably the best understood among all RTKs. Ligand-enhanced endocytosis of EGFRs requires intrinsic tyrosine kinase activity and endocytic sequence motifs located in the cytoplasmic domain of the receptor and also depends on numerous signals in the endosomes that can direct receptors for lysosomal degradation (6, 9–11). The addition of polyubiquitin chains to EGFRs has been recognized as a critical determinant in controlling EGFR endocytosis (6). The Cbl family of RING-type ubiquitin ligases was shown to facilitate polyubiquitination of EGFRs and thereby promote their destruction in the lysosome (12–15). However, more recent reports have indicated that monoubiquitination rather than polyubiquitination is required for endosomal sorting of proteins into MVBs and their targeting for degradation (16). In yeast, ubiquitinated cargo proteins bind to the endosomal sorting complex required for transport (ESCRT-1) via Vps23, a protein containing a ubiquitin conjugating-like domain (17). Cells lacking TSG101, the mammalian orthologue of Vps23, were shown to be impaired in their ability to down-regulate and degrade EGFRs (17). Furthermore, several proteins involved in receptor endocytosis, including eps15, Hrs, and epsins, are monoubiquitinated after EGF stimulation (18). These proteins contain ubiquitin-interacting motifs that were shown to be required for their monoubiquitination (18).
CIN85 (Cbl-interacting protein of 85 kDa) is a multiadaptor protein containing three Src homology (SH)3 domains at the amino terminus and a proline-rich region and a coiled-coil domain in the carboxyl terminus (19). CIN85 was shown recently to link Cbl–EGFR complexes with endophilin-dependent receptor internalization (20). CIN85 binds to the distal carboxyl terminus of Cbl/Cbl-b after growth factor stimulation while it is constitutively associated with endophilin (ref. 20 and unpublished data). In addition, Cbl and CIN85 were colocalized with activated EGFRs in endosomes (20). Here we show that Cbl and Cbl-b mediate monoubiquitination of the adaptor proteins CIN85 and its homologue CMS (p130Cas ligand with multiple SH3 domains) after EGF stimulation. CIN85 was also found in the complex with Cbl and polyubiquitinated EGFRs, leading to their common degradation in the lysosome. These results indicate that Cbl-mediated polyubiquitination of EGFRs as well as monoubiquitination of receptor-associated accessory proteins are coordinated events during degradation of RTKs.
Materials and Methods
Products and Cloning.
EGF was purchased from Intergen (Purchase, NY), and MG132 was purchased from Calbiochem. Goat polyclonal antiubiquitin and rabbit polyclonal anti-ERK2 antibodies were from Santa Cruz Biotechnology. Mouse monoclonal anti-FLAG M2 and M5 antibodies were from Sigma, and rabbit polyclonal anti-Nedd4 antibodies were from Becton Dickinson–PharMingen. Rabbit polyclonal anti-EGFR (RK2) antibodies were provided by Joseph Schlessinger (Yale University, New Haven, CT). Rabbit polyclonal anti-CIN85 (SH3) antibodies were raised against a glutathione S-transferase fusion protein containing all three SH3 domains of CIN85. Rabbit polyclonal anti-Cbl (RF) and anti-CIN85 (CT) have been described (20). Constructs of EGFR, Cbl, CIN85, CIN85-3SH3, CIN85-PCc, and FLAG-tagged ubiquitin have been described (20). FLAG-CMV2-CMS was provided by Kathrin Kirsch (The Rockefeller University, New York); pcDNA3-Nedd4 was provided by Pier Paolo Di Fiore (European Institute of Oncology, Milan); pSRαneo-Cbl-70Z and pSRαneo-Cbl-G306E were provided by Wallace Y. Langdon (University of Western Australia, Crawley); and pCEFL-Cbl-b constructs were provided by Stanley Lipkowitz (National Cancer Institute, Bethesda, MD).
Site-Directed Mutagenesis.
pcDNA-FLAG-CIN85 mutant constructs were generated by PCR using QuickChange (Stratagene). CIN85-ΔCc was made by using the primer pair 5′-CTGTTCGGCACGGAATGAAAACCAAAGATGGAG-3′ and 5′-CTCCATCTTTGGTTTTCATTCCGTGCCGAACAG-3′, introducing a stop codon at amino acid 595. CIN85-ΔP, lacking amino acids 319–588, was generated by using 5′-CCGATAACTTCGTGAAGGGACACAGAGCCAACTCC-3′ and 5′-GGAGTTGGCTCTGTGTCCCTTCACGAAGTTATCGG-3′. CIN85- K627,631,635,645,646,659,660,665R (Cc-K-less) was created in six-step site-directed mutagenesis by PCR using QuickChange. CIN85-W36,135,306A (3SH3-M) was made by three-step mutagenesis. The constructs were verified by sequencing.
Cell Culture and Transfections.
HEK293T cells were transfected by using the Lipofectamine reagent (Invitrogen) following the manufacturer's instructions. Thirty hours after transfection, the cells were starved for 12 h and stimulated with 100 ng/ml EGF for indicated times. Cells were lysed in ice-cold 1% Triton X-100 lysis buffer (pH 7.4/50 mM Hepes/150 mM NaCl/1 mM EDTA/1 mM EGTA/10% glycerol) with protease inhibitors (leupeptin, aprotinin, and PMSF), NaF, ZnCl2, and sodium orthovanadate. The lysates were cleared by centrifugation at 16,000 × g for 20 min at 4°C.
Immunoprecipitation and Immunoblotting.
Lysates with adjusted protein concentration (Bradford assay, Bio-Rad) were incubated with antibody for 2 h at 4°C. The following antibodies were used: rabbit polyclonal anti-CIN85 (CT), rabbit polyclonal anti-CIN85 (SH3), and mouse monoclonal anti-FLAG (M2). Immune complexes were precipitated after 1-h incubation with protein A Sepharose beads. After washing in cold lysis buffer, the complexes were resuspended in Laemmli sample buffer (Bio-Rad), boiled, and resolved by SDS/PAGE. Proteins were transferred to nitrocellulose membranes and probed with indicated antibodies as described (20).
CIN85 Monoubiquitination Assays.
HEK293T cells were transfected with pRK5-EGFR and pcDNA3-FLAG-ubiquitin together with pCEFL-Cbl-b, pRK5-Cbl, or pcDNA3-Nedd4 alone or pCEFL-Cbl-b with pcDNA3-FLAG-CIN85, pcDNA3-FLAG-CIN85-3SH3, pcDNA3-FLAG-CIN85-PCc, pcDNA3-FLAG-CIN85-ΔCc, pcDNA3-FLAG-CIN85-ΔP, or FLAG-CMV2-CMS constructs. The constructs pCEFL-Cbl-b-N1/2, pCEFL-Cbl-b-C2/3, or pSRαneo-Cbl-70Z were transfected instead of pCEFL-Cbl-b where indicated. Serum-starved cells were mock-treated or stimulated with EGF (100 ng/ml) for indicated time points at 37°C, and immunoprecipitates of CIN85 or total cell lysates (TCLs) were subjected to Western blotting with anti-FLAG (M5), anti-CIN85 (CT), anti-Cbl (RF), anti-EGFR (RK2), and antiubiquitin antibodies as indicated. For all experiments the levels of transfected Cbl and CIN85 were analyzed in the cell lysates. The proteasome inhibitor MG132 was used at a concentration of 20 μM for 1 h before EGF stimulation where indicated.
Results
CIN85 and CMS Are Monoubiquitinated by Cbl and Cbl-b After EGF Stimulation.
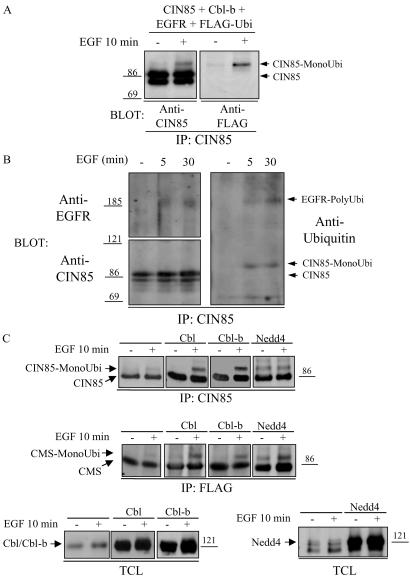
Endocytosis of EGFRs is regulated by the formation of receptor-associated protein complexes and by their posttranslational modifications including phosphorylation and ubiquitination (6). We therefore tested whether the adaptor protein CIN85, which is recruited rapidly in the complex with Cbl and activated EGFRs, is posttranslationally modified and whether these modifications are important for the ability of CIN85 to regulate receptor down-regulation. Up to this point we could not detect EGF-induced tyrosine phosphorylation of CIN85 in several cell types expressing endogenous CIN85 or in HEK293T cells overexpressing both CIN85 and EGFRs (data not shown). However, we have observed an increase in the apparent relative molecular mass of CIN85 of ≈8 kDa after ligand stimulation (Fig. 1A). By immunoblotting with specific antibodies for tagged ubiquitin, we determined that the observed shift of overexpressed CIN85 is caused by the attachment of a single ubiquitin molecule (Fig. 1A). We next tested whether EGF stimulation leads to monoubiquitination of endogenous CIN85 in cultured fibroblasts. EGF treatment of NIH 3T3-EGFR cells for 5 or 30 min led to ligand-inducible monoubiquitination of CIN85 and its coprecipitation with polyubiquitinated EGFRs (Fig. 1B). Previous work indicated a critical role for HECT-type ubiquitin ligases, Rsp5 and Nedd4, in promoting monoubiquitination of endocytic proteins (16, 18), and RING-type Cbl ubiquitin ligase in mediating polyubiquitination of activated EGFRs (12–15). Because both Cbl and Nedd4 are expressed endogenously in HEK293T cells (Fig. 1C), we next tested the effect of Cbl, Cbl-b, or Nedd4 overexpression on CIN85 monoubiquitination. EGF stimulation led to a modest monoubiquitination of CIN85 in HEK293T cells (Fig. 1C), whereas EGF-induced CIN85 monoubiquitination was increased significantly after overexpression of Cbl or Cbl-b (Fig. 1C). In addition, overexpression of Cbl or Cbl-b facilitated ligand-induced monoubiquitination of CMS (Fig. 1C), a CIN85 homologue that was shown previously to bind to Cbl after EGF stimulation (21). On the other hand, overexpression of Nedd4 induced multiubiquitination of both CIN85 and CMS independently of EGF stimulation (Fig. 1C). It therefore is likely that endogenous Cbl present in HEK293T cells (Fig. 1C) is sufficient to mediate EGF-dependent monoubiquitination of CIN85, and overexpression of Cbl further increases CIN85 and CMS monoubiquitination (Fig. 1C). Similar findings were observed for Cbl-induced polyubiquitination of EGFRs after overexpression of Cbl (data not shown and ref. 20). Taken together, these results suggest that Cbl mediates ligand-induced monoubiquitination of the CIN85/CMS family of adaptor proteins as well as polyubiquitination of EGFRs.
Fig 1.
CIN85 and CMS are monoubiquitinated (MonoUbi) after EGF stimulation. (A) HEK293T cells were transiently transfected with expression vectors coding for CIN85, EGFR, Cbl-b, and FLAG-tagged ubiquitin (FLAG-Ubi). Thirty hours after transfection, cells were starved for 12 h and mock-treated (−) or stimulated with 100 ng/ml EGF (+) for 10 min. CIN85 was immunoprecipitated (IP) with anti-CIN85 (CT) antibodies, and monoubiquitination was detected with anti-FLAG (M5) antibodies. Levels of precipitated CIN85 were determined with anti-CIN85 (CT) antibodies. Transfection of pcDNA3-CIN85 into HEK293T cells led to expression of two isoforms migrating with the relative molecular mass of ≈78 and 85 kDa. (B) NIH 3T3-EGFR cells, transfected with FLAG-tagged ubiquitin, were starved for 12 h and mock-treated (−) or stimulated with 100 ng/ml EGF for 5 or 30 min. Immunoprecipitates of CIN85 were analyzed by Western blotting with antiubiquitin antibodies, anti-CIN85 (CT), and anti-EGFR (RK2) antibodies. Two CIN85 isoforms (p78 and p85) may represent previously described alternatively spliced forms of CIN85 (32, 33). (C) HEK293T cells overexpressing EGFRs and FLAG-tagged ubiquitin without or with Cbl, Cbl-b, or Nedd4, and FLAG-CIN85 or FLAG-CMS were mock-treated (−) or stimulated with 100 ng/ml EGF (+) for 10 min. CIN85 was immunoprecipitated with anti-CIN85 antibodies (Top) and CMS with anti-FLAG (M2) antibodies (Middle), and their monoubiquitination was analyzed by blotting with anti-FLAG (M5) antibodies. Levels of Cbl, Cbl-b, and Nedd4 in representative TCLs are shown (Bottom).
Requirements for Cbl-Mediated Monoubiquitination of CIN85.
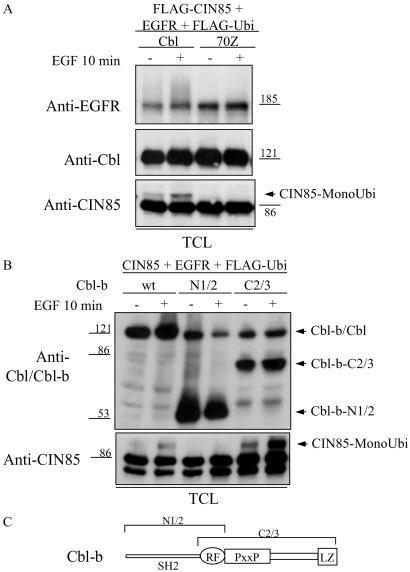
The above results indicated that Cbl may be critical for EGF-induced monoubiquitination of CIN85 in mammalian cells. To further study the importance of Cbl domains required for monoubiquitination of CIN85, we tested the ability of a ligase-deficient mutant of Cbl (Cbl-70Z) or various deletion mutants of Cbl-b to ubiquitinate CIN85. Wild-type Cbl promoted monoubiquitination of CIN85 and polyubiquitination of EGFRs, whereas Cbl-70Z was impaired in the ability to ubiquitinate either CIN85 or EGFRs (Fig. 2A). In addition, a deletion mutant of Cbl-b (N1/2), that contains the SH2 domain and RING finger but lacks a binding site for CIN85 (Fig. 2C), was impaired in its ability to monoubiquitinate CIN85 (Fig. 2B). On the other hand, a Cbl-b mutant (C2/3) containing the RING domain and the carboxyl terminus that is able to bind to CIN85 strongly promoted monoubiquitination of CIN85 even in the absence of EGF stimulation (Fig. 2 B and C). These data demonstrated that the intact RING domain and the distal carboxyl terminus of Cbl are required for EGF-induced monoubiquitination of CIN85.
Fig 2.
Monoubiquitination (MonoUbi) of CIN85 is mediated by Cbl and Cbl-b. (A) HEK293T cells overexpressing EGFRs, FLAG-tagged ubiquitin, FLAG-tagged CIN85 and Cbl, or Cbl-70Z were stimulated with EGF as described for Fig. 1A. Levels of EGFR, Cbl, Cbl-70Z, CIN85, and monoubiquitinated CIN85 were determined in the TCLs. (B) HEK293T cells expressing EGFR, FLAG-tagged ubiquitin (FLAG-Ubi), CIN85 and Cbl-b, Cbl-b-N1/2, or Cbl-b-C2/3 were stimulated with EGF as described in Fig. 1A. TCLs were analyzed for the presence of monoubiquitinated CIN85 by blotting with anti-CIN85 antibodies (CT), and levels of Cbl-b and Cbl-b deletion forms are shown (Upper). wt, wild type. (C) Cbl-b-N1/2 contains the SH2 domain and RING finger domain (RF) of Cbl-b. Cbl-b-C2/3 consists of the RING finger domain, proline-rich region (PxxP), and leucine zipper (LZ) domain of Cbl-b.
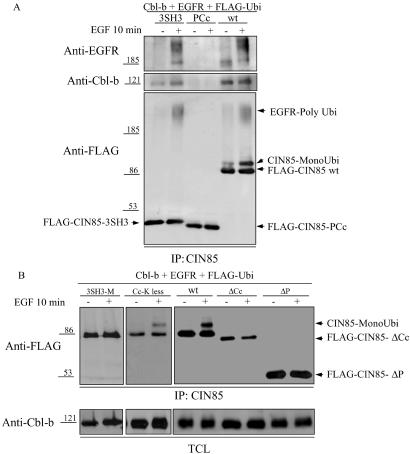
To define the region of CIN85 that is necessary for the attachment of ubiquitin, several deletion mutants of CIN85 were created and analyzed for their ability to undergo Cbl-mediated monoubiquitination. Overexpression of FLAG-tagged CIN85 together with Cbl-b and EGFRs led to monoubiquitination of CIN85 and its association with Cbl-b and polyubiquitinated EGFRs (Fig. 3A). The basal level of monoubiquitination of CIN85 and association between CIN85, Cbl-b, and EGFRs seen in unstimulated cells were caused by activation of EGFRs because of its overexpression (Fig. 3A). On the other hand, the truncated form of CIN85 containing three SH3 domains (CIN85-3SH3) strongly bound Cbl-b and polyubiquitinated EGFRs but was not monoubiquitinated (Fig. 3A). In addition, the carboxyl-terminal part of CIN85 containing the proline-rich region and the coiled-coil domain of CIN85 (CIN85-PCc) did not interact with Cbl-b or EGFRs and was not ubiquitinated by Cbl-b after EGF stimulation (Fig. 3A). Furthermore, a CIN85 mutant with point mutations in all three SH3 domains (CIN85-3SH3-M), yielding a CIN85 protein unable to bind to Cbl/Cbl-b (data not shown), was not monoubiquitinated by Cbl-b after EGF stimulation (Fig. 3B). Thus, binding of CIN85 to Cbl-b is critical for monoubiquitination of full-length CIN85 (Figs. 2B and 3B). Because coiled-coil domains have been found in several proteins involved in endosomal sorting (22–24) and are responsible for oligomerization of CIN85 (25), we tested whether deletion of the coiled-coil domain of CIN85 would affect its monoubiquitination. A truncated form of CIN85 without the coiled-coil domain (CIN85-ΔCc) was not monoubiquitinated by Cbl after EGF stimulation (Fig. 3B). Moreover, a mutant of CIN85, lacking the proline-rich region between the SH3 domains and the coiled-coil domain (CIN85-ΔP), was also impaired in Cbl-induced monoubiquitination (Fig. 3B). These results suggest that the intact structure of the carboxyl terminus of CIN85 is required for CIN85 monoubiquitination. Moreover, our data indicate that the amino-terminal part of CIN85 recruits Cbl, whereas its carboxyl terminus provides an acceptor site for ubiquitin. Because both CIN85 and its homologue CMS were monoubiquitinated by Cbl, we searched for conserved lysines in their carboxyl termini. Individual mutations of 19 conserved lysines, mutation of seven clustered lysine pairs, or individual substitution of all remaining nonconserved lysines in the carboxyl terminus of CIN85 did not abolish EGF-induced monoubiquitination of CIN85 (data not shown). The fact that deletion of the coiled-coil domain inhibited monoubiquitination of CIN85 (Fig. 3B) suggested that monoubiquitination could take a place on one of the eight lysines present in the structure of the coiled-coil domain. However, exchange of all lysines to arginine residues (K627,631,635,645,646,659,660,665R) in the coiled-coil domain of CIN85 yielding lysine-less CIN85-Cc (CIN85-Cc-K-less) did not abolish EGF-induced monoubiquitination of CIN85 (Fig. 3B). Taken together, these extensive studies indicated that Cbl-directed monoubiquitination of CIN85 is not specific for a particular lysine residue but rather utilizes various lysine residues in the context of the intact carboxyl terminus of CIN85.
Fig 3.
Monoubiquitination (MonoUbi) of the carboxyl terminus of CIN85. (A) HEK293T cells overexpressing EGFRs, Cbl-b, and FLAG-tagged ubiquitin (FLAG-Ubi) together with FLAG-tagged CIN85-3SH3 (3SH3), CIN85-PCc (PCc), or wild-type CIN85 (wt) were stimulated with EGF as described for Fig. 1A. CIN85 and CIN85 mutants were immunoprecipitated (IP) with anti-CIN85 (CT or SH3) antibodies, and a Western blot was probed with anti-EGFR (RK2), anti-Cbl-b (RF), and anti-FLAG (M5) antibodies as indicated. (B) HEK293T cells overexpressing EGFR, Cbl-b, and FLAG-tagged ubiquitin together with FLAG-tagged wild-type CIN85 (wt), CIN85-3SH3-M (3SH3-M), CIN85-Cc-lysine-less (Cc-K less), CIN85-ΔCc (ΔCc), or CIN85-ΔP (ΔP) were stimulated with EGF as described for Fig. 1A. CIN85 and CIN85 mutants were immunoprecipitated by using anti-CIN85 (SH3) antibodies, and their monoubiquitination was analyzed by Western blotting with anti-FLAG (M5) antibodies. Levels of Cbl-b in TCLs are shown (Lower).
Monoubiquitinated CIN85 Is Degraded Together with Cbl/Cbl-b and EGFRs.
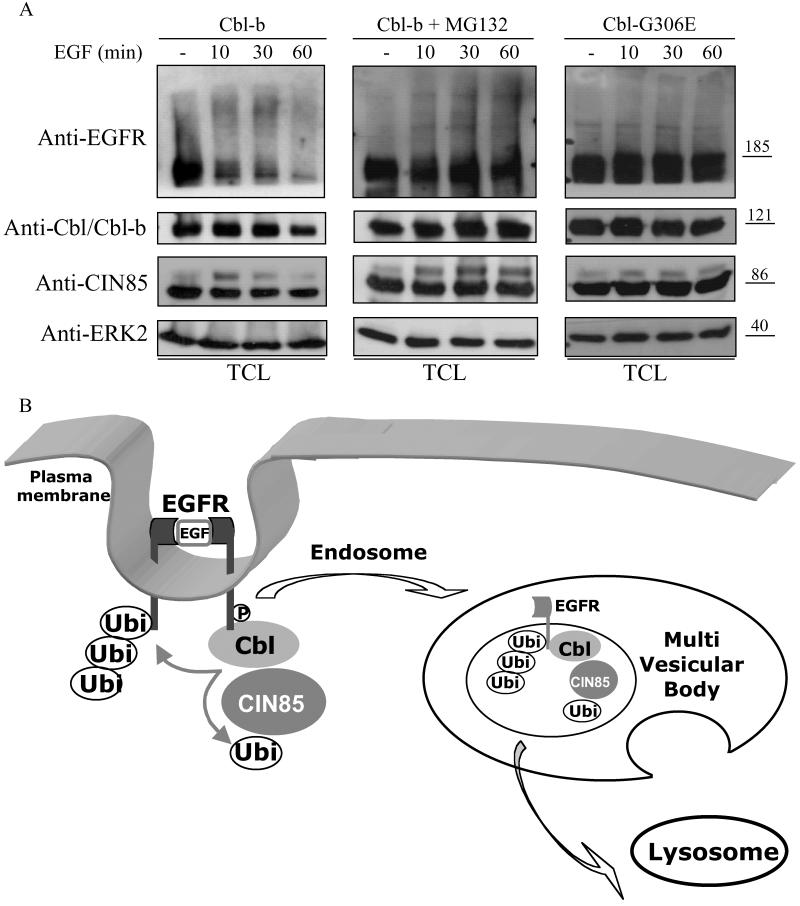
Previous studies have shown that CIN85 remains associated with Cbl and EGFRs for a prolonged time, leading to their colocalization in endocytic vesicles (20, 26, 27). The EGFR kinase activity and the ubiquitin ligase activity of Cbl were shown to be critical for EGFR sorting in the endosome, because blocking either of these functions is sufficient to increase recycling of the receptor to the plasma membrane (11, 26). Because we observed that CIN85 is associated with polyubiquitinated EGFRs after ligand stimulation (Figs. 1B and 3A), we further analyzed whether these proteins are targeted for common destruction in the lysosome. Prolonged EGF stimulation of HEK293T cells expressing CIN85, Cbl-b, and EGFRs led to a sustained monoubiquitination of CIN85 and polyubiquitination of EGFRs and their common degradation (Fig. 4A). Degradation of EGFRs was almost complete, whereas degradation of CIN85 and Cbl-b was partial (Fig. 4A). In addition, degradation of EGFR and CIN85 was sensitive to treatment with the proteasome inhibitor MG132 (Fig. 4A). MG132 was shown to act as a potent inhibitor of lysosomal degradation of EGFRs by blocking proteasomal activity and sorting of EGFRs in the MVB (28). Interestingly, monoubiquitinated CIN85 seems to accumulate with time in MG132-treated cells (Fig. 4A). Moreover, overexpression of Cbl-G306E, which is unable to interact with phosphotyrosines of activated EGFRs (15, 29, 30), did not induce significant degradation of EGFRs or CIN85 (Fig. 4A), suggesting that interactions between EGFRs and Cbl are critical for efficient down-regulation of EGFRs and CIN85. Taken together, these data indicate that CIN85 and EGFRs are concomitantly degraded along the same pathway and suggest that their ubiquitination may be involved in the control of down-regulation of EGFRs. These results are consistent with the finding that dominant interfering forms of CIN85, which are not monoubiquitinated by Cbl-b (Fig. 3A), are sufficient to delay ligand-induced EGFR degradation (20). It remains unclear whether interfering functions of CIN85-3SH3 and CIN85-PCc are only caused by their competition with endogenous CIN85 for binding to Cbl or endophilin, respectively (20), or if the lack of their monoubiquitination can also affect endosomal sorting and thus contribute to delay in receptor degradation.
Fig 4.
CIN85 is degraded together with Cbl-b and EGFRs. (A) HEK293T cells overexpressing EGFRs, FLAG-tagged ubiquitin (Ubi), FLAG-tagged CIN85, and Cbl-b or Cbl-G306E were starved and either mock-treated or treated for 1 h with 20 μM of the proteasome inhibitor MG132 as indicated, followed by mock treatment or EGF (100 ng/ml) stimulation for indicated times. TCLs were analyzed for EGFR, Cbl-b, CIN85, and ERK2 levels with specific antibodies. ERK2 was used as a control for equal loading, because it is not degraded in the complex with EGFRs. (B) A schematic model of a Cbl–CIN85 role in EGFR down-regulation. After ligand binding, EGFRs dimerize and autophosphorylate, leading to recruitment of Cbl to phosphorylated EGFRs. Cbl polyubiquitinates the EGFR and recruits CIN85, which also becomes monoubiquitinated by Cbl. The EGFR–Cbl–CIN85 complex is sorted along the endocytic pathway and targeted to the MVB and lysosome for degradation. Cbl thus mediates ubiquitination of both EGFRs and CIN85 in the same complex. CIN85 also has a dual role in controlling EGFR down-regulation, because it both promotes receptor internalization and participates in endosomal sorting and subsequent degradation of activated receptors.
Discussion
Cbl proteins play important roles in RTK down-regulation by acting as ubiquitin ligases and multiadaptor proteins (12). Previous reports have shown that Cbl mediates ubiquitination of receptors as well as recruits CIN85-endophilin in complexes with activated receptors, thus controlling RTK endocytosis (14, 15, 20, 31). Here we show that Cbl and Cbl-b also promote monoubiquitination of CIN85 and its association with polyubiquitinated EGFRs, leading to their common destruction in the lysosome (Fig. 4B). Our data indicate that CIN85 has a dual role in controlling EGFR down-regulation by promoting receptor internalization and participating in endosomal sorting and subsequent degradation of activated receptors and that Cbl-mediated mono- and polyubiquitination are critical determinants in regulating degradation of EGFRs (Fig. 4B).
Different ubiquitin ligases may be involved in mediating ubiquitination of CIN85 in mammalian cells (Fig. 1C). Ligand-induced monoubiquitination of CIN85 and CMS seems to be mediated by Cbl/Cbl-b (Figs. 1 and 2). This conclusion is supported by findings that overexpression of Cbl or Cbl-b leads to an EGF-induced increase in monoubiquitination of CIN85 (Fig. 1C), whereas expression of Cbl-b-N1/2 that is unable to bind to CIN85 partially inhibits the ability of endogenous Cbl to monoubiquitinate CIN85 (Fig. 2B). On the other hand, expression of Nedd4 ubiquitin ligase leads to multiubiquitination of CIN85 in the absence of EGF stimulation (Fig. 1C). These data indicate that Cbl monoubiquitinates CIN85 after endocytosis of activated EGFRs, whereas Nedd4 may function as a CIN85 ubiquitin ligase independent of growth factor stimulation. Furthermore, EGF-induced monoubiquitination of CIN85 was not dependent on a specific lysine-based recognition sequence as several lysine residues could be tagged by ubiquitin (Fig. 3B). However, it is not clear yet how monoubiquitination of CIN85 and polyubiquitination of EGFRs are controlled by Cbl in a trimeric complex. These processes can occur as parallel events where Cbl molecules can simultaneously ubiquitinate CIN85 and EGFRs or as sequential events where Cbl ubiquitinates first EGFRs and subsequently CIN85. In either scenario, there must be a mechanism that is specific for CIN85 but not EGFRs that prevents addition of ubiquitin chains after the first ubiquitin molecule is attached to a lysine residue. It is also a saturable mechanism, because high overexpression of Cbl ligases in mammalian cells leads to the appearance of two or three ubiquitin molecules attached to CIN85 (data not shown). It was suggested recently that ubiquitin-interacting motifs can inhibit further ubiquitin chain assembly by a direct binding to ubiquitin (18). It will be interesting to investigate further whether ubiquitin-interacting motif-containing proteins such as epsins, eps15, and Hrs are found in the complex with monoubiquitinated CIN85 after EGF stimulation.
Protein ubiquitination is also a reversible process that is controlled actively by the deubiquitination machinery in the cell. We have observed that the addition of inhibitors of deubiquitination such as N-ethylmaleimide (NEM) in the lysis buffer increases monoubiquitination of CIN85 (data not shown), suggesting that CIN85 may be regulated also by ubiquitination-deubiquitination cycles. This dynamic process may ensure that only a portion of CIN85 is monoubiquitinated at a given moment in the cell and raises an interesting possibility that only monoubiquitinated CIN85 is targeted together with polyubiquitinated receptors for destruction in the lysosome (Fig. 4A), whereas deubiquitinated CIN85 is recycled back to the cytoplasmic pool. In addition, treatment of cells with proteasome inhibitors blocked degradation of EGFRs and CIN85 (Fig. 4A), suggesting that the proteasome function is critical for proper targeting of these complexes to lysosomes. In accordance, recent findings have shown that not only ubiquitination but also proteasome activity is required for sorting of the EGFR to inner membranes of MVBs and its subsequent degradation (28). Moreover, spatial formation of EGFR-associated complexes seems to be critical for their common degradation. For example, a direct binding of Cbl to activated EGFRs is required for degradation of both EGFRs and CIN85 (Fig. 4A). Furthermore, other EGFR-bound proteins such as the adaptor proteins Grb2 and Shc were shown to be degraded together with EGFRs in a Cbl-b-dependent fashion (13). Taken together, our results revealed an important role for Cbl/Cbl-b in mediating ubiquitination and formation of CIN85–EGFR complexes leading to their common degradation. Visualization of these complexes in living cells as well as their dynamic interactions along the endocytic route remains a challenge for future investigations.
Acknowledgments
We thank K. Kirsch for the CMS expression vector; W. Y. Langdon for Cbl-G306E and Cbl-70Z constructs; J. Schlessinger for anti-EGFR antibodies; P. P. Di Fiore for the Nedd4 expression vector; and S. Lipkowitz for Cbl-b constructs. We are also thankful to other members of the Dikic laboratory for helpful discussions, critical comments on the manuscript, and providing various CIN85 reagents. This work was supported by a grant from the Swedish Strategic Funds (to I.D.). I.D. is a research fellow of the Boehringer Ingelheim Fonds.
Abbreviations
RTK, receptor tyrosine kinase
MVB, multivesicular body
EGF, epidermal growth factor
EGFR, EGF receptor
CIN85, Cbl-interacting protein of 85 kDa
SH, Src homology
CMS, p130Cas ligand with multiple SH3 domains
TCL, total cell lysate
References
- 1.Ullrich A. & Schlessinger, J. (1990) Cell 61, 203-212. [DOI] [PubMed] [Google Scholar]
- 2.Hunter T. (1997) Cell 88, 333-346. [DOI] [PubMed] [Google Scholar]
- 3.Lemmon M. A. & Schlessinger, J. (1998) Methods Mol. Biol. 84, 49-71. [DOI] [PubMed] [Google Scholar]
- 4.Sorkin A. & Waters, C. M. (1993) BioEssays 15, 375-382. [DOI] [PubMed] [Google Scholar]
- 5.Hopkins C. R., Gibson, A., Shipman, M. & Miller, K. (1990) Nature (London) 346, 335-339. [DOI] [PubMed] [Google Scholar]
- 6.Waterman H. & Yarden, Y. (2001) FEBS Lett. 490, 142-152. [DOI] [PubMed] [Google Scholar]
- 7.Di Fiore P. P. & Gill, G. N. (1999) Curr. Opin. Cell Biol. 11, 483-488. [DOI] [PubMed] [Google Scholar]
- 8.Floyd S. & De Camilli, P. (1998) Trends Cell Biol. 8, 299-301. [DOI] [PubMed] [Google Scholar]
- 9.Kornilova E., Sorkina, T., Beguinot, L. & Sorkin, A. (1996) J. Biol. Chem. 271, 30340-30346. [DOI] [PubMed] [Google Scholar]
- 10.Sorkin A., Mazzotti, M., Sorkina, T., Scotto, L. & Beguinot, L. (1996) J. Biol. Chem. 271, 13377-13384. [DOI] [PubMed] [Google Scholar]
- 11.Felder S., Miller, K., Moehren, G., Ullrich, A., Schlessinger, J. & Hopkins, C. R. (1990) Cell 61, 623-634. [DOI] [PubMed] [Google Scholar]
- 12.Thien C. B. & Langdon, W. Y. (2001) Nat. Rev. Mol. Cell Biol. 2, 294-307. [DOI] [PubMed] [Google Scholar]
- 13.Ettenberg S. A., Magnifico, A., Cuello, M., Nau, M. M., Rubinstein, Y. R., Yarden, Y., Weissman, A. M. & Lipkowitz, S. (2001) J. Biol. Chem. 276, 27677-27684. [DOI] [PubMed] [Google Scholar]
- 14.Joazeiro C. A., Wing, S. S., Huang, H., Leverson, J. D., Hunter, T. & Liu, Y. C. (1999) Science 286, 309-312. [DOI] [PubMed] [Google Scholar]
- 15.Levkowitz G., Waterman, H., Ettenberg, S. A., Katz, M., Tsygankov, A. Y., Alroy, I., Lavi, S., Iwai, K., Reiss, Y., Ciechanover, A., et al. (1999) Mol. Cell 4, 1029-1040. [DOI] [PubMed] [Google Scholar]
- 16.Hicke L. (2001) Cell 106, 527-530. [DOI] [PubMed] [Google Scholar]
- 17.Babst M., Odorizzi, G., Estepa, E. J. & Emr, S. D. (2000) Traffic 1, 248-258. [DOI] [PubMed] [Google Scholar]
- 18.Polo S., Sigismund, S., Faretta, M., Guidi, M., Capua, M. R., Bossi, G., Chen, H., De Camilli, P. & Di Fiore, P. P. (2002) Nature (London) 416, 451-455. [DOI] [PubMed] [Google Scholar]
- 19.Take H., Watanabe, S., Takeda, K., Yu, Z. X., Iwata, N. & Kajigaya, S. (2000) Biochem. Biophys. Res. Commun. 268, 321-328. [DOI] [PubMed] [Google Scholar]
- 20.Soubeyran P., Kowanetz, K., Szymkiewicz, I., Langdon, W. Y. & Dikic, I. (2002) Nature (London) 416, 183-187. [DOI] [PubMed] [Google Scholar]
- 21.Kirsch K. H., Georgescu, M. M., Shishido, T., Langdon, W. Y., Birge, R. B. & Hanafusa, H. (2001) J. Biol. Chem. 276, 4957-4963. [DOI] [PubMed] [Google Scholar]
- 22.Kranz A., Kinner, A. & Kolling, R. (2001) Mol. Biol. Cell 12, 711-723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Katzmann D. J., Babst, M. & Emr, S. D. (2001) Cell 106, 145-155. [DOI] [PubMed] [Google Scholar]
- 24.Salcini A. E., Chen, H., Iannolo, G., De Camilli, P. & Di Fiore, P. P. (1999) Int. J. Biochem. Cell Biol. 31, 805-809. [DOI] [PubMed] [Google Scholar]
- 25.Watanabe S., Take, H., Takeda, K., Yu, Z. X., Iwata, N. & Kajigaya, S. (2000) Biochem. Biophys. Res. Commun. 278, 167-174. [DOI] [PubMed] [Google Scholar]
- 26.Levkowitz G., Waterman, H., Zamir, E., Kam, Z., Oved, S., Langdon, W. Y., Beguinot, L., Geiger, B. & Yarden, Y. (1998) Genes Dev. 12, 3663-3674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.de Melker A. A., van Der Horst, G., Calafat, J., Jansen, H. & Borst, J. (2001) J. Cell Sci. 114, 2167-2178. [DOI] [PubMed] [Google Scholar]
- 28.Longva K. E., Blystad, F. D., Stang, E., Larsen, A. M., Johannessen, L. E. & Madshus, I. H. (2002) J. Cell Biol. 156, 843-854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bonita D. P., Miyake, S., Lupher, M. L., Jr., Langdon, W. Y. & Band, H. (1997) Mol. Cell. Biol. 17, 4597-4610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Thien C. B. & Langdon, W. Y. (1997) Oncogene 14, 2239-2249. [DOI] [PubMed] [Google Scholar]
- 31.Petrelli A., Gilestro, G. F., Lanzardo, S., Comoglio, P. M., Migone, N. & Giordano, S. (2002) Nature (London) 416, 187-190. [DOI] [PubMed] [Google Scholar]
- 32.Borinstein S. C., Hyatt, M. A., Sykes, V. W., Straub, R. E., Lipkowitz, S., Boulter, J. & Bogler, O. (2000) Cell Signal. 12, 769-779. [DOI] [PubMed] [Google Scholar]
- 33.Gout I., Middleton, G., Adu, J., Ninkina, N. N., Drobot, L. B., Filonenko, V., Matsuka, G., Davies, A. M., Waterfield, M. & Buchman, V. L. (2000) EMBO J. 19, 4015-4025. [DOI] [PMC free article] [PubMed] [Google Scholar]