Abstract
Pre-mRNA splicing takes place in a large and highly dynamic complex known as the spliceosome. Here we report the optimization of a maltose-binding protein (MBP) affinity-purification method to isolate functional spliceosomes for electron microscopic analysis. Visualization of the spliceosome preparations revealed distinct 40–60 nm particles. Immunogold-conjugated antibodies to spliceosome components specifically label these particles, which are eliminated by treatment with either RNase or protease. Moreover, spliceosomes assembled on two different pre-mRNAs are indistinguishable. This first visualization of purified functional spliceosomes assembled in vitro reveals striking structural features, including one or more central cavities and multiple elongate lobes.
The spliceosome, which is highly conserved from yeast to mammals, contains five small nuclear RNAs (snRNAs) and numerous distinct proteins (1–4). Early studies led to a model in which the spliceosome assembles on pre-mRNA by sequential binding of U1, U2, and U4/5/6 small nuclear ribonucleoprotein particles (snRNPs) to form the E, A, B, and C intermediate complexes (2). However, recent studies in yeast showed that a single penta-snRNP, containing all five snRNPs, is a functional precursor to the spliceosome (5). In mammals, all five snRNPs are associated with both early and late functional spliceosomes (6). These and other data have led to the proposal that the spliceosome may be a preformed entity that associates with the pre-mRNA and then remains bound throughout the splicing reaction (5, 7). This spliceosome undergoes numerous dynamic changes that may only be detectable (as the A, B, and C complexes) when different stages of splicing are analyzed under stringent conditions (5, 7).
Significant progress in determining the structure of the spliceosome by electron microscopy was provided by analyses of purified snRNPs, including U1, U2, U4/U6, U5, and the U4/U5/U6 tri-snRNP (4). These studies revealed that U1 snRNP possesses an almost circular main body (8 nm in diameter) with two adjacent protuberances (4–7 nm) (8, 9), U2 snRNP appears dumbbell shaped (12–22 nm) (10, 11), and U4/U5/U6 tri-snRNP has a triangle-like structure (20–25 nm) (12). Additional progress toward understanding the structure of the spliceosome has come from studies of the C complex isolated under stringent conditions (high salt and heparin treated). This complex, which contains only U2, U5, and U6 snRNPs, has an asymmetric structure with average dimensions of 24 × 27 nm (13). Finally, “native spliceosomes,” which may correspond to mammalian penta-snRNPs or to spliceosomes assembled on endogenous pre-mRNAs present in splicing extracts, have been visualized by electron microscopy (14). These complexes, isolated by gel filtration under nonstringent conditions, are relatively homogenous in size (40–60 nm) with a morphology characterized by multiple globular subunits (14).
To date, functional spliceosomes assembled in vitro have not been visualized by electron microscopy (EM) because it has not been possible to purify them free in solution. In this study, we have optimized a method for obtaining highly purified functional spliceosomes for visualization by EM. Our analysis revealed 40–60 nm particles with striking similarity to the native endogenous spliceosomes previously described (14).
Methods and Materials
Plasmids encoding AdML pre-mRNA or Ftz pre-mRNA have been described (6). A fragment containing three MS2 hairpins was generated by annealing and extension of the following oligos: 5′-CGGGATCCGATATCCGTACACCATCAGGGTACGAGCTAGCCCATGGCGTACACCATCAGGGTACGA-3′; 5′-GCTCTAGAGAATTCCGTACCCTGATGGTGTACGAGATCTACTAGTCGTACCCTGATGGTGTACGC-3′.
The double-stranded DNA products were subcloned into the pCR2.1 plasmid (Invitrogen), confirmed by sequencing, and further inserted into the 3′ end of AdML pre-mRNA or Ftz pre-mRNA. The MS2-MBP (maltose-binding protein) construct was a gift from M. Moore (Brandeis University, Waltham, MA). The MS2-MBP fusion protein was expressed in E. coli, and isolated by binding to amylose beads according to the manufacturer (NEB, Beverly, MA), and further purified over a Heparin Hi-trap column by using a NaCl gradient (Pharmacia; ref. 13). The fusion proteins and pre-mRNAs were incubated in a buffer containing 20 mM Hepes, pH 7.9, and 60 mM NaCl, on ice for 30 min, and the binding was assayed on a 1.5% native agarose gel. Spliceosomes were assembled on the MS2-MBP/pre-mRNA-M3 complex by using standard conditions and isolated by gel filtration (15, 16). Subsequently, the spliceosomes were affinity-selected on amylose beads by rotating for 4 h at 4°C and were eluted with 12 mM maltose, 20 mM Hepes, pH 7.9, 60 mM NaCl, 10 mM β-mercaptoethanol, and 1 mM PMSF. For identification of snRNAs, total RNA was prepared from equivalent amounts of each purified complex and end-labeled with [32P]pCp and RNA ligase. The spliceosome samples were prepared for EM by fixation with 0.6% glutaraldehyde for 5 min at room temperature, chromatography through 2-ml columns of BiogelA5m (Bio-Rad), and mounting onto thin carbon foils in a solution containing spermidine followed by rotary shadowcasting with tungsten (14). For immunogold labeling, the spliceosomes bound to the amylose resin were incubated with purified primary antibodies (anti U5–40kD or trimethyl cap) followed by goat-anti-rabbit 6-nm gold conjugate or goat-anti-mouse 10-nm gold conjugate (Aurion), respectively, at 4°C for 2 h. The spliceosomes were then eluted after extensive washing as described above and mounted for EM. Micrographs were taken on a Philips CM12 instrument and images on sheet film scanned with a Nikon LS4500 film scanner and arranged using photoshop software (Adobe Systems, Mountain View, CA).
Results and Discussion
Optimization of MBP-Affinity Purification for EM Analysis.
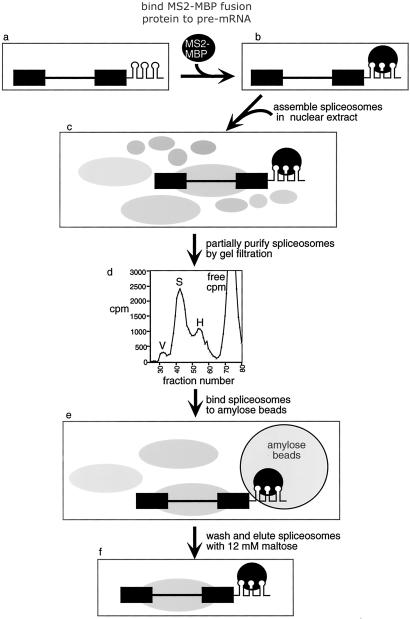
To carry out EM analysis of functional spliceosomes assembled in vitro, it was first necessary to establish conditions for obtaining the complexes free in solution, highly purified, and concentrated. We recently established an affinity purification strategy for isolating functional spliceosomes (refs. 6 and 17; Fig. 1). In this method, the pre-mRNA contains bacteriophage R17/MS2 coat protein binding sites (hairpins; refs. 18–20) at the 3′ end of exon 2 (ref. 21; Fig. 1a). A fusion protein of the MS2 coat protein and MBP is bound to the hairpins (Fig. 1b). This pre-mRNA/MS2-MBP complex is then assembled into spliceosomes by incubation in HeLa cell nuclear extract under splicing conditions (Fig. 1c). Spliceosomes are partially purified by Sephacryl S-500 gel filtration (Fig. 1d) and then affinity selected by binding to Amylose beads (Fig. 1e). After washing, the spliceosomes are eluted with maltose (Fig. 1f).
Fig 1.
Strategy for MBP-affinity purification of the spliceosome. Spliceosomes are isolated by gel filtration followed by affinity purification (see text for details).
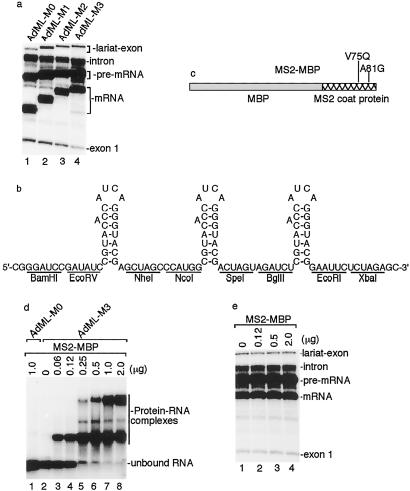
To optimize the affinity purification strategy, we first asked whether the number of hairpins affects the efficiency of splicing and the spliceosome purification. As demonstrated in Fig. 2a, the presence of one, two, or three hairpins at the 3′ end of AdML exon 2 does not significantly affect splicing. The spliceosome purification efficiency using these pre-mRNAs is typically 10, 18, and 35%, respectively. Increasing the number of hairpins to five or nine adversely affects the resolution of the spliceosome on the gel filtration column (data not shown). Thus, we chose three hairpins for further optimization (the sequence of the cassette containing the three hairpins is shown in Fig. 2b).
Fig 2.
Optimization of MBP-affinity purification. (a) Insertion of MS2 hairpins at the 3′ end of exon 2 does not affect splicing efficiency. AdML pre-mRNA containing 0, 1, 2, or 3 hairpins are spliced in nuclear extract for 40 min, and total RNAs were analyzed on a 16% denaturing polyacrylamide gel. (b) Sequence of the cassette containing three MS2 RNA binding sites (hairpins). The secondary structure and the bulged adenosine characteristic of the hairpins are shown. The restriction enzyme sites of the corresponding DNA sequence are indicated. (c) Schematic of MS2-MBP fusion protein. The mutations that prevent MS2 oligomerization are indicated. (d) The MS2-MBP protein specifically binds to AdML-M3 pre-mRNA in a dose-dependent manner. AdML (lane 1) or AdML-M3 (lanes 2–8) pre-mRNA was incubated with increasing amounts of MS2-MBP for 30 min. The RNA-protein complexes were separated on a 1.5% agarose gel. (e) Binding of MS2-MBP to AdML-M3 does not affect splicing efficiency. Increasing amounts of MS2-MBP protein were incubated with AdML-M3 pre-mRNA for 30 min, followed by incubation under splicing conditions in nuclear extract for 40 min. Total RNAs were analyzed on a 16% denaturing polyacrylamide gel.
In initial EM studies, we found that the MS2-MBP protein alone forms multimeric complexes that coelute with spliceosomes by gel filtration and are present in the maltose elution (data not shown). Other tags that were tested for spliceosome purification resulted in either high background (protein A tag) or low binding efficiency of the spliceosome (GST or Histidine tag). Recently, it was reported that a combination of purifying MS2-MBP by heparin column chromatography and mutation in the MS2 coat protein (V75Q and A81G) prevents MS2 oligomerization (13, 22). Thus, we tested this new preparation of MS2-MBP (see Fig. 2c for protein schematic) in our studies. To determine the minimal amount of MS2-MBP that binds quantitatively to AdML-M3 pre-mRNA, we titrated the protein and analyzed complex formation by using a gel shift assay. The MS2-MBP protein is specific for the hairpin-containing RNAs, because no complexes are formed on AdML pre-mRNA lacking the hairpins (Fig. 2d, lane 1). In the presence of AdML-M3 pre-mRNA, MS2-MBP was assembled into complexes in a dose-dependent manner (lanes 2–8). In the range of a 50–200-fold molar excess of MS2-MBP to AdML-M3 pre-mRNA, quantitative binding of MS2-MBP to one or more hairpins was observed (lanes 6–8). As shown in Fig. 2e, AdML-M3 pre-mRNA splicing was not inhibited by a 200-fold molar excess of MS2-MBP. On the basis of these data, we chose a 100-fold molar excess for spliceosome purification.
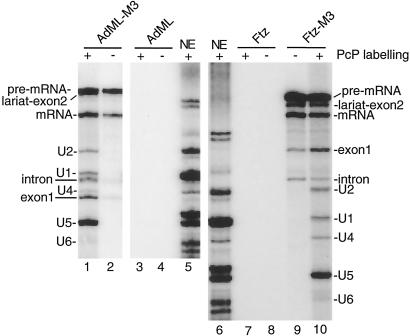
For EM studies, we isolated spliceosomes assembled on AdML-M3 or Ftz-M3 pre-mRNAs. As negative controls, spliceosomes were also assembled on AdML or Ftz pre-mRNAs lacking the hairpins. The splicing products and spliceosomal snRNPs were detected in AdML-M3 or Ftz-M3 elution, but not in the AdML or Ftz elution (Fig. 3). In addition, we found that spliceosomal proteins specifically associate with the AdML-M3 and Ftz-M3 pre-mRNAs (ref. 23 and data not shown). Refractionation of the purified spliceosomes over a gel filtration column showed that they elute in the same column fractions before and after affinity purification (ref. 23 and data not shown). These data indicate that the purified spliceosomes are intact.
Fig 3.
Analysis of snRNAs in purified spliceosomes. Spliceosomes were purified using AdML-M3, AdML, Ftz-M3, or Ftz pre-mRNAs as substrates (AdML or Ftz pre-mRNAs lacking hairpins were used for negative controls). Total RNAs extracted from the nuclear extract (NE), AdML, AdML-M3, Ftz, or Ftz-M3 elutions were 32P-end-labeled (+) or not labeled (−) and fractionated on an 8% polyacrylamide gel. Note that the snRNAs are 3′-end-labeled differentially with RNA ligase (15).
EM Visualization of Spliceosomes.
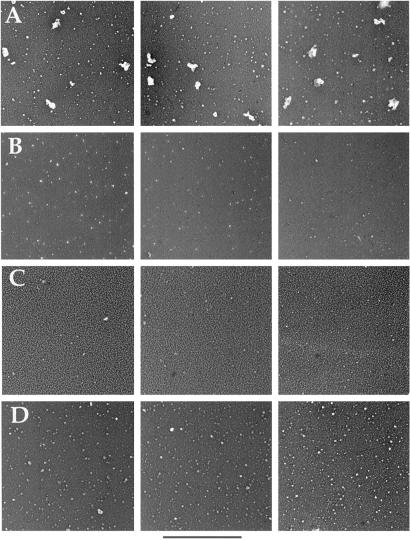
The highly purified spliceosomes were then visualized by EM using rotary tungsten shadowing. Representative fields of the AdML spliceosome preparation are shown in Fig. 4A. Multiple particles ranging in size from 40–60 nm were observed. In contrast, these large particles are not present in the AdML negative control purification (Fig. 4B). Moreover, the particles are sensitive to Proteinase K (Fig. 4C) or RNase A (Fig. 4D) treatment. The smaller particles observed in all samples are MS2-MBP protein complexes (data not shown). These data are consistent with the possibility that the RNP complexes visualized by EM are spliceosomes.
Fig 4.
Visualization of purified functional spliceosomes by electron microscopy. (A) Three different fields of purified spliceosomes assembled on AdML-M3 pre-mRNA. (B) Fields of control purification. (C) Fields of spliceosomes after treatment with Proteinase K. (D) Fields of spliceosomes after treatment with RNase A. The scale bar represents 360 nm.
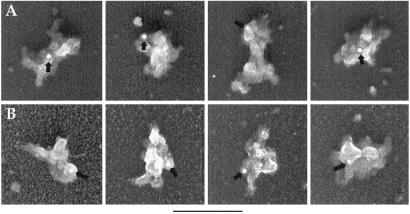
To determine directly whether the particles that we visualize correspond to spliceosomes, we carried out immunogold labeling. As shown in Fig. 5A, the particles are specifically labeled with gold conjugates by using antibodies that recognize the trimethyl cap at the 5′ end of snRNPs (4). In addition, gold conjugates were detected when antibodies to the U5 snRNP 40kD protein (24) were used (Fig. 5B) (indicated with arrows). In contrast, no gold labeling was detected in the negative controls in which the primary antibodies were omitted or the primary and the secondary antibodies were mismatched (data not shown). We conclude that the RNP particles visualized by EM are spliceosomes.
Fig 5.
Immunogold labeling of spliceosomes. Spliceosomes assembled on AdML-M3 pre-mRNA were incubated with antitrimethyl cap antibody followed by a secondary antibody conjugated with 10 nm gold (A) or with anti-U5–40kD antibody followed by a secondary antibody conjugated with 6 nm gold (B). Arrows indicate the positions of gold labels. The 3-cm bars are equal to 180 nm (A) and 110 nm (B), respectively.
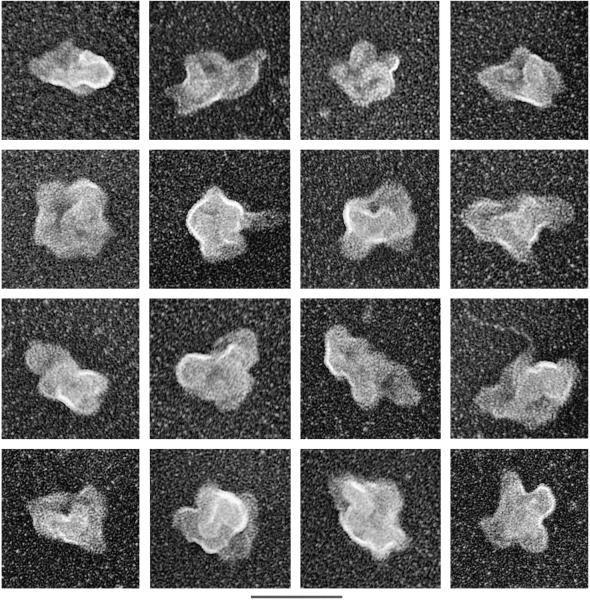
Analysis of individual spliceosomes at higher magnification revealed that they are highly structured particles with multiple elongate, globular domains (Fig. 6). Another striking feature is the presence of one or more large central cavities. Significantly, spliceosomes assembled on AdML-M3 pre-mRNA (Figs. 4 and 5 and data not shown) or Ftz-M3 pre-mRNA (Fig. 6) have the same types of morphologies. Moreover, the AdML and Ftz spliceosomes are similar in size, averaging about 40–60 nm, which is the same as that previously observed for the native endogenous spliceosomes (14). Our purified spliceosomes also appear remarkably similar in morphology to the native spliceosomes (14). We conclude that the spliceosomes assembled in vitro are highly related to the native endogenous spliceosomes that exist in nuclear extracts.
Fig 6.
EM of individual spliceosomes assembled on Ftz-M3 pre-mRNA. Size bars are indicated at the bottom. The 3-cm bar is equal to 45 nm.
As shown in Fig. 3, our purified spliceosome preparation consists of a mixture of unspliced pre-mRNA, splicing intermediates, and products. All five snRNAs are also present in the preparation. At present, it is not possible to distinguish between the possibility that the different shapes and sizes of the particles observed by EM are due to different views of a single “holospliceosome” structure (see the introduction) or whether we are visualizing a mixture of different spliceosomal complexes (i.e., A, B, C, and spliced mRNP complexes). This question also remains unanswered for the endogenous native spliceosomes (14).
On the basis of the dimensions of native U1, U2, and U4/U5/U6 snRNPs (see the introduction), the size of our purified spliceosome (40 × 60 nm) could accommodate at least one each of these snRNPs. The central cavities present in the spliceosomes are not observed in the individual snRNPs, and thus must form when all of the snRNPs and other proteins associate with each other in the spliceosome. As previously suggested, the globular domains may correspond to individual snRNPs (14). Recent analysis of the proteome of MBP-purified spliceosomes identified ≈145 distinct proteins (23). As many as half of these may be non-snRNP proteins (23). Thus, the large size of the purified spliceosome visualized by EM may be explained not only by the snRNPs but also by numerous non-snRNP proteins.
Acknowledgments
We thank A. Moopen for assistance on analysis of the AdML-MS2 pre-mRNAs and Dr. N. Dorman for critical comments on the manuscript. This work was supported by National Institutes of Health Grants GM31819 (to J.G.) and GM43375 (to R.R.).
Abbreviations
EM, electron microscopy
MBP, maltose-binding protein
snRNA, small nuclear RNA
snRNP, small nuclear ribonucleoprotein particle
References
- 1.Staley J. P. & Guthrie, C. (1998) Cell 92, 315-326. [DOI] [PubMed] [Google Scholar]
- 2.Burge C. B., Tuschl, T. H. & Sharp, P. A. (1999) in The RNA World, eds. Gesteland, R. F., Cech, T. R. & Atkins, J. F. (Cold Spring Harbor Lab. Press, Plainview, NY), pp. 525–560.
- 3.Hastings M. L. & Krainer, A. R. (2001) Curr. Opin. Cell Biol. 13, 302-309. [DOI] [PubMed] [Google Scholar]
- 4.Will C. L. & Luhrmann, R. (2001) Curr. Opin. Cell Biol. 13, 290-301. [DOI] [PubMed] [Google Scholar]
- 5.Stevens S. W., Ryan, D. E., Ge, H. Y., Moore, R. E., Young, M. K., Lee, T. D. & Abelson, J. (2002) Mol. Cell 9, 31-44. [DOI] [PubMed] [Google Scholar]
- 6.Zhou Z., Luo, M. J., Straesser, K., Katahira, J., Hurt, E. & Reed, R. (2000) Nature (London) 407, 401-405. [DOI] [PubMed] [Google Scholar]
- 7.Nilsen T. W. (2002) Mol. Cell 9, 8-9. [DOI] [PubMed] [Google Scholar]
- 8.Kastner B., Kornstadt, U., Bach, M. & Luhrmann, R. (1992) J. Cell Biol. 116, 839-849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stark H., Dube, P., Luhrmann, R. & Kastner, B. (2001) Nature (London) 409, 539-542. [DOI] [PubMed] [Google Scholar]
- 10.Kastner B., Bach, M. & Luhrmann, R. (1990) Proc. Natl. Acad. Sci. USA 87, 1710-1714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Behrens S. E., Tyc, K., Kastner, B., Reichelt, J. & Luhrmann, R. (1993) Mol. Cell. Biol. 13, 307-319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fabrizio P., Esser, S., Kastner, B. & Luhrmann, R. (1994) Science 264, 261-265. [DOI] [PubMed] [Google Scholar]
- 13.Jurica M. S., Licklider, L. J., Gygi, S. R., Grigorieff, N. & Moore, M. J. (2002) RNA 8, 426-439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Reed R., Griffith, J. & Maniatis, T. (1988) Cell 53, 949-961. [DOI] [PubMed] [Google Scholar]
- 15.Bennett M., Michaud, S., Kingston, J. & Reed, R. (1992) Genes Dev. 6, 1986-2000. [DOI] [PubMed] [Google Scholar]
- 16.Reed R. (1990) Proc. Natl. Acad. Sci. USA 87, 8031-8035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Das R., Zhou, Z. & Reed, R. (2000) Mol. Cell 5, 779-787. [DOI] [PubMed] [Google Scholar]
- 18.Carey J., Cameron, V., de Haseth, P. L. & Uhlenbeck, O. C. (1983) Biochemistry 22, 2601-2610. [DOI] [PubMed] [Google Scholar]
- 19.Witherell G. W., Wu, H. N. & Uhlenbeck, O. C. (1990) Biochemistry 29, 11051-11057. [DOI] [PubMed] [Google Scholar]
- 20.Bardwell V. J. & Wickens, M. (1990) Nucleic Acids Res. 18, 6587-6594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Graveley B. R. & Maniatis, T. (1998) Mol. Cell 1, 765-771. [DOI] [PubMed] [Google Scholar]
- 22.LeCuyer K. A., Behlen, L. S. & Uhlenbeck, O. C. (1995) Biochemistry 34, 10600-10606. [DOI] [PubMed] [Google Scholar]
- 23.Zhou, Z., Licklider, L. J., Gygi, S. P. & Reed, R. (2002) Nature (London), in press. [DOI] [PubMed]
- 24.Achsel T., Ahrens, K., Brahms, H., Teigelkamp, S. & Luhrmann, R. (1998) Mol. Cell. Biol. 18, 6756-6766. [DOI] [PMC free article] [PubMed] [Google Scholar]