Abstract
Epidermal growth factor receptor (EGFr) is a key mediator of cell communication during animal development and homeostasis. In Drosophila, the signaling event is commonly regulated by the polytopic membrane protein Rhomboid (RHO), which mediates the proteolytic activation of EGFr ligands, allowing the secretion of the active signal. Until very recently, the biochemical function of RHO had remained elusive. It is now believed that Drosophila RHO is the founder member of a previously undescribed family of serine proteases, and that it could be directly responsible for the unusual, intramembranous cleavage of EGFr ligands. Here we show that the function of RHO is conserved in Gram-negative bacteria. AarA, a Providencia stuartii RHO-related protein, is active in Drosophila on the fly EGFr ligands. Vice versa, Drosophila RHO-1 can effectively rescue the bacterium's ability to produce or release the signal that activates density-dependent gene regulation (or quorum sensing). This study provides the first evidence that prokaryotic and eukaryotic RHOs could have a conserved role in cell communication and that their biochemical properties could be more similar than previously anticipated.
Cell communication is central to the development and physiology of multicellular organisms. Metazoan signaling molecules can be roughly grouped into two classes: freely diffusible, membrane permeable signals, of which steroid hormones are a prime example, and molecules that need to be intercepted by specific receptors at the cell surface, such as growth factors. Signal release is tightly regulated; for instance, growth factors are often synthesized as inactive polypeptide precursors. Posttranslational modifications such as proteolytic cleavage and/or glycosylation convert the precursor into the active receptor ligand (1). Prokaryotes seem to have adopted similar solutions to accommodate the need for intercellular communication. In the bacterial world, Gram-negative species use small diffusible molecules, typically modified amino acids, to regulate a variety of responses in a density dependent manner. Gram-positive bacteria seem to favor oligopeptidic signals, synthesized as inactive precursors that are then proteolytically activated and secreted, to be recognized by specific receptors at the cell surface (reviewed in ref. 2).
The epidermal growth factor (EGF) family comprises small proteins widely used as signals by animal cells (3). EGF-receptor (EGFr) signaling is required for cell fate specification, growth, and survival at multiple steps of animal development. In addition, activation of EGFr is often associated with tumorigenesis in humans and mice (4). In vertebrates, multiple genes encode different EGF receptors and ligands, whereas the genome of invertebrates seem to encode a single receptor and a variable number of ligands (reviewed in ref. 5). EGFr signaling has been extensively studied in the model organism Drosophila melanogaster, where it is required, for example, in determining the correct organization of the fly compound eye and the number and positioning of veins in the insect wing (6). Four proteins have so far been indicated as EGFr ligands in Drosophila, but the number is likely to increase as the existing ligands are not sufficient to account for all of the aspects of EGFr signaling so far described (7, 8). The spitz (spi) and gurken (grk) genes encode transforming growth factor α-related EGFr ligands, both initially produced as inactive transmembrane precursors. The neuregulin homologue VEIN, and Argos, the only known antagonistic EGFr ligand, are instead likely to be directly secreted (reviewed in ref. 9). The Drosophila polytopic transmembrane protein Rhomboid (RHO) is often the time-setter for activation of EGFr signaling in flies. It is known that RHO is required in the signal-emitting cell, rather than signal-receiving cell, and that it functions by activating the membrane-tethered precursor of EGFr ligands (10). Despite this, it proved difficult to resolve which biochemical mechanism lay behind the RHO-mediated signal activation, also because of the lack of homologues with a defined function in other organisms. It was recently shown that RHO has indeed the hallmarks of a protease and that it directly catalyses the cleavage of the fly EGFr ligand Spitz (11). Regulated intramembrane proteolysis (also termed Rip) was originally described as a conserved mechanism of releasing membrane-tethered transcription factors (12). Hence, the discovery that RHO acts as an intramembrane serine protease to release the extracellular portion of SPI, was described as the first example of Rip-mediated growth factor release (11, 12). A total of seven RHO related (RHOr) sequences is encoded in the Drosophila genome (13, 14), but it is at present not clear whether they all function as EGF signal activators or are devoted to different cellular functions. RHO-3, uncovered by the classical mutation roughoid, was shown to cooperate with RHO (now RHO-1) in activating EGFr during eye development (14). More recently RHO-2, also known as Brother-of-Rhomboid (BRHO), was reported to activate GRK during oogenesis (13, 15). Hence, the evidence available at this point seems to suggest that multiple RHOs are required in different tissues and/or for the activation of different EGFr ligands.
Besides animals, RHOr sequences are also found in diverse organisms among archaea, eubacteria, yeasts, and plants (13, 14, 16). The overall sequence similarity is generally low, but they all share the structure of a polytopic membrane protein as well as a conserved set of amino acids. Interestingly, the only RHOr function that has been characterized outside the animal kingdom has revealed its involvement in a cell communication mechanism. Providencia stuartii is a Gram-negative bacterium responsible for nosocomial and opportunistic infections in humans. During bacterial growth, the accumulation of an extracellular factor regulates a number of loci in a density-dependent manner (17). aarA mutant P. stuartii are defective in the biosynthesis or export of the activating signal, whose biochemical properties are consistent with those of a small peptide (17). In a previous publication, we pointed to the hitherto overlooked functional similarity, in genetic terms, between the rhomboid genes of Drosophila and the aarA gene of P. stuartii (16). In both systems, a polytopic membrane protein with a conserved amino acidic signature, the RHO domain, is central to the release of an extracellular signal. Here we present evidence that AarA could indeed substitute for the fly RHO in a number of in vivo assays, activating the fly EGFr ligands. Conversely, a fly RHO could rescue the cell communication phenotype associated with aarA deletion, namely the production of an extracellular signal that activated density dependent gene regulation during bacterial growth. Our results, coupled with the wide distribution of RHOr sequences, indicate that RHO signaling could represent a key step in a widely conserved mechanism for cell communication.
Methods
Sequence Analysis and Phylogenetic Reconstruction.
Protein sequences were obtained from the SWALL (SPRT) database (accessible as SWALL on the European Bioinformatic Institute SRS server) or from GenBank at the National Center for Biotechnology Information (NCBI). For the accession numbers of individual entries included in our analysis and a link to the database, see Table 1, which is published as supporting information on the PNAS web site, www.pnas.org. Sequence alignment was performed by using clustalw within the clustalx platform (NCBI). The phylogenetic relationship among the retrieved sequences was evaluated by using the neighbor joining method. Neighbor joining trees were then tested by bootstrap analysis with 1,000 replicates.
Fly Strains and Genetics.
A genomic ClaI fragment containing the P. stuartii aarA gene was cloned into a pUAST vector, under the control of the yeast upstream activation sequence (UAS) promoter (18). Five independent transgenic lines were produced. Mutant strains and Gal-4-expressing lines were from stock centers and include UAS-EGFrDN, UAS-S, and Gal-4 10968 and GMR-Gal4 expressed in developing wing and eyes, respectively. The ru/rho3pLLb allele was obtained from M. Freeman, and the UAS-mSPI and UAS-mGRK strains were from E. Bier's lab. Anti-activated mitogen-activated protein kinase (MAPK) antibody was from Sigma and was used to stain imaginal tissues as in ref. 13.
Plasmid Constructions.
To construct a plasmid with the RHO-1 cDNA under control of the lacZ promoter a 2.5-kb EcoRI fragment was excised from pBluescript SK+rho1 and cloned into the EcoRI site of pBCSK. Colonies containing RHO-1, inserted such that transcription would initiate from the lac promoter, were selected and a representative plasmid was designated pBCSK+Rho-1. Other plasmids used include the empty vector, designated pBCSK, pAARA (17).
Preparation of Conditioned Medium.
P. stuartii strains were inoculated at low density in 30 ml of LB broth containing chloramphenicol (50 μg/ml) and shaken at 37°C in 250-ml flasks at 20 × g. At an optical density of A600 = 1.3, cells were pelleted at 3,000 × g for 10 min. The resulting supernatants were adjusted to pH 7.5 and filter sterilized by using a 0.22-μm filter attached to a disposable syringe. The first 5 ml of supernatant was discarded to wash any contaminants off the filter. These preparations were frozen at −80°C before use.
β-Galactosidase Assays.
Strain XD37/pACYC184 was used as a biosensor for extracellular signals (17). Cultures (3 ml) of conditioned medium or control LB were inoculated to an OD600 of ≈0.05 with a freshly concentrated suspension of XD37/pACYC184 cells that were at early-log phase. Tubes were shaken at 20 × g at 37°C and cells were harvested at early-log phase A600 = 0.25. This typically involved 3–4 h of growth. Cells were assayed for β-galactosidase by the method of Miller (19) using SDS/chloroform-permeabilized cells. All reported values represent the mean of quadruplicate samples from two independent experiments. The standard deviations were less than 12% of the mean in all cases.
Results
The Prokaryotic RHOr Genes.
It has been previously reported that sequences related to Drosophila RHO are encoded in the genome of many prokaryotes (13, 14, 16). To gain insight into the distribution of RHOr sequences in the microbial world, we searched nonredundant databases for proteins matching the features of a RHO domain (PF01694 or IPR002610). In addition, we searched all publicly available sequence databases with eukaryotic and prokaryotic RHO sequences. The SWALL database (as of the February 16, 2002, update) proved to contain a representative set of high-quality sequences, which was selected as a starting point for the analysis presented below. In total, 50 microbial RHOr sequences were found in the database: 9 sequences deriving from 8 archaeal species (the genome of Pyrobacillum aerophilum encoding 2 RHOr), 18 sequences from Gram-positive bacteria, and 23 sequences from Gram-negative bacteria. Within bacteria, a few genomes seemed to encode more than a single RHOr sequence; in the gram-positives, 4 sequences belonged to Streptomyces coelicolor, and 2 each belonged to Bacillus subtilis, Bacillus halodurans, and Mycobacterium tuberculosis. In the gram-negatives, 3 sequences were found in the Xylella fastidiosa genome, and 2 each were found in Thermotoga maritima, Sinorhizobium meliloti, and Mesorhizobium loti. RHOr sequences are therefore widespread, and their presence is not apparently associated with any particular microbial function or lifestyle. Interestingly, RHOr sequences are present in both primitive (Aquifex aeolicus, Thermotoga maritima) and derived (Gram-positive) species. It should be noted that the above list is representative but not exhaustive, as additional sequences that were exclusively found by direct homology searches in other databases are not included in it. In contrast, we were unable to find a consistent RHO signature in the complete genome sequence of several microbial species. For instance, we could find no RHO homologues in the fully completed genome of a number of human pathogens, including Borrelia and various Mycoplasma as well as some obligate intracellular bacteria, such as Chlamydia, Rickettsia, and Buchnera, perhaps reflecting gene loss that accompanied the adaptation to life within the animal host.
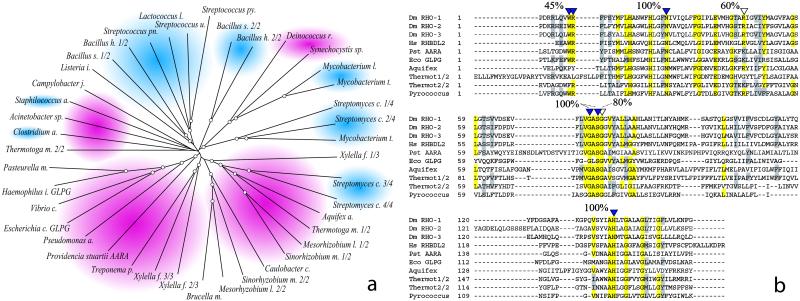
To further study the distribution and phylogeny of the bacterial RHOr, a phylogenetic tree was assembled from an alignment of the above-mentioned 41 bacterial sequences (Fig. 1a). The low level of similarity that characterizes the RHO family is apparent from the length of the tree branches and does not allow for detailed phylogenetic inferences. Nevertheless, the Gram-positive and Gram-negative sequences grouped separately with but a few exceptions, perhaps representing horizontal gene transfer events. Within each group, at least two subclasses of sequences were resolved, so that most bacterial species that possess more than a single RHOr gene had a representative in each of the subclasses, indicating functional divergence or an ancient event of gene duplication. The P. stuartii AarA protein appeared loosely related to the GlpG group of homologues. A very similar tree was obtained aligning exclusively the RHO domain of the above sequences (data not shown), consistent with the notion that the RHO domain is likely to represent the core function of the protein.
Fig 1.
RHOr sequences are widespread among bacteria. (a) Phylogenetic tree derived from the alignment of 41 bacterial RHOr. Each sequence is represented by the name of the bacterial genus from which it derives. To facilitate distinction of different species of the same genus, the first letter of the species name is also reported. Gram-negative groups are shaded in red, Gram-positive groups are shaded in blue. Tree nodes with a bootstrap support greater than 50% are encircled. With a few exceptions, Gram-negative and positive sequences group separately. In addition, within each group at least two subfamilies seem to resolve indicating perhaps an ancient event of gene duplication. (b) The RHO-domain of all of the proteins that have been shown able to activate SPI is aligned to the one of AarA and of a few representative prokaryotic sequences. Residues that are essential for RHO-1 function are indicated by a full arrowhead. Residues that, when mutated, decrease RHO-1 activity without abolishing it, are designated by empty arrowheads. A total of 41 bacterial RHOr where aligned as above, and the conservation of catalytic residues determined. The percent value reported above the alignment represents the approximate level of conservation of the key residues, a positive score was assigned if residues where found absolutely conserved (in yellow) or very similar (in gray). In AarA and GlpG all of the catalytic residues are absolutely conserved.
Finally, the RHO domain of the bacterial RHOr was directly aligned and compared with Drosophila and human RHO domains to asses the conservation of the proposed serine-protease catalytic residues (Fig. 1b shows a representative set of sequences). Most bacterial proteins showed a striking degree of conservation, with almost all of the essential residues absolutely conserved or highly similar. For instance, in AarA and GlpG, all of the key residues were absolutely conserved or highly similar (Fig. 1b).
In about half of the sequences although, a WR motif (Tryptophan-Arginine, in the single letter amino acid code) that was shown essential for the fly RHO-1 catalysis (11) was not conserved. It is not clear whether this change represents an effective change in the catalytic proprieties of the protein or a different adaptation.
P. stuartii AarA Can Activate EGFr Signaling in Drosophila.
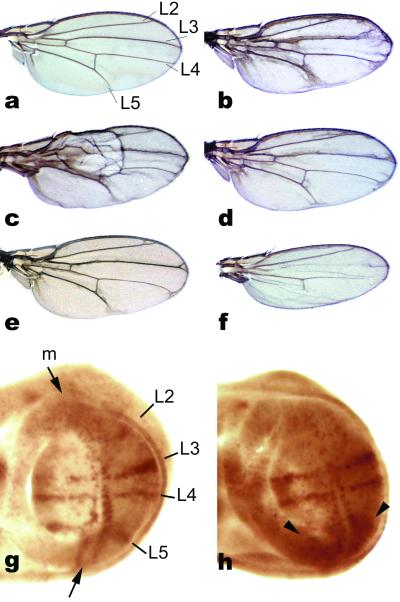
To determine whether the structural and functional similarities between Drosophila and bacterial RHOs implicated similar biochemical proprieties, we set out to determine their activities in the reciprocal host. P. stuartii AarA was chosen as the only bacterial RHOr experimentally characterized, as well as for its involvement in cell communication (see Introduction). We produced transgenic flies expressing aarA by using the Gal4/UAS system for ectopic gene expression (see Methods and ref. 18). Expression of aarA in Drosophila under the control of a wing-specific promoter resulted in phenotypes that were indistinguishable from overexpression of endogenous RHOs (Fig. 2). EGFr signaling is required in wing development to promote wing vein formation (20). Overexpression of any of the three Drosophila RHOs characterized to date results in the formation of variable amounts of ectopic vein tissue, vein thickening, and in an overall dark and blistered appearance of the wing (13, 14, 20). Wings of flies expressing P. stuartii aarA were most similar to those deriving from mild overexpression of RHO-1 from a heat-inducible promoter (Fig. 2c and ref. 20), or from the ectopic expression of bRHO/RHO-2 (13), showing all of the hallmarks of EGFr ectopic activation. The AarA-induced ectopic vein phenotype was consistently reduced by halving the dose of EGFr normally present in the wing (in a heterozygous EGFr mutant background, Fig. 2e). Furthermore, as previously reported for the fly RHOs, coexpression of a dominant-negative EGFr construct (21) was sufficient to entirely suppress the aarA induced wing phenotype, resulting in narrow wings with missing veins typical of the dominant-negative construct (Fig. 2f). Taken together, these results suggest that the phenotype induced by expression of aarA is mediated by the fly EGFr. In addition, EGFr activation was confirmed by increased levels of activated MAPK, the main signal transducer used by EGFr, detected in developing wings by a specific antibody (Fig. 2h).
Fig 2.
Providencia stuartii AarA can activate EGFr signaling in developing wings. (a) Wild-type wing, the names of the major longitudinal veins are indicated to facilitate comparison with developing wings (g and h). (b) Wing-directed expression of AarA results in ectopic wing vein tissue formation and wing blistering. (c) This phenotype is very similar, for example, to mild RHO-1 overexpression achieved by an heat-shock promoter. (d) Halving the dosage of SPI, an endogenous EGFr ligand normally expressed in the wing, consistently suppressed the AarA induced phenotype (compare with b). (e and f) In addition, the AarA-induced phenotype is (e) partially suppressed in an heterozygote EGFr mutant background, and (f) completely suppressed by coexpression of a dominant-negative EGFr construct (resulting in wings indistinguishable to DN-EGFr expressing ones). (g and h), Activation of EGFr signaling is also confirmed by activated MAPK staining. (g) In wild-type developing wings, activated MAPK is restricted to vein-precursor territories. (h) After AarA expression, ectopic signal is typically detected in the regions most affected in the adult wing (arrowheads).
AarA Expression Functions Through the Activation of the Fly EGFr Ligands.
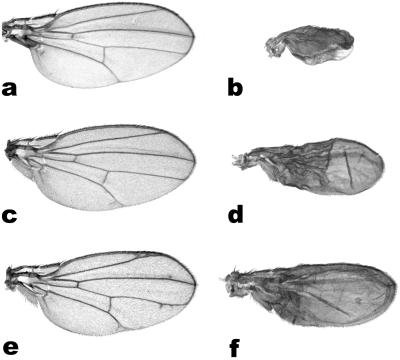
As discussed, RHO is needed to process membrane bound, inactive EGFr ligand precursors into secreted, active forms. As expected, overexpression of full-length SPI or GRK, the two known EGFr ligands requiring RHO mediated activation, is not sufficient to induce a visible wing phenotype (Fig. 3 and ref. 13). However, coexpression of endogenous RHOs with full-length SPI or GRK results in strong wing phenotypes (13). Likewise, the aarA-induced ectopic vein phenotype was strongly enhanced by coexpression of SPI or GRK, the former showing the strongest synergy (Fig. 3 b and d). Therefore, AarA seems capable of activating the fly EGFr ligands in a manner similar to the endogenous RHOs. Moreover, halving the dose of active SPI normally present in the wing (in a heterozygous spi mutant background) consistently reduced the AarA-mediated ectopic vein phenotype (Fig. 2d and compare with b), indicating that aarA expression functions, at least in part, through the activation of SPI. Next, the aarA induced phenotype was tested for modulation by Star (S). Star is a single pass transmembrane protein recently shown to chaperone SPI out of the endoplasmic reticulum and into the Golgi apparatus, where SPI becomes exposed to the activity of RHO (22). As reported for the fly RHOs, coexpression of Star with aarA in Drosophila wings had a strong synergistic effect (Fig. 3f), wing-specific expression of S alone showing an extremely mild ectopic vein phenotype (Fig. 3e and ref. 8). Conversely, halving the dose of S had no detectable effect on the aarA induced phenotype, perhaps an indication that the bacterial RHO, when expressed in flies, might not be confined to the Golgi apparatus.
Fig 3.
P. stuartii AarA can activate endogenous EGFr ligands in Drosophila. (a–c) Wing-directed expression of full length SPI (a) or GRK (c) has no effect on wing development. In contrast, coexpression of AarA with SPI (b) or GRK (d) shows a strong synergistic effect, transforming most of the wing into vein-like tissue. (e and f) A similar interaction is observed with Star. (e) Wing-directed expression of Star produces a very weak extra-vein phenotype. (f) Coexpression of Star with AarA is strongly synergistic. Note that all wings are shown at the same scale.
AarA Can Activate EGFr in Different Developmental Contexts.
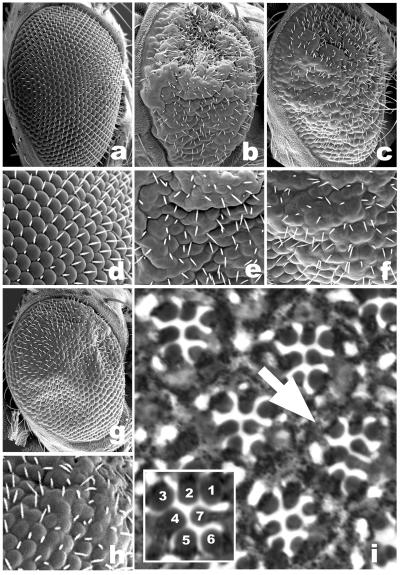
EGFr signaling is central to many aspects of Drosophila development. One of the best understood examples is the development of the fly compound eye, where EGFr signaling is required for the recruitment of all of the cell types that constitute each unit eye or ommatidium (6, 23). Eye-directed expression of aarA was not sufficient to induce any visible eye phenotype, possibly because of low expression levels. Similarly, as reported for the wing, eye-directed expression of full-length SPI or GRK had no detectable effect on eye morphology (ref. 24, and data not shown). In contrast, coexpression of full-length SPI or GRK with aarA had a striking effect on eye morphology, resulting in malformed and fused ommatidia that closely resemble RHO-1 overexpressing ones (Fig. 4). As expected, the AarA-induced eye phenotype was correlated with an over-recruitment of photoreceptor neurons (Fig. 4i) and accessory cells (data not shown), consistent with an over-activation of EGFr signaling during eye development. AarA is therefore able to activate EGFr signaling in at least two different developmental contexts in the fly.
Fig 4.
AarA can activate EGFr signaling in developing eyes. (a–c and g) Adult compound eyes. (d–f and h) Detail. (a and d) Eye-directed expression of full-length SPI or GRK has no effect on developing eyes. In contrast, coexpression of AarA with SPI (c and f) or GRK (g and h) results in highly compromised compound eyes with malformed and fused ommatidia, closely resembling RHO-1 overexpressing ones (b and e). (i) Tangential sections through adult eyes reveal recruitment of extra photoreceptor neurons (arrow, a m-GRK/AarA eye is shown). (Inset) The wild-type pattern of seven photoreceptor cells normally visible in apical sections.
AarA Can Directly Substitue for Drosophila RHOs.
To strengthen the functional similarities between AarA and RHO, we tested the ability of AarA to rescue phenotypes derived from available mutations in different fly rhomboids. Wing-directed expression of P. stuartii AarA could rescue the loss of vein phenotype typical of veinlet alleles (Fig. 5), which specifically reduce RHO-1 activity in the wing (20). Similarly, eye-directed expression of AarA was sufficient to rescue in part the eye phenotype deriving from deletion of roughoid/RHO-3 (Fig. 5), the second fly RHO to be described, which cooperates with RHO-1 in activating EGFr signaling during eye development (14). The rescue of wing venation defects of rho-1veinlet alleles was much more pronounced than the one of the eye phenotype typical of rho-3roughoid ones. It is not clear whether this difference is to be ascribed to lower aarA expression levels achieved by the eye-specific promoter, or to some intrinsic difference between the two systems.
Fig 5.
Expression of AarA in Drosophila can directly rescue phenotypes associated with viable mutations in different fly rhomboids. (a) rho-1veinlet alleles are characterized by extensive loss of wing venation. (c) Wing-directed expression of AarA in the mutant completely rescues this phenotype (the mutant wings shown also carry the genetic marker hairy). (b) rho-3roughoid deletion results in malformed eyes with collapsed and blistered ommatidia. (d) Expression of AarA in the mutant partially rescues the retinal patterning defects.
Drosophila RHO-1 Can Substitute for AarA in P. stuartii.
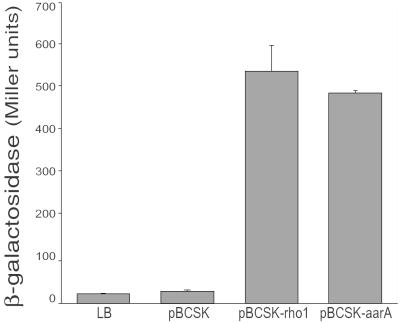
To test whether the Drosophila RHO could in turn substitute for P. stuartii AarA, we expressed the fly RHO-1 in the bacterium. Strikingly, expression of the fly RHO-1 was sufficient to rescue the cell communication phenotype associated with aarA mutations. As mentioned, aarA mutant P. stuartii are severely delayed in the production or release of a diffusible factor that regulates several loci in a density dependent manner (17). As a biological readout for aarA activity, we used a reporter gene fused to an extracellular factor-sensitive promoter. One such sensor, cma37 (for conditioned medium activated), shows more than a 20-fold activation when incubated in medium conditioned by wild-type P. stuartii as compared with unconditioned medium, or to medium derived from aarA mutant P. stuartii (Fig. 6 and ref. 17). Expression of the fly rho-1 in the mutant strain restored its ability to produce the extracellular activating signal, resulting in levels of cma37 activation comparable to those achieved by aarA expression in the mutant (Fig. 6). Importantly, the fly rho was confirmed to function in the signal-emitting cell because the experiments described above were performed by using filtered growth medium. In agreement with previous results (25), the signal produced by rho-1 expressing aarA mutant bacteria was sensitive to the potent protease pronase (1 mg/ml, for 2 h), confirming its peptidic nature (data not shown).
Fig 6.
Drosophila RHO-1 can effectively rescue the cell communication phenotype associated with aarA deletion in P. stuartii. Namely, RHO-1 expression rescues the mutant's ability to produce an extracellular activating signal. Conditioned medium from an aarA deletion strain (XD37.A) containing an empty vector activates sensor expression to a level comparable to LB medium alone (27 ± 2 and 23 ± 1 units, respectively). In contrast, medium conditioned by XD37.A/pBCSK+Rho-1 activated the sensor to a level 24-fold above medium alone and 20-fold above that seen for XD37.A/pBCSK (541 ± 59 units). Medium conditioned by XD37.A/pAARA activated the sensor to similar levels (490 ± 6 units).
Discussion
We report a remarkable level of functional conservation for a signal-releasing mechanism that relies on Regulated intramembranous proteolysis (26) in bacteria and eukaryotes. A bacterial RHO-related protein, encoded by the P. stuartii gene aarA, was shown to substitute for the well known Drosophila EGF activator RHO, acting on endogenous fly targets. Conversely, the fly RHO-1 restored the ability of aarA mutant bacteria to activate the signal that mediates quorum sensing in the microbe. Our results have important implications for the biology of both prokaryotic and eukaryotic RHOs.
AarA Is an Intramembrane-Cleaving Protease (I-CliP).
Regulated intramembrane proteolysis was originally described as a means to release transcription factors from the cytoplasmic side of the cell membrane. The discovery that the fly RHO acts as a protease, freeing the luminal portion of SPI (11), prompted to expand the definition of Rip to include the release of any functional protein domain from its transmembrane anchor (12). Enzymes that catalyze this unusual reaction are termed I-CliPs, and belong to different families (27). In eukaryotes, the best known examples of I-CliPs are presenilins, which are aspartyl-proteases, and S2P (or site-2 protease), a Zn2+ metalloprotease. RHO was described as the first intramembrane-cleaving serine protease (11). I-CliPs belonging to the same families are also present in prokaryotes, but the functional relationship to their eukaryotic counterparts is not known. Examples include TFPP (or type four prepilin peptidase) (28), a membrane-embedded aspartyl protease, and SpoIVFB, a metalloprotease related to S2P (26). We have shown in a number of in vivo assays that P. stuartii AarA, a protein related to the fly RHO, could substitute for RHO in activating endogenous EGFr ligands in the fly. In turn, the glpG gene from Escherichia coli could substitute for AarA in P. stuartii (P.R., unpublished data). This finding, coupled with the striking conservation of overall structure and catalytic residues within the prokaryotic rhor genes, suggests that most, if not all rhor could encode intramembrane serine proteases. Rip has been previously reported in bacteria (26), but AarA is the first bacterial example of an intramembrane serine protease.
Peptide Signaling in Gram-Negative Bacteria.
As discussed, peptide-mediated signaling in bacteria has long been considered unique to Gram-positives (2). Several lines of evidence now suggest that it could be more widespread than anticipated, perhaps representing an ancient means of cell communication. We have shown that Drosophila RHO-1 could restore the ability of aarA mutant bacteria to release the signal that activates quorum sensing in P. stuartii. The signal has been previously reported to have properties consistent with those of a small peptide, most notably it was sensitive to a number of peptidases (25). In addition, peptide-receptor like genes have long been known to be present in the genome of Gram-negative bacteria (29). Taken together, these results suggest that AarA works in P. stuartii to activate a signal peptide by regulated intramembrane proteolysis. This represents a clear suggestion that Gram-negative bacteria, like the Gram-positive, also sense population density by using peptide signals. It is worth noticing that also plants have recently been found to possess peptide mediated signaling (30). The use of peptides as signaling molecules is therefore likely to be universal.
A Widely Conserved Mechanism for Signal Release.
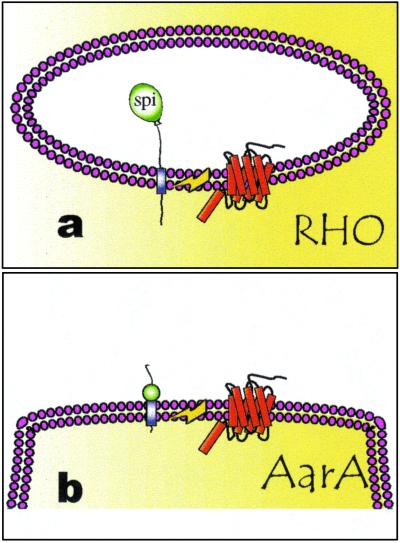
A single example of Rip-mediated extracellular signal release has been reported outside of Drosophila. In the Gram-positive bacterium Enterococcus faecalis, an 8-aa peptide pheromone called cAD1 is derived from a 143-aa transmembrane precursor (31). The processing is step-wise, after the removal of a signal peptide, an I-CliP, a protein termed Eep, releases the mature pheromone (32). Remarkably, Eep is recognizably similar to the human site-2 protease, the first I-CliP to be described (26). Sequences related to Eep are also found in B. subtilis and in the Gram-negative E. coli, but their function is unknown (26). RHBL2, a human RHO homologue, behaves as the fly RHO in a SPI cleaving assay (11). This finding indicates that receptor ligands, which are activated in a corresponding manner, are likely to be found in humans (11). Our evidence suggests a similar model for signal release in the Gram-negative bacterium P. stuartii (Fig. 7). By analogy with Eep and RHO, we suggest that AarA works as an intramembranous serine protease to release a signaling peptide from an inactive transmembrane precursor. Rip seems therefore to emerge as a general control mechanism for peptide-signal release. The case of RHO/AarA provides an additional interesting insight in the evolution of peptide-mediated signaling: not only the signal releasing mechanism seems conserved, but also the molecule that carries it out. The involvement of the fly RHOs and AarA in a similar process of cell communication suggests the intriguing possibility that these molecules could share a common ancestry. If this is the case, we anticipate that more signaling molecules will be discovered which are released by RHO-mediated regulated intramembrane proteolysis. Importantly, this does not imply that the signals themselves will be conserved, as the evolutionary constrains acting on an extracellular signal are likely to be very different from the ones acting on the signal-releasing protease. The clarification of RHO signaling in species as distant as Drosophila and Providencia might unveil a very ancient language for cell communication.
Fig 7.
Mechanism of RHO and AarA action. (a) RHO acts in the fly as an I-CliP to release the luminal portion of SPI. (b) In analogy, and in a similar manner to the release of the cAD1 in E. faecalis, we propose that AarA works as an I-CliP to release a peptide pheromone in P. stuartii.
Supplementary Material
Acknowledgments
M.G. is grateful to Carlos Ribeiro, Annabel Guichard, and Sinisa Urban for fly strains and materials. P.R. was supported by National Science Foundation Grant MCB9904766.
Abbreviations
Rip, regulated intramembrane proteolysis
I-CliP, intramembrane-cleaving protease
EGF, epidermal growth factor
EGFr, EGF receptor
RHO, rhomboid
RHOr, RHO related
MAPK, mitogen activated protein kinase
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Blobel C. P. (2000) Curr. Opin. Cell Biol. 12, 606-612. [DOI] [PubMed] [Google Scholar]
- 2.Schauder S. & Bassler, B. L. (2001) Genes Dev. 15, 1468-1480. [DOI] [PubMed] [Google Scholar]
- 3.Moghal N. & Sternberg, P. W. (1999) Curr. Opin. Cell Biol. 11, 190-196. [DOI] [PubMed] [Google Scholar]
- 4.Casci T. & Freeman, M. (1999) Cancer Metastasis Rev. 18, 181-201. [DOI] [PubMed] [Google Scholar]
- 5.Stein R. A. & Staros, J. V. (2000) J. Mol. Evol. 50, 397-412. [DOI] [PubMed] [Google Scholar]
- 6.Schweitzer R. & Shilo, B. Z. (1997) Trends Genet. 13, 191-196. [DOI] [PubMed] [Google Scholar]
- 7.Dominguez M., Wasserman, J. D. & Freeman, M. (1998) Curr. Biol. 8, 1039-1048. [DOI] [PubMed] [Google Scholar]
- 8.Guichard A., Biehs, B., Sturtevant, M. A., Wickline, L., Chacko, J., Howard, K. & Bier, E. (1999) Development (Cambridge, U.K.) 126, 2663-2676. [DOI] [PubMed] [Google Scholar]
- 9.Bogdan S. & Klambt, C. (2001) Curr. Biol. 11, R292-R295. [DOI] [PubMed] [Google Scholar]
- 10.Klambt C. (2000) Curr. Biol. 10, R388-R391. [DOI] [PubMed] [Google Scholar]
- 11.Urban S., Lee, J. R. & Freeman, M. (2001) Cell 107, 173-182. [DOI] [PubMed] [Google Scholar]
- 12.Huppert S. & Kopan, R. (2001) Dev. Cell 1, 590-592. [DOI] [PubMed] [Google Scholar]
- 13.Guichard A., Roark, M., Ronshaugen, M. & Bier, E. (2000) Dev. Biol. 226, 255-266. [DOI] [PubMed] [Google Scholar]
- 14.Wasserman J. D., Urban, S. & Freeman, M. (2000) Genes Dev. 14, 1651-1663. [PMC free article] [PubMed] [Google Scholar]
- 15.Ghiglione C., Bach, E. A., Paraiso, Y., Carraway, K. L., III, Noselli, S. & Perrimon, N. (2002) Development (Cambridge, U.K.) 129, 175-186. [DOI] [PubMed] [Google Scholar]
- 16.Gallio M. & Kylsten, P. (2000) Curr. Biol. 10, R693-R694. [DOI] [PubMed] [Google Scholar]
- 17.Rather P. N., Ding, X., Baca-DeLancey, R. R. & Siddiqui, S. (1999) J. Bacteriol. 181, 7185-7191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brand A. H. & Perrimon, N. (1993) Development (Cambridge, U.K.) 118, 401-415. [DOI] [PubMed] [Google Scholar]
- 19.Miller J. H., (1972) Experiments in Molecular Genetics (Cold Spring Harbor Lab. Press, Plainview, NY).
- 20.Sturtevant M. A., Roark, M. & Bier, E. (1993) Genes Dev. 7, 961-973. [DOI] [PubMed] [Google Scholar]
- 21.Buff E., Carmena, A., Gisselbrecht, S., Jimenez, F. & Michelson, A. M. (1998) Development (Cambridge, U.K.) 125, 2075-2086. [DOI] [PubMed] [Google Scholar]
- 22.Lee J. R., Urban, S., Garvey, C. F. & Freeman, M. (2001) Cell 107, 161-171. [DOI] [PubMed] [Google Scholar]
- 23.Freeman M. (1997) Development (Cambridge, U.K.) 124, 261-270. [DOI] [PubMed] [Google Scholar]
- 24.Hsiung F., Griffis, E. R., Pickup, A., Powers, M. A. & Moses, K. (2001) Mech. Dev. 107, 13-23. [DOI] [PubMed] [Google Scholar]
- 25.Rather P. N., Parojcic, M. M. & Paradise, M. R. (1997) Antimicrob. Agents Chemother. 41, 1749-1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Brown M. S., Ye, J., Rawson, R. B. & Goldstein, J. L. (2000) Cell 100, 391-398. [DOI] [PubMed] [Google Scholar]
- 27.Wolfe M. S., De Los Angeles, J., Miller, D. D., Xia, W. & Selkoe, D. J. (1999) Biochemistry 38, 11223-11230. [DOI] [PubMed] [Google Scholar]
- 28.LaPoinite C. F. & Taylor, R. K. (2000) J. Biol. Chem. 275, 1502-1510. [DOI] [PubMed] [Google Scholar]
- 29.Shapiro J. A. (1998) Annu. Rev. Microbiol. 52, 81-104. [DOI] [PubMed] [Google Scholar]
- 30.Lindsey K., Casson, S. & Chilley, P. (2002) Trends Plant Sci. 7, 78-83. [DOI] [PubMed] [Google Scholar]
- 31.Dunny G. M. & Leonard, B. A. (1997) Annu. Rev. Microbiol. 51, 527-564. [DOI] [PubMed] [Google Scholar]
- 32.An F. Y., Sulavik, M. C. & Clewell, D. B. (1999) J. Bacteriol. 181, 5915-5921. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.