Abstract
Intracellular iron homeostasis is regulated posttranscriptionally by iron regulatory proteins 1 and 2 (IRP1 and IRP2). In the absence of iron in the labile pool, IRPs bind to specific nucleotide sequences called iron responsive elements (IREs), which are located in the 5′ untranslated region of ferritin mRNA and the 3′ untranslated region of transferrin receptor mRNA. IRP binding to the IREs suppresses ferritin translation and stabilizes transferrin receptor mRNA, whereas the opposite scenario develops in iron-replete cells. Binding of IRPs to the IREs is also affected by nitrogen monoxide (NO), but there are conflicting reports regarding the effect of NO on ferritin synthesis. In this study, we demonstrated that a short exposure of RAW 264.7 cells (a macrophage cell line) to the NO+ donor, sodium nitroprusside (SNP), resulted in a dramatic increase in ferritin synthesis. The SNP-mediated increase of ferritin synthesis could be blocked by MG132, an inhibitor of proteasome-dependent protein degradation, which also prevented the degradation of IRP2 caused by SNP treatment. Moreover, treatment of RAW 264.7 cells with IFN-γ and lipopolysaccharide caused IRP2 degradation and stimulated ferritin synthesis, changes that could be prevented by specific inhibitors of inducible nitric oxide synthase. Furthermore, the SNP-mediated increase in ferritin synthesis was associated with a significant enhancement of iron incorporation into ferritin. These observations indicate that NO+-mediated modulation of IRP2 plays an important role in controlling ferritin synthesis and iron metabolism in murine macrophages.
Ferritin is a multimeric protein whose only clearly defined function is the sequestration and storage of excess cellular iron. It is composed of a protein shell that can accommodate up to 4,500 atoms of iron in its cavity (1, 2). The protein shell has a molecular mass between 430 and 460 kDa and is made up of 24 symmetrically arranged subunits of two types, a light subunit (L) and a heavy subunit (H) with sizes of 19 and 21 kDa, respectively (2). Both recombinant H- and L-homopolymers can incorporate iron, but H-ferritin is several times more efficient than L-ferritin. This difference seems to be caused by a ferroxidase center associated with the H-ferritin subunit that promotes the oxidation of ferrous iron (3). On the other hand, the L-subunit has a higher capacity than the H-subunit to induce iron-core nucleation (4, 5), suggesting that both ferritin chains cooperate in the overall uptake and storage of iron.
Ferritin synthesis is controlled by a mechanism that is part of a remarkable regulatory system that tightly controls iron levels in the “labile iron pool” (LIP), i.e., iron in transit among various intracellular compartments. Pivotal players in this regulation are iron regulatory proteins 1 and 2 (IRP1 and -2) that sense iron levels in the LIP. IRPs control the synthesis of ferritin and transferrin receptor (TfR) by binding to iron-responsive elements (IREs) that are located in the 5′ untranslated region (UTR) and the 3′ UTR of their respective mRNAs. When levels of iron in the LIP are low, IRPs bind to IREs and this association blocks the translation of ferritin mRNA and stabilizes TfR mRNA. On the other hand, expansion of the LIP inactivates the RNA- binding activity of IRP1 and leads to degradation of IRP2, resulting in efficient translation of ferritin mRNA and rapid degradation of TfR mRNA (reviewed in refs. 6–9).
Binding activity of IRPs is controlled not only by iron but also by other factors such as hydrogen peroxide, hypoxia, phosphorylation, and nitrogen monoxide (NO) (10–19). (NO is used here as a generic expression encompassing all NO species.) NO is an important physiological regulator that, in its reduced form (NO⋅), interacts with iron in the heme of guanylyl cyclase or in the [Fe-S] centers of nonheme iron proteins (20, 21). NO is well known to modulate the activity of mitochondrial aconitase (ref. 22; reviewed in ref. 23), an [Fe-S] protein that is homologous to IRP1 (6, 24). Several laboratories (13–17) reported that NO enhanced the IRE-binding activity of IRP1 in various cell types. Moreover, the NO-mediated increase in IRP1-binding activity was coincident with the translational repression of ferritin synthesis in fibroblasts (10), macrophages (13), and hepatoma cells (17). However, these results are in apparent conflict with many other studies showing that inflammation and inflammatory cytokines, which are known to induce NO production in macrophages (25), stimulate ferritin synthesis in these cells (refs. 26, 27; reviewed in ref. 28). These paradoxical results may possibly be explained by the fact that NO has markedly different biological effects depending on its redox state. In contrast to NO•, the oxidized form of NO, NO+ (nitrosonium ion), causes S-nitrosylation of thiol groups of numerous proteins (21, 29, 30). Recently, we demonstrated that sodium nitroprusside (SNP), a compound releasing NO+ with S-nitrosylating character, decreased IRP2 protein levels (18) in RAW 264.7 cells (a macrophage cell line), a response that was remarkably similar to the IRP2 decrease in IFN-γ/lipopolysaccharide (LPS)-treated cells (19). In this study, we investigated how the IFN-γ/LPS- or SNP-mediated decrease of IRP2 in RAW 264.7 cells affects ferritin synthesis.
Materials and Methods
Cells.
RAW 264.7 murine macrophages, a cell line established from a tumor induced by Abelson murine leukemia virus (31), were obtained from American Type Culture Collection. These cells have phagocytic activity (31) and can lyse erythrocytes (32). Cells were grown in 100-cm2 plastic culture dishes (from Life Technologies, Burlington, ON, Canada) in a humidified atmosphere of 95% air and 5% CO2 at 37°C in DMEM containing 10% FCS, extra l-glutamine (300 μg/ml), sodium pyruvate (110 μg/ml), penicillin (100 units/ml), and streptomycin (100 μg/ml).
Chemicals.
DMEM was obtained from Mediatech (Washington, DC); FCS, penicillin, streptomycin, and glutamine were from Life Technologies. BSA, SNP, ferric ammonium citrate, LPS, IFN-γ, and transferrin (Tf) were from Sigma; S-nitroso-N-acetylpenicillamine (SNAP) was from Precision Biochemicals (Vancouver, BC, Canada); N-[3-(aminomethyl)benzoyl]acetamide and l-N6-(1-iminoethyl)lysine [inducible nitrous-oxide synthase (iNOS)-specific inhibitors] were from Alexis Biochemicals (Vancouver, BC, Canada); MG132 and lactacystin (proteasome-specific inhibitors) were from Biomol (Plymouth Meeting, PA); and [35S]methionine and 59FeCl3 were from Mandel (Boston). The iron chelator salicylaldehyde isonicotinoyl hydrazone was synthesized as described (33).
Gel Retardation Assay.
A gel retardation assay was used to measure the interaction between IRPs and IREs by using established techniques (18, 19, 34).
Western Blot Analysis.
About 5 × 107 cells were lysed with extraction buffer (see above), and 60 μg of protein was resolved by using 6% SDS/PAGE. Protein was transferred to a nitrocellulose membrane, which was subsequently incubated with rabbit anti-IRP2 antibodies (a generous gift from E. A. Leibold, University of Utah). After a 1-h incubation, the membranes were washed and incubated with alkaline phosphatase-conjugated goat anti-rabbit IgG (Sigma) for 1 h. The protein was then visualized with an enhanced chemiluminescence Western blotting detection system (Bio-Rad) according to the manufacturer's manual.
Immunoprecipitation Analysis.
Cells (≈5 × 107) were incubated with [35S]methionine in methionine-deficient minimum essential medium for 1 h. The cells were washed three times with cold PBS, after which they were lysed with RIPA buffer (50 mM Tris⋅HCl/150 mM NaCl/1% Nonidet P-40/0.5% sodium deoxycholate/0.1% SDS), and the lysate was cleaned with protein A Sepharose (Amersham Pharmacia Biotech). Three micrograms of antiferritin antibody was added to the lysate and incubated for 3 h at 4°C, then 60 μl of protein A Sepharose was added to immunoprecipitate the immune complexes. The beads were washed three times with cold RIPA and then boiled with SDS loading dye. Immunoprecipitated protein was resolved by using 15% SDS/PAGE. The gel was dried and analyzed by fluorography.
Assay of Ferritin Levels.
RAW 264.7 cells were lysed with RIPA buffer, and the insoluble fraction was removed by centrifugation. Ferritin content was determined by immunoturbidimetric assay with the Tine quant kit (Roche Diagnostics), according to the manufacturer's recommendations, in a Hitachi 917 (Tokyo) turbidimeter. Data are means ± SD of triplicate determinations in a typical experiment from five experiments performed.
Iron Uptake by Cells and Its Incorporation into Ferritin.
RAW 264.7 cells were labeled for a period of 150 min at 37°C in a 5% CO2 atmosphere with 5 μM 59Fe2-Tf that was prepared as previously described (19). The incubation medium consisted of DMEM supplemented with 2% BSA. After labeling, cells were washed three times with cold PBS, and radioactivity was measured in a gamma counter [LKB Wallac (Gaithersburg, MD) 1282 Compugamma]. After counting, the cells were lysed with lysis buffer (50 mM Tris/300 mM NaCl/1% Triton X-100, pH 7.4), and an equal amount of protein in each sample was cleaned with protein A Sepharose (Amersham Pharmacia Biotech). Three micrograms of antiferritin antibody was added to the lysate and incubated for 3 h at 4°C, and then 60 μl of protein A Sepharose was added (1 h). The Sepharose beads were washed three times with cold lysis buffer and their radioactivities measured in a gamma counter to determine the amount of 59Fe incorporated into ferritin. Data shown are the means ± SD of triplicate determinations in a typical experiment that was performed five times.
Results
NO+ Mimics the Effect of IFN-γ/LPS on Ferritin Synthesis in RAW 264.7 Cells.
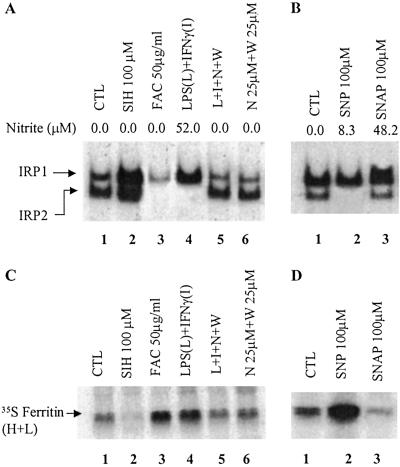
It has been well established that inflammation leads to increased ferritin synthesis in macrophages (refs. 26 and 27; reviewed in ref. 28). Because NO is the major effector molecule in murine macrophages activated by IFN-γ/LPS (25), we examined ferritin synthesis and IRP-binding activities in RAW 264.7 cells exposed to IFN-γ/LPS as compared with those treated with two different NO donors, SNAP (NO⋅ donor), and SNP (NO+ donor). A large production of NO in IFN-γ/LPS-treated cells led to an increase in RNA-binding activity of IRP1 and a decrease in IRP2 levels (Fig. 1A, lane 4), and these changes were associated with an increase in ferritin synthesis (Fig. 1C, lane 4). It is pertinent to mention that, on the basis of the concentration of nitrite in the media, 100 μM SNP generated (3 h) a lower amount of NO than RAW 264.7 cells treated with IFN-γ/LPS (10 h) (Fig. 1 A and B). Hence, the NO generated chemically in this system is in the physiologically relevant range. To determine whether the increase in ferritin synthesis was due to NO production, IFN-γ/LPS-treated cells were simultaneously incubated with iNOS-specific inhibitors. Fig. 1A shows that inhibition of NO production prevented the IFN-γ/LPS-induced increase in IRP1-binding activity as well as the decrease in IRP2-binding/levels. Moreover, we found that iNOS-specific inhibitors attenuated the IFN-γ/LPS-mediated increase in ferritin synthesis. These data indicate that the changes in IRPs and ferritin synthesis, caused by IFN-γ/LPS, were NO-mediated.
Fig 1.
Effects of NO donors and IFN-γ/LPS on RNA-binding activities of IRPs (A and B) and ferritin synthesis (C and D). RAW 264.7 cells were incubated (10 h) with various agents as indicated. (A) Cytosolic protein extracts (10 μg) were subjected to gel retardation assays (see Materials and Methods). CTL, control; L, LPS; I, IFN-γ; W, N-(3-(aminomethyl)-benzyl)acetamide and N, L-N6-(1-iminoethyl)-lysine are specific inhibitors of iNOS. (B) Cells were incubated without or with SNP (100 μM) and SNAP (100 μM) for 3 h. (C and D) Ferritin synthesis in RAW 264.7 cells. After 10 h of preincubation with the reagents, cells were pulse-labeled (2 h) with [35S]methionine and harvested. Ferritin was immunoprecipitated by using antiferritin antibodies and analyzed by 12.5% SDS.PAGE followed by autoradiography. Nitrate was assayed by using the Griess reagent as described by Green et al. (35).
We found that synthetic NO⋅ and NO+ donors have opposite effects on IRPs and ferritin synthesis. The NO⋅ donor SNAP (Fig. 1 B and D, lane 3) as well as other NO⋅ donors (NONOates, not shown) increased IRP1-binding activity and decreased ferritin synthesis. In contrast, the NO+ donor SNP decreased IRP2 levels and dramatically increased ferritin synthesis (Fig. 1 B and D, lane 2), associated with a slight increase in total ferritin levels in RAW 264.7 cells. Control and SNP-treated (3 h) cells contained 256 ± 12 and 293 ± 27 ng of ferritin per milligram of total protein, respectively. The relatively modest increase in the total amount of ferritin after SNP treatment (3 h) can be explained by the fact that the cells contain a large pool of preexisting ferritin. Therefore, even a dramatic increase in de novo synthesis of ferritin would not be expected to substantially increase the total ferritin level. Data in Fig. 1 clearly indicate that exposure of RAW 264.7 cells to the NO+-donor produces changes in IRPs and ferritin synthesis that are very similar to those seen after treatment with IFN-γ/LPS. As expected, the iron chelator salicylaldehyde isonicotinoyl hydrazone caused a significant increase in RNA-binding activity of IRPs and inhibited ferritin synthesis. On the other hand, treatment of RAW 264.7 cells with ferric ammonium citrate stimulated ferritin synthesis, associated with a decrease in the binding activity of IRPs (Fig. 1 A and C).
SNP-Mediated IRP2 Degradation Causes Induction of Ferritin Synthesis.
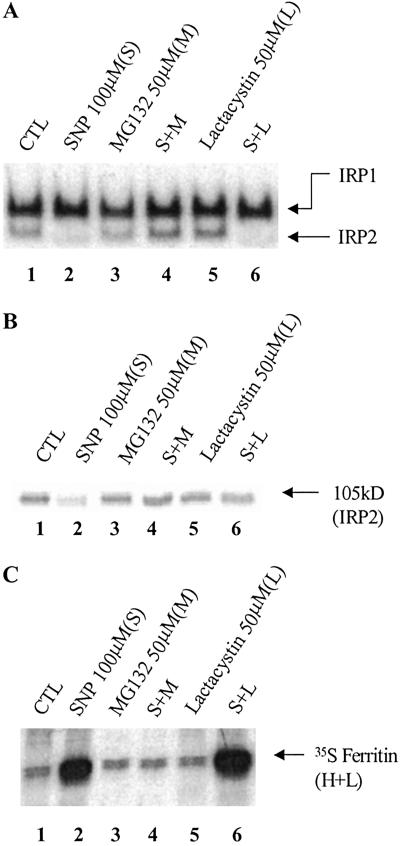
In SNP-treated cells, IRP2 is degraded via the ubiquitin–proteasome pathway, and this degradation causes a decrease in TfR mRNA levels (18). Hence, we examined whether IRP2 degradation is also responsible for the increase in ferritin synthesis in RAW 264.7 cells. To do so, we exploited the proteasome-specific inhibitors, MG132 and lactacystin. RAW 264.7 cells were pretreated with either MG132 or lactacystin for 30 min, after which SNP was added to the cells for an additional 3 h. These inhibitors did not affect either the RNA-binding activity of IRPs or IRP2 protein levels (Fig. 2 A and B, lanes 3 and 5). MG132 totally prevented the SNP-mediated loss of IRP2-binding activity (Fig. 2A, lane 4) as well as its degradation (Fig. 2B, lane 4) in SNP-treated cells and also prevented the SNP-mediated increase in ferritin synthesis (Fig. 2C, lane 4). Although lactacystin failed to attenuate the SNP-mediated decrease in RNA-binding activity of IRP2, it prevented IRP2 degradation (Fig. 2 A and B, lane 6), confirming our previous observations (18). Lactacystin did not prevent the SNP-mediated increase in ferritin synthesis (Fig. 2C, lane 6), likely because of the lack of IRP2 binding to the IRE (Fig. 2A, lane 6).
Fig 2.
Effects of proteasome inhibitors on RNA-binding activities of IRPs (A), IRP2 protein levels (B), and ferritin synthesis. RAW 264.7 cells were grown in control medium (lane 1, CTL), or were preincubated (lanes 3–6) with MG132 (50 μg/ml) or lactacystin (50 μM) for 30 min, after which the cells were incubated for 3 h with SNP (100 μM), as indicated. (A) Gel retardation assays of protein (10 μg) extracted from RAW 264.7 cells after different treatments. (B) Western blot analysis of protein (50 μg) extracted from RAW 264.7 cells (as in A), by using anti-IRP2 antibodies. (C) After 3 h of preincubation with the reagents, cells were pulse-labeled (2 h) with [35S]methionine, and 35S-ferritin was immunoprecipitated and subjected to SDS/PAGE autoradiography.
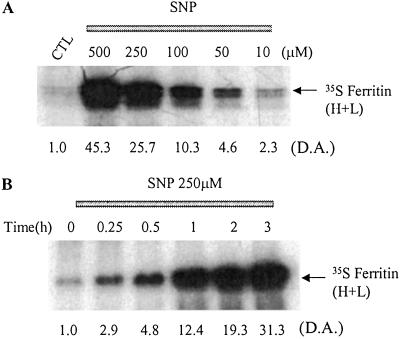
The Effects of SNP on Ferritin Synthesis Are Concentration- and Time- Dependent.
The above studies indicate that SNP causes a dramatic increase in ferritin synthesis. In the following experiments, we investigated how this increase is affected by SNP concentrations and the time of exposure to this agent. Treatment of RAW 264.7 cells with SNP for 3 h increased ferritin synthesis in a dose-dependent manner (Fig. 3A), and the induction of ferritin synthesis was noticeable with SNP concentrations as low as 10 μM (Fig. 3A, lane 6). Five hundred micromolar SNP yielded 40 μM nitrite in 3 h, a value that was comparable to nitrite production in IFN-γ/LPS-treated (10 h) RAW 264.7 cells (Fig. 1A). Additionally, RAW 264.7 cells were incubated with 250 μM SNP for different time intervals, after which the cells were thoroughly washed and incubated with [35S]methionine for 1 h. Fig. 3B shows that an exposure time as short as 15 min leads to a significant increase in [35S]methionine incorporation into ferritin in RAW 264.7 cells. Because ferritin synthesis may also be induced transcriptionally during inflammation, we examined ferritin mRNA levels by Northern blot analysis but did not observe any change in ferritin mRNA in SNP-treated cells (not shown). This finding further supports the conclusion that SNP induces ferritin synthesis by a mechanism involving the IRP/IRE system.
Fig 3.
The effect of SNP on ferritin synthesis. RAW 264.7 cells were grown in control medium (lane 1, CTL) or were incubated (3 h) with different concentrations of SNP (A) added for different time intervals (B), as indicated. D.A., densitometric analysis, in arbitrary units.
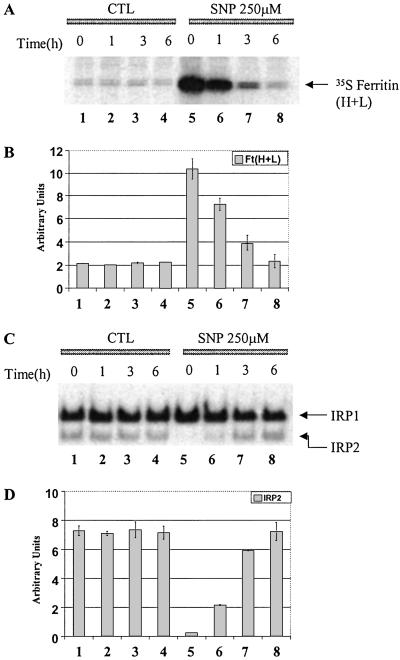
IRP2 Plays a Major Role in Regulation of Ferritin Synthesis.
In the following experiments, we examined whether the increase in ferritin synthesis in SNP-treated cells is transient and, if so, whether the reduction in ferritin synthesis correlates with a recovery in IRP2 binding. After pretreatment with 250 μM SNP for 3 h, RAW 264.7 cells were washed and reincubated for different time intervals in control medium. The cells were divided into two groups: one group was labeled with [35S]methionine to measure ferritin synthesis, whereas cells in the other group were lysed to measure IRP-binding activities. As expected, the rate of ferritin synthesis and RNA-binding activity did not change during 6 h of incubation of control cells (Fig. 4 A–D, lanes 1–4). A 3-h treatment of RAW 264.7 cells with SNP caused the total disappearance of IRP2-binding activity (Fig. 4 C and D, lane 5), associated with a dramatic increase in ferritin synthesis (Fig. 4 A and B, lane 5). Importantly, on removal of SNP from the incubation medium, the rate of ferritin synthesis gradually decreased (Fig. 4 A and B, lanes 5–8), so that at 6 h the rate was as low as in control (untreated) cells; the decline in ferritin synthesis was associated with a gradual restoration of IRP2 binding (Fig. 4 C and D, lanes 5–8). Interestingly, changes in IRP2-binding activity after SNP treatment were not associated with a significant change in IRP1-binding activity (Fig. 4 C and D, lanes 5–8), suggesting that IRP2 is the major regulator controlling ferritin synthesis in RAW 264.7 cells.
Fig 4.
Effects of pretreatment with SNP on ferritin synthesis (A) and IRP binding (B). RAW 264.7 cells were grown in control medium (lane 1–4, CTL) or were preincubated with 250 μM SNP (lanes 5–8, SNP 250 μM) for 3 h. The cells were washed with PBS (3×) and incubated in the control medium for indicated time intervals. (A) Autoradiogram of 35S-ferritin; (B) densitometric analysis of 35S-ferritin (n = 3); error bars represent SD; (C) gel retardation assay; (D) densitometric analysis of IRP2-binding activity (n = 3); error bars represent SD.
SNP and IFN-γ/LPS Increase Iron Accumulation in Ferritin.
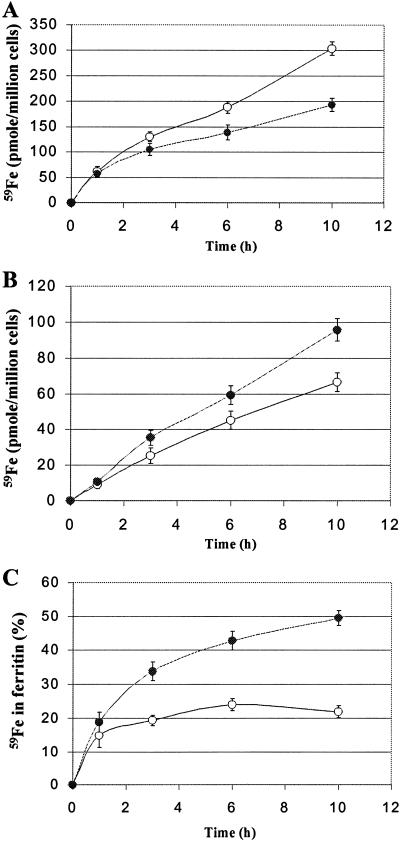
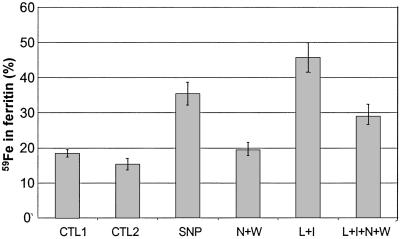
We next examined whether the observed SNP-mediated increase in ferritin synthesis can affect iron accumulation in ferritin. RAW 264.7 cells were incubated with or without 500 μM SNP in the presence of 59Fe2-Tf for various time intervals, after which 59Fe uptake by the cells and 59Fe incorporation into ferritin were measured. The amount of 59Fe taken up by RAW 264.7 cells was slightly reduced in the presence of 500 μM SNP (Fig. 5A), in agreement with a previous report from this laboratory (36). Nevertheless, SNP treatment was associated with a significant increase in 59Fe incorporation into ferritin (Fig. 5B) so that after 10 h of incubation, SNP-treated and control cells contained 50 and 20% of their 59Fe in ferritin, respectively (Fig. 5C). Importantly, IFN-γ/LPS also altered 59Fe distribution in RAW 264.7 cells, resulting in a higher accumulation of 59Fe in ferritin (Fig. 6). Specific inhibitors of iNOS attenuated the IFN-γ/LPS-mediated increase in 59Fe incorporation into ferritin, suggesting that this increase was NO mediated.
Fig 5.
Effects of SNP on 59Fe uptake by RAW 264.7 cells (A) and 59Fe incorporation into ferritin (B and C). RAW 264.7 cells were incubated with 5 μM 59Fe2-Tf in the absence (○) or presence (•) of 500 μM SNP for indicated time intervals, after which the cells were washed with cold PBS (3×). (A) 59Fe uptake by control and SNP-treated RAW 264.7 cells. (B) 59Fe incorporation into ferritin was determined (see Materials and Methods). (C) 59Fe associated with ferritin is expressed as a percentage of total cellular 59Fe content. The results are means ± SD of triplicate determinations in a typical experiment performed.
Fig 6.
Effect of SNP and IFN-γ/LPS on 59Fe incorporation into ferritin. RAW 264.7 cells were incubated with 5 μM 59Fe-Tf for 3 h, after which the cells were washed three times with cold PBS. 59Fe-labeled cells were then incubated with either 100 μM SNP or 10 μg/ml of LPS (L) and 100 units/ml of IFN-γ (I), in the presence or absence of iNOS-specific inhibitors (N, W; see Fig. 1) for 18 h. 59Fe in ferritin (see Materials and Methods) is expressed as a percentage of total cellular 59Fe content. CTL1, percentage of 59Fe in ferritin after 3-h incubation with 59Fe-Tf, CTL2, percentage of 59Fe in ferritin following 18-h reincubation in control medium.
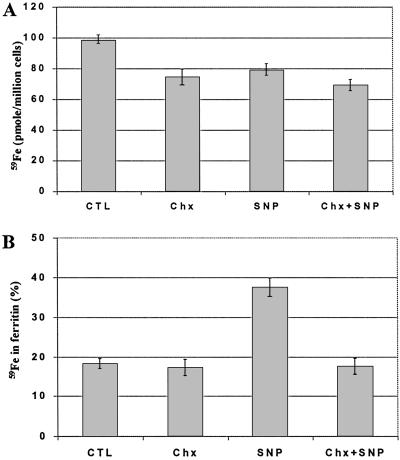
Fig. 7 shows that the protein synthesis inhibitor, cycloheximide, causes only slight inhibition of 59Fe uptake by RAW 264.7 cells (Fig. 7A) but can fully prevent the SNP-mediated increase in 59Fe incorporation into ferritin (Fig. 7B). Collectively, these data strongly suggest that de novo synthesized ferritin, as compared with ferritin already present in the cell, has a significantly higher efficiency in sequestering iron.
Fig 7.
Effect of cycloheximide on SNP-induced 59Fe incorporation into ferritin. RAW 264.7 cells were incubated with 5 μM 59Fe2-Tf for 3 h, after which the cells were washed with cold PBS (3×). After counting 59Fe in cell pellets (A), the cells were lysed, and 59Fe incorporation into ferritin (B) was measured (see Materials and Methods). 59Fe associated with ferritin is expressed as a percentage of total cellular 59Fe content. The results are means ± SD of triplicate determinations in a typical experiment performed. Chx, cycloheximide (100 μM).
Discussion
IRPs are cytosolic proteins that play a major role in preventing the expansion of the LIP while maintaining sufficient levels of iron in this pool for the synthesis of essential heme and nonheme iron proteins. The sequence-specific binding of IRPs to IREs coordinately regulates iron uptake and storage and, consequently, keeps iron in the LIP at appropriate levels. The level of iron in the LIP is sensed by IRPs, which are responsible for the opposite regulation of ferritin and TfR expression.
However, iron is not the only player that controls IRPs, because IRP activities/levels can also be affected by various forms of oxidative stress and NO (10–19). IRP1 is homologous to mitochondrial aconitase, an [Fe-S] protein whose activity is modulated by NO (6, 7, 16, 22). Hence, it is not surprising that IRP1-binding activity can also be affected by NO, which was shown to increase TfR mRNA levels (13–17). However, these results are in conflict with findings that TfR levels are decreased in IFN-γ- and/or LPS-treated macrophages (37–39), a condition known to produce a large quantity of NO (25). These paradoxical results may be explained by the fact that NO has markedly different biological effects depending on its redox state. NO in its reduced form, NO⋅, has high affinity for iron, whereas NO in its oxidized form, the nitrosonium ion (NO+), causes S-nitrosylation of various proteins (21, 29, 30). Hence, it is conceivable that NO⋅ affects the [Fe-S] cluster of IRP1, whereas NO+ may regulate the RNA-binding activity of IRPs by S-nitrosylation of their key sulfhydryl groups. Indeed, we previously demonstrated that the NO+ generator, SNP, in contrast to the NO⋅ donor SNAP, markedly decreased IRP1 RNA-binding activity and TfR mRNA levels (15). More recently, we found that IRP2 is also a very important target of NO+ (18). We showed that a 1-h exposure of RAW 264.7 cells to SNP resulted in a significant decrease in IRP2 binding, followed by a decrease in IRP2 protein levels, associated with a dramatic decrease in TfR mRNA (18).
In this study, we demonstrated that IFN-γ/LPS-mediated degradation of IRP2 in RAW 264.7 cells is associated with a dramatic increase in ferritin synthesis (Fig. 1). Interestingly, a slight increase in IRP1 RNA-binding activity could not overcome the lack of IRP2 and block ferritin mRNA translation. Hence, IRP2 alone can play a significant role in controlling ferritin synthesis, a conclusion also supported by a recent report by Schalinske et al. (40). Importantly, the iNOS-specific inhibitors N-[3-(aminomethyl)-benzyl]acetamide and L-N6-(1-iminoethyl)lysine prevented the decrease in IRP2 levels, resulting in the suppression of ferritin synthesis in IFN-γ/LPS-treated RAW 264.7 cells (Fig. 1). This provides strong evidence that IFN-γ/LPS exert their effects via an NO-mediated mechanism.
We also found that the treatment of RAW 264.7 cells with SNP caused changes that were remarkably similar to those seen after IFN-γ/LPS treatment (Fig. 1). Treatment of RAW 264.7 cells with SNP resulted in dramatic stimulation of ferritin synthesis, and this increase correlated with a decrease in IRP2 levels. Moreover, we demonstrated that the proteasome inhibitor MG132, which can prevent the SNP-mediated degradation of IRP2 (which regains normal IRE binding), blocked the increase in ferritin synthesis (Fig. 2). Although IRP2 was not degraded in cells treated with SNP and lactacystin, its modification by SNP prevented its binding to the IRE, allowing efficient ferritin translation. We also showed that the suppression of ferritin synthesis after SNP removal was associated with a recovery of IRP2 binding to IRE (Fig. 4). These experiments extend our previous findings (18, 19) and indicate that IRP2 alone plays a key role, not only in regulating TfR mRNA levels, but also in controlling ferritin synthesis. Further research is needed to explain why an active IRP1 in SNP- or IFN-γ/LPS-treated macrophages is unable to suppress ferritin synthesis when IRP2 is absent. In this context, it is pertinent to mention that IRP2 knockout mice exhibit a dramatic increase in ferritin levels in various tissues (41).
There is no doubt that under certain conditions, transcriptional mechanisms are involved in ferritin synthesis (28, 42), but transcriptional activity is unlikely to explain the increase in ferritin synthesis in SNP-treated RAW 264.7 cells. We did not notice any change in ferritin mRNA levels under conditions when SNP stimulated ferritin synthesis (data not shown), suggesting that SNP enhances ferritin synthesis posttranscriptionally.
Importantly, our data also suggest that de novo synthesized ferritin, as compared with preexisting ferritin, sequesters iron more efficiently. First we found that, whereas RAW 264.7 cells exposed to SNP (3 h) exhibited only about a 15% increase in ferritin levels, they accumulated twice as much 59Fe in ferritin compared with untreated cells (Fig. 5C). Second, the SNP-mediated increase in iron accumulation in ferritin can be attenuated by cycloheximide. To the best of our knowledge, this is the first report showing a positive correlation between de novo ferritin synthesis and its iron accumulation. This finding also indicates that SNP is unlikely to increase cytosolic “free” iron levels, and this conclusion is further supported by the following arguments. First, we have previously demonstrated that a potent iron chelating agent, desferrioxamine, was unable to attenuate SNP-induced degradation of IRP2 (18). Second, the exposure of RAW 264.7 cells to SNP for up to 10 h did not decrease IRP1 RNA-binding activity (18), which would be expected if intracellular iron levels increased. Hence, it can be concluded that SNP stimulates ferritin synthesis by an iron-independent mechanism.
Recent research supports the conclusion that cellular iron homeostasis appears to be regulated not only by iron levels but also by other factors that manifest during inflammation, a condition associated with increased NO production. We demonstrated that nitrogen monoxide-mediated degradation of IRP2 leads to a dramatic up-regulation of ferritin synthesis in macrophages. Moreover, this is the first report, to our knowledge, showing that NO-mediated increase of ferritin synthesis leads to increased sequestration of intracellular iron in ferritin. These observations explain, at least in part, the molecular pathogenesis of anemia of chronic disorders that commonly accompanies infectious disease, inflammatory disease, and malignancies. Macrophages of the reticuloendothelial system phagocytose senescent erythrocytes, after which iron is liberated from heme by heme oxygenase 1. Normally, iron is very efficiently released to the circulation (43) but in anemia of chronic disorders hypoferremia, caused by a defect in the release of iron from macrophages (44), develops. Hence, increased sequestration of iron in ferritin, when its synthesis is stimulated in macrophages of patients with chronic inflammation, explains hypoferremia and, consequently, anemia. On the basis of earlier studies (45), it can be argued that levels of iNOS expression and NO production in human macrophage lineage cells do not reach the high levels observed in murine macrophages. However, later studies using human macrophages have convincingly documented iNOS protein and mRNA expression as well as NO production under a variety of activation conditions and in several disease states (46). Hence, our data obtained on murine macrophages may provide insight into human pathophysiology, although studies exploiting human macrophages are necessary.
Acknowledgments
Dr. Lukas Kühn (Swiss Institute for Experimental Cancer Research) kindly supplied pST18-fer plasmid. We thank Kostas Pantopoulos for valuable advice, Joan Buss and Alex Sheftel for reading the manuscript and for useful comments, and Sandy Fraiberg for excellent editorial assistance. This work was supported by a grant from Canadian Institutes of Health Research (CIHR) (formerly the Medical Research Council of Canada).
Abbreviations
LIP, labile iron pool
IRP, iron regulatory protein
Tf, transferrin
TfR, Tf receptor
IRE, iron-responsive element
NO, nitrogen monoxide
SNP, sodium nitroprusside
LPS, lipopolysaccharide
SNAP, S-nitroso-N-acetylpenicillamine
iNOS, inducible nitrous-oxide synthase
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Munro H. N. & Linder, M. C. (1978) Physiol. Rev. 58, 317-396. [DOI] [PubMed] [Google Scholar]
- 2.Harrison P. M. & Arosio, P. (1996) Biochim. Biophys. Acta 1275, 161-203. [DOI] [PubMed] [Google Scholar]
- 3.Levi S., Luzzago, A., Cozzi, A., Franceschinelli, F., Albertini, A. & Arosio, P. (1988) J. Biol. Chem. 263, 18086-18092. [PubMed] [Google Scholar]
- 4.Santambrogio P., Levi, S., Arosio, P., Palagi, L., Vecchio, G., Lawson, D. M., Yewdall, S. J., Artymium, P. J., Harrison, P. M., Jappelli, R. & Cesareni, G. (1992) J. Biol. Chem. 267, 14077-14083. [PubMed] [Google Scholar]
- 5.Levi S., Santambrogio, P., Cozzi, A., Rovida, E., Corsi, B., Tamborini, E., Spada, S., Albertini, A. & Arosio, P. (1994) J. Mol. Biol. 238, 649-654. [DOI] [PubMed] [Google Scholar]
- 6.Rouault T. & Klausner, R. D. (1997) Curr. Top. Cell Regul. 35, 1-19. [DOI] [PubMed] [Google Scholar]
- 7.Richardson D. R. & Ponka, P. (1997) Biochim. Biophys. Acta 1331, 1-40. [DOI] [PubMed] [Google Scholar]
- 8.Mikulits W., Schranzhofer, M., Beug, H. & Mullner, E. W. (1999) Mutat. Res. 437, 219-230. [DOI] [PubMed] [Google Scholar]
- 9.Eisenstein R. S. (2000) Annu. Rev. Nutr. 20, 627-662. [DOI] [PubMed] [Google Scholar]
- 10.Pantopoulos K. & Hentze, M. W. (1995) Proc. Natl. Acad. Sci. USA 92, 1267-1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hanson E. S. & Leibold, E. A. (1998) J. Biol. Chem. 273, 7588-7593. [DOI] [PubMed] [Google Scholar]
- 12.Eisenstein R. S., Tuazon, P. T., Schalinske, K. L., Anderson, S. A. & Traugh, J. A. (1993) J. Biol. Chem. 268, 27363-27370. [PubMed] [Google Scholar]
- 13.Weiss G., Goossen, B., Doppler, W., Fuchs, D., Pantopoulos, K., Werner-Felmayer, G., Wachter, H. & Hentze, M. W. (1993) EMBO J. 12, 3651-3657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Drapier J., Hirling, H., Wietzerbin, J., Pierre, K. & Kühn, L. C. (1993) EMBO J. 12, 3643-3649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Richardson D. R., Neumannova, V., Nagy, E. & Ponka, P. (1995) Blood 86, 3211-3219. [PubMed] [Google Scholar]
- 16.Hentze M. W. & Kühn, L. C. (1996) Proc. Natl. Acad. Sci. USA 93, 8175-8182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Phillips J. D., Kinikini, D. V., Yu, Y., Guo, B. & Leibold, E. A. (1996) Blood 87, 2983-2992. [PubMed] [Google Scholar]
- 18.Kim S. & Ponka, P. (1999) J. Biol. Chem. 274, 33035-33042. [DOI] [PubMed] [Google Scholar]
- 19.Kim S. & Ponka, P. (2000) J. Biol. Chem. 275, 6220-6226. [DOI] [PubMed] [Google Scholar]
- 20.Ignarro L. J. (1994) Adv. Pharmacol. 26, 35-65. [DOI] [PubMed] [Google Scholar]
- 21.Stamler J. S., Singer, J. D. & Loscalzo, J. (1992) Science 258, 1898-1902. [DOI] [PubMed] [Google Scholar]
- 22.Kennedy M. C., Antholine, W. E. & Beinert, H. (1997) J. Biol. Chem. 272, 20340-20347. [DOI] [PubMed] [Google Scholar]
- 23.Richardson D. R. & Ponka, P. (1997) Methods Neurosci. 31, 329-345. [Google Scholar]
- 24.Kennedy M. C., Mende-Mueller, L., Blondin, G. A. & Beinert, H. (1992) Proc. Natl. Acad. Sci. USA 89, 11730-11734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.MacMicking J., Xie, Q. W. & Nathan, C. (1997) Annu. Rev. Immunol. 15, 323-350. [DOI] [PubMed] [Google Scholar]
- 26.Konijn A. M. & Hershko, C. (1997) Br. J. Haematol. 37, 7-16. [PubMed] [Google Scholar]
- 27.Konijn A. M., Carmel, N., Levy, R. & Hershko, C. (1981) Br. J. Haematol. 49, 361-370. [DOI] [PubMed] [Google Scholar]
- 28.Ponka P., Beamont, C. & Richardson, D. R. (1998) Semin. Hematol. 35, 35-54. [PubMed] [Google Scholar]
- 29.Stamler J. S. (1994) Cell 78, 931-936. [DOI] [PubMed] [Google Scholar]
- 30.Jaffrey S. R., Erdjument-Bromage, H., Ferris, C. D., Tempst, P. & Snyder, H. (2001) Nat. Cell Biol. 3, 193-197. [DOI] [PubMed] [Google Scholar]
- 31.Raschke W. C., Baird, S., Ralph, P. & Nakoinz, I. (1978) Cell 15, 261-267. [DOI] [PubMed] [Google Scholar]
- 32.Ralph P. & Nakoinz, I. (1977) J. Immunol. 119, 950-954. [PubMed] [Google Scholar]
- 33.Ponka P., Borova, J., Neuwirt, J., Fuchs, O. & Necas, E. (1979) Biochim. Biophys. Acta 586, 278-297. [DOI] [PubMed] [Google Scholar]
- 34.Mullner E. W., Neupert, B. & Kühn, L. C. (1989) Cell 58, 373-382. [DOI] [PubMed] [Google Scholar]
- 35.Green L. C., Wagner, D. A., Glogowski, J., Skipper, P. L., Wishnok, J. S. & Tannenbaum, S. R. (1982) Anal. Biochem 126, 131-138. [DOI] [PubMed] [Google Scholar]
- 36.Richardson D. R., Neumannova, V. & Ponka, P. (1995) Biochim. Biophys. Acta 1266, 250-260. [DOI] [PubMed] [Google Scholar]
- 37.Hamilton T. A., Gray, P. W. & Adams, D. O. (1984) Cell. Immunol. 89, 478-488. [DOI] [PubMed] [Google Scholar]
- 38.Byrd T. F. & Horwitz, M. A. (1989) J. Clin. Invest. 83, 1457-1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wardrop S. L. & Richardson, D. R. (2000) Eur. J. Biochem. 267, 6586-6593. [DOI] [PubMed] [Google Scholar]
- 40.Schalinske K. L., Blemings, K. P., Steffen, D. W., Chen, O. S. & Eisenstein, R. S. (1997) Proc. Natl. Acad. Sci. USA 94, 10681-10688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.LaVaute T., Smith, S., Cooperman, S., Iwai, K., Land, W., Meyron-Holtz, E., Drakje, S. K., Miller, G., Abu-Asab, M., Tsokos, M., et al. (2001) Nat. Genet. 27, 209-214. [DOI] [PubMed] [Google Scholar]
- 42.Tsuji Y., Ayaki, H., Whitman, S. P., Morrow, C. S., Torti, S. V. & Torti, F. M. (2000) Mol. Cell. Biol. 20, 5818-5827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hershko C. (1977) Progr. Hematol. 10, 105-148. [PubMed] [Google Scholar]
- 44.Fillet G., Beguin, Y. & Badelli, L. (1989) Blood 74, 844-851. [PubMed] [Google Scholar]
- 45.Denis M. (1994) J. Leukocyte Biol. 55, 682-684. [DOI] [PubMed] [Google Scholar]
- 46.Weinberg J. B. (1998) Mol. Med. 4, 557-591. [DOI] [PMC free article] [PubMed] [Google Scholar]