Abstract
Chloroplasts were once free-living cyanobacteria that became endosymbionts, but the genomes of contemporary plastids encode only ≈5–10% as many genes as those of their free-living cousins, indicating that many genes were either lost from plastids or transferred to the nucleus during the course of plant evolution. Previous estimates have suggested that between 800 and perhaps as many as 2,000 genes in the Arabidopsis genome might come from cyanobacteria, but genome-wide phylogenetic surveys that could provide direct estimates of this number are lacking. We compared 24,990 proteins encoded in the Arabidopsis genome to the proteins from three cyanobacterial genomes, 16 other prokaryotic reference genomes, and yeast. Of 9,368 Arabidopsis proteins sufficiently conserved for primary sequence comparison, 866 detected homologues only among cyanobacteria and 834 other branched with cyanobacterial homologues in phylogenetic trees. Extrapolating from these conserved proteins to the whole genome, the data suggest that ≈4,500 of Arabidopsis protein-coding genes (≈18% of the total) were acquired from the cyanobacterial ancestor of plastids. These proteins encompass all functional classes, and the majority of them are targeted to cell compartments other than the chloroplast. Analysis of 15 sequenced chloroplast genomes revealed 117 nuclear-encoded proteins that are also still present in at least one chloroplast genome. A phylogeny of chloroplast genomes inferred from 41 proteins and 8,303 amino acids sites indicates that at least two independent secondary endosymbiotic events have occurred involving red algae and that amino acid composition bias in chloroplast proteins strongly affects plastid genome phylogeny.
Chloroplasts arose from cyanobacteria through endosymbiosis (1), but molecular studies have yet to link plastids robustly with any particular group of contemporary cyanobacteria, leaving the precise lineage of cyanobacteria that gave rise to plastids unknown (2–4). The evolutionary process that transformed the cyanobacterial symbiont into a contemporary organelle involved both inheritance and invention. Such inhertances include photosynthesis, 70S ribosomes, cell division proteins, and, in some primitive plastids, a peptidoglycan wall (5–10). Important inventions include the protein import machinery, which permits the plastid to import nuclear-encoded proteins (11), and hence to donate genes to the nucleus over evolutionary time (12–15).
Contemporary chloroplast genomes encode between 60–200 proteins in various photosynthetic lineages and have thus undergone a process of severe genome reduction during the course of endosymbiosis (13), because contemporary cyanobacteria encode several thousand proteins (16). But plastids contain roughly just as many proteins as their free-living cyanobacterial cousins, current estimates suggesting that between 1,000 and 5,000 proteins in higher plants are targeted to plastids (15, 17, 18).
Previous work has shown that many gene transfers to the nucleus have occurred during plastid evolution (19, 20), but estimates for the total number of genes that were transferred have been elusive. Previous calculations based on blast surveys and subsamples of the Arabidopsis genome data have suggested that between 800 and perhaps as many as 2,000 genes in the Arabidopsis genome might come from cyanobacteria (15, 17, 18, 21). Here we report the phylogenetic analysis of proteins from Arabidopsis (21), three cyanobacterial genomes [Synechocystis sp. PCC6803 (16), Prochlorococcus marinus, and Nostoc punctiforme (22)], 16 other prokaryotic reference genomes, and yeast, in addition to the phylogeny of 15 sequenced chloroplast genomes and the identification of transferred nuclear homologues of genes still encoded in at least one plastid genome.
Methods
Analysis of 24,990 Arabidopsis Proteins.
Proteins were retrieved from GenBank or from the U.S. Department of Energy web site (Nostoc and Prochlorococcus; www.jgi.doe.gov/JGI_ microbial/html). blast comparisons (23), filtering, retrieval, alignments (24), removal of gapped sites, and maximum likelihood (ML) (25) analyses were performed as described (17) by using the neighbor-joining (NJ) (26) tree of ML-distances as the starting topology. Homologues were retrieved from blast tables as described (17) by using a drop-off point of 10−6. Protein sorting prediction was performed with targetp (27) as described (15).
Analysis of 15 Chloroplast Genomes.
The set of proteins common to 15 sequenced chloroplast genomes—the nine previously analyzed (19) plus Guillardia theta, Cyanidium caldarum, Chlorella vulgaris, Nephroselmis olivacea, Mesostigma viridis, and Oenothera elata (28–33)—were identified and assembled into a concatenated data set. Presence or absence of proteins was determined by sequence comparison. The chloroplast-encoded subunits of the RNA polymerase were previously shown to be problematic in early chloroplast phylogeny (19, 34) and were excluded from analysis, leaving 8,308 amino acid positions from 41 proteins for phylogenetic inference. Many proteins among these 41 were missing in the yet incomplete Prochlorococcus and Nostoc (22) data. Phylogenies were inferred with the complete data set (8,308-site data), after excluding gapped sites (7,474-site data), after excluding constant sites (5,153-site data), and after excluding both constant and gapped sites (4,319-site data) using NJ (26) with uncorrected (NJP), Dayhoff (NJD), and Kimura (NJK) distances (35), with protml (25) (ML) and puzzle (36) using the JTT-F matrix and with parsimony (MP). Protein log determinant (LogDet, LD) (37) and spectral analysis (38) was performed with the 8,308-site data after excluding gapped sites and under iterative down-weighting of constant sites in steps of 10%. Amino acid composition equilibrium was tested with puzzle (35). Topologies were compared with the Shimodaira–Hasegawa test (39). Alignments, data, and results are available at www.molevol.de/people/martin/projects/how_many/.
Results and Discussion
Cyanobacterial Genes in the Arabidopsis Genome.
Given sufficient sequence conservation (35, 40), genes that were transferred from chloroplasts to the nucleus should share a common branch with their cyanobacterial homologues in a phylogenetic tree (17). To see how many genes in the Arabidopsis genome satisfy this criterion, 24,990 nonredundant Arabidopsis protein sequences were first compared individually with blast (23) to all proteins from the 20 reference genomes shown in Fig. 1. The 9,368 Arabidopsis proteins that detected a homologue in one of the other genomes at a probability threshold (E value) of better than 10−10 were considered further, because less conserved proteins are unalignable for phylogenetic inference. Among the 9,368 blast tables, 7,304 contained a cyanobacterial homologue with an E value better than 10−4. Among these, the cyanobacterial homologue was the best match in 2,363 cases; in 1,265 other cases, a cyanobacterial homologue was among the best matches. For these 3,628 Arabidopsis proteins, the homologues so identified were extracted, aligned, purged of gapped positions to reduce the effects of poorly aligned regions, and subjected to phylogenetic analysis using NJ and protml (Fig. 1).
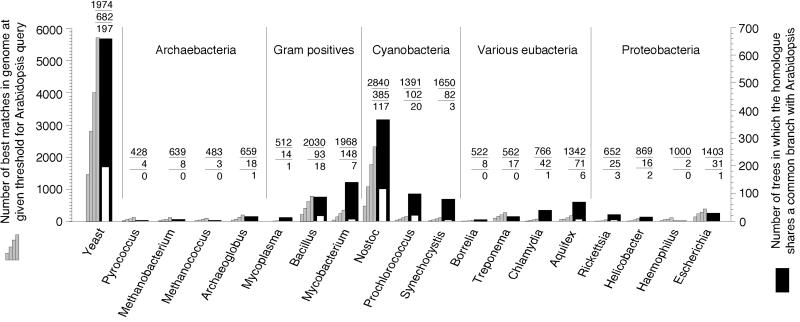
Fig 1.
Similarity of 24,990 Arabidopsis proteins to 51,361 proteins from 20 reference genomes (two Mycoplasma genome sequences are treated as one species). Gray columns: number of times that the genome gave the best match against Arabidopsis when blast was used (23) at four E value thresholds, from left to right 10−40, 10−20, 10−10, and 10−4. The number of times that a homologue from the genome occurred in any tree is indicated (top number above columns). Black columns indicate the number of times that proteins from the genome indicated gave a common branch with the Arabidopsis homologue in protml (25) analyses using the JTT-F matrix (middle number), white columns therein indicate the number of those trees in which the branch was supported at BP ≥ 0.95 (bottom number).
Many Arabidopsis proteins investigated were most similar to their yeast homologues (Fig. 1). These genes were probably present in the host cell that acquired plastids (2, 3, 17, 18) and have been retained in both yeast and Arabidopsis. The second largest fraction of Arabidopsis genes are cyanobacterial acquisitions.
For 677 Arabidopsis proteins, blast detected a homologue in one cyanobacterium but in no other genome; for 133 proteins, homologues were detected in two cyanobacteria only; for 56 proteins, homologues were detected in all three cyanobacteria only, making 866 proteins that are shared by Arabidopsis and cyanobacteria among the genomes sampled, and hence are likely of cyanobacterial origin. An additional 834 proteins branch specifically with cyanobacterial homologues in phylogenetic analysis: 513 Arabidopsis proteins shared a common branch with one cyanobacterium, 179 were the sister to two cyanobacteria, and 142 branched as the sister to all three cyanobacteria sampled. In NJ trees of ML distances, the corresponding numbers were similar, comprising 680 total.
Based on their similarity patterns, these 1,700 (834 + 866) Arabidopsis proteins are encoded by genes that were transferred to the nucleus from plastids. Expressed as a proportion of the 9368 genes investigated by virtue of sufficient sequence conservation, this makes 18.1% of the total. However, an additional 354 Arabidopsis proteins were equivocal because blast detected either (i) only two homologues, one from cyanobacteria and one from another genome (300 cases), or (ii) only three homologues, two from cyanobacteria and one from another genome (54 cases). Many of these 354 equivocal genes, which always give an (Arabi,cyano) branch, are also probably cyanobacterial, but they were not counted. At the same time, Arabidopsis branched on average 32 times with each noncyanobacterial prokaryote sampled (Fig. 1), probably due to chance (see below). Conservatively allowing 354 false negatives (the equivocals) to counterweigh 32 false positves (due to chance) leaves an estimate of ≈1,700 genes among the 9,368 investigated, or ≈18% of the total that come from cyanobacteria.
Notably, this analysis encompasses only those 9,368 proteins in the Arabodopsis genome with sufficient sequence conservation to yield a match of at least 10−10 in blast analysis. This is only ≈37% of the 24,990 Arabidopsis proteins. There is no a priori reason to suspect that the Arabidopsis genes that come from cyanobacteria should preferentially belong to this conservatively evolving fraction of proteins more so than, for example, the proteins that come from the Arabidopsis host lineage do. Furthermore, the remaining 63% of Arabidopsis genes that did not meet the 10−10 criterion must have come from somewhere. Either they arose de novo from noncoding DNA, which is very improbable, or, more likely, they arose through sequence divergence, recombination, and duplication involving preexisting coding sequences, the cyanobacterial component of which should reflect that demonstrable in the conserved fraction of genes analyzed here. Hence, with some caution, our estimate of 18%, which is based on the phylogenetically analyzable fraction of sequences only, can be extrapolated to the genome as a whole, which would indicate a total of ≈4,500 cyanobacterial genes in the Arabidopsis genome.
Is 18% an Underestimate or an Overestimate?
One possibility that might suggest this value to be an overestimate concerns the use of yeast as the only nonphotosynthetic reference eukaryote. Yeast has a rather small genome, hence the inclusion of other eukaryotes could increase the number of genes that identify a homologue at the 10−10 threshold, thereby increasing our reference sample of 9,368 proteins. In a similar case involving eubacterial genes in the human genome, increasing the eukaryotic sample by five lineages increased the reference sample by only a few hundred additional homologues (41), so this factor is probably not too severe. Another factor that might lead to an overestimate concerns the relationship between cyanobacteria and plastids. If the cyanobacteria sampled here diverged from the cyanobacterium that gave rise to plastids more recently than the divergence of the yeast and Arabidopsis lineages, then the cyanobacterial genes in Arabidopsis would share a more recent common ancestor with their homologues in free-living cyanobacteria than the “host” genes in the Arabidopsis lineage would share with yeast homologues. If so, there would be a bias in our data making cyanobacterial homologues easier to detect with blast and easier to correctly tree relative to their yeast homologues. This would yield an overestimate. Conversely, however, if the cyanobacteria sampled here diverged from the cyanobacterium that gave rise to plastids before the yeast–Arabidopsis divergence, the converse bias would lead to an underestimate. We know of no evidence that would unambiguously indicate the cyanobacteria sampled here to have diverged from the ancestor of plastids after the yeast–Arabidopsis divergence, and given the antiquity of the cyanobacterial lineage, the converse bias may even be more likely. On the other hand, at least two lines of evidence suggest that the value of 18% is probably an underestimate.
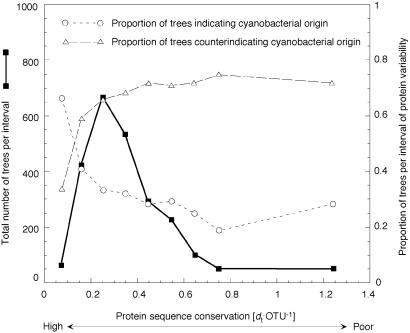
First, the efficiency of phylogenetic inference decreases with increasing sequence divergence (35, 40). Thus, many additional Arabidopsis genes analyzed here may have entered the Arabidopsis nuclear lineage via the cyanobacterial ancestor of plastids, but their (Arabi,cyano) branch was not recovered because of the poor performance of phylogenetic methods with poorly conserved proteins (35, 40). Fig. 2 depicts the fraction trees indicating a cyanobacterial origin of Arabidopsis genes plotted against conservation of the proteins investigated and reveals that the (Arabi,cyano) branch is indeed recovered much more frequently among conservatively evolving proteins. So it is quite likely that our trees failed to detect many genuinely cyanobacterial genes in Arabidopsis.
Fig 2.
Freqency distribution of protml results vs. protein variability, expressed as protml tree length in substitutions per site per taxon [dt⋅OTU−1] (abcissa). Highly conserved proteins are at the left, highly variable proteins at the right. Bin intervals of 0.1 were used except the last interval, which contains all trees with dt⋅OTU−1 > 0.8 (plotted at abcissa mean). Squares indicate the number of trees per interval (left ordinate). Circles indicate the proportion of trees per interval (right ordinate) that yield an (Arabi,cyano) branch. Triangles indicate the proportion of trees per interval that do not. Equivocal trees were excluded.
Second, we found a surprisingly large fraction of Arabidopsis proteins that branch with their homologues from Gram-positive (G+ve) bacteria. For example, more Arabidopsis proteins branched with their homolgues from Mycobacterium (148 proteins) than did with either Prochlorococcus (102) or Synechocystis (82) (Fig. 1). Naively, this might be interpreted as suggesting that the Arabidopsis lineage acquired genes specifically from a G+ve donor subsequent to its divergence from the yeast lineage. But by that same measure, the data in Fig. 1 would suggest at face value that the Arabidopsis lineage acquired genes from all organisms sampled in this study. Such interpretations can hardly be true and are at odds with the finding that the data in Fig. 1 would suggest at face value the Arabidopsis lineage to have acquired genes not from one cyanobacterium, but from all three sampled [even at a bootstrap probability (BP) ≥ 0.95], whereby that view contradicts independent evidence suggesting a single origin of plastids from one cyanobacterium (42, 43), not three or more in the Arabidopsis lineage. The G+ve signal in the Arabidopsis data most likely reflects an overall similarity of many proteins in G+ve genomes to homologues in cyanobacteria. Data from rRNA (44) and protein trees (45–47), operon organization (48), and lipoprotein components (49) phylogenetically link G+ves and cyanobacteria. In our view, the G+ve signal in the Arabidopsis data are most easily attributed to genes that entered the plant lineage through the ancestors of plastids, even though the gene trees recover a G+ve branch, either because of shared ancestry or lateral transfer of G+ve and cyanobacterial genes (17). Importantly, this G+ve signal—though substantial and probably cyanobacterial in origin—was not counted in our estimate of 18%.
In a previous study involving fewer proteins and Synechocystis as the only cyanobacterium (17), topology tests revealed that about 2% of the proteins sampled indicated a cyanobacterial origin at P = 0.05, whereas 9% did not exclude same at P = 0.05. That margin of uncertainty was caused by the poorly conserved proteins (17), whose phylogenies do not discriminate. In the present study, Fig. 1 reveals that only 377 topologies supported a common branch for Arabidopsis with the homologue from any genome sampled at BP ≥ 0.95, 197 (52%) of which indicated Arabidopsis as the sister to yeast and 140 (37%) of which indicated Arabidopsis as the sister to one cyanobacterium. Yet the main factor underlying the difference between the present (18%) and previous (2–9%) estimate (17) is not topology testing, but rather the complete Arabidopsis data and inclusion of Nostoc (Fig. 1). Excluding 4 spp. cases, the mean BP for the (Arabi, cyano) branch was 0.87 with a median of 0.95. If we count only those 446 trees that support branching of Arabidopsis with cyanobacteria at BP ≥ 90, the estimate becomes 14% (or ≈3,500 genes). Clearly, sampling of both cyanobacterial and reference species, protein conservation, and topology support all bear on this estimate.
Protein Compartmentation, Functional Categories, and Gene Families.
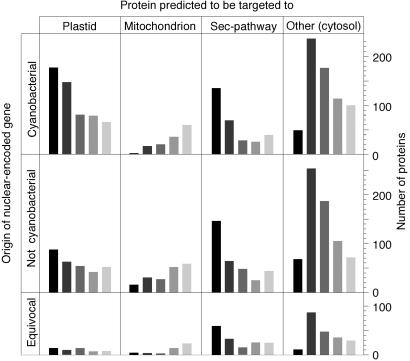
Despite numerous findings to the contrary (15, 50), it is still widely held that the products of nuclear genes that were donated by organelles are, as a rule, targeted back to the donor organelle, in other words, that protein compartmentation and gene origin correspond (51). Previous findings have indicated that plant proteins encoded by genes of cyanobacterial origin are not, as a rule, targeted to the chloroplast, but rather to various compartments, and furthermore that proteins that were not acquired from cyanobacteria can be targeted to plastids (13, 15, 50). Protein-targeting predictions at five significance thresholds (Fig. 3) for the 3,628 proteins in question indicate that more than half of the cyanobacterial proteins are not targeted to the plastid, whereas many noncyanobacterial proteins are. Furthermore, many proteins of cyanobacterial origin appear to enter the secretory pathway. Clearly, gene origin and protein compartmentation do not strictly correspond (50).
Fig 3.
Targeting predictions for 3,628 Arabidopsis proteins examined. Columns indicate the number of genes predicted to be targeted to the compartment shown at five significance thresholds (27). Dark bars (left) indicate the highest threshold, light bars (right) indicate the lowest significance threshold.
The 1,700 genes of cyanobacterial origin encompass all functional categories (Table 1), and many are involved in functions that are not typically cyanobacterial, for example disease resistance and intracellular protein routing, indicating that genes acquired from the ancestor of plastids were a rich source of genetic raw material for the evolution of new functions. Furthermore, once translocated to the nucleus, acquired genes can undergo duplication and diversification like any preexisting gene, and many Arabidopsis genes are indeed recent duplicates (21). When 90% amino acid identity was used as the threshold to define a gene family, the Arabidopsis genes of cyanobacterial descent fall into 1,392 gene families (Table 4, which is published as supporting information on the PNAS web site, www.pnas.org). At the very low 30% amino acid identity level, they still fall into 572 gene families, providing a much too conservative lower boundary—among the 9,363 genes investigated that satisfy the 10−10 criterion—for the number of individual gene transfer-and-fixation events.
Table 1.
Functional categories for Arabidopsis proteins of cyanobacterial origin
| Functional category | No. |
|---|---|
| Biosynthesis and metabolism | 562 |
| Energy generation | 93 |
| Cell growth and division | 31 |
| Transcription | 54 |
| Protein synthesis | 68 |
| Protein destination | 63 |
| Transport facilitators | 35 |
| Intracellular transport | 12 |
| Biogenesis | 38 |
| Signal transduction | 189 |
| Cellular response | 137 |
| Homeostasis | 5 |
| Cell organization | 71 |
| Classification not clear-cut | 39 |
| Unclassified | 303 |
Functional categories from ref. 21.
Number of proteins per category among 1,700 identified.
Plastid Ancestry in Nuclear Genes.
Because they possess chlorophyll b, the prochlorophytes (e.g., Prochlorococcus) were once suspected to be the closest living relatives of plastids, but more recent findings have cast doubt on that view (4, 52, 53). Proteins of the filamentous cyanobacterium Nostoc showed much greater overall similarity to Arabidopsis nuclear-encoded proteins than did those of Prochlorococcus or Synechocystis. Nostoc, for which 7,479 proteins were analyzed, possesses homologues of many Arabidopsis proteins that Synechocystis (3,168 proteins) and Prochlorococcus (2,156 proteins analyzed) lack. Nostoc proteins gave the (Arabi,cyano) branch in 372 trees containing homologues from Synechocystis (211 trees) or Prochlorococcus (165 trees). Keeping in mind that lateral gene transfer between free-living prokaryotes occurs to a great extent (54, 55), our data suggest that relative to the other two cyanobacteria studied here, Nostoc's overall complement of genes is more similar to that which the ancestor of plastids possessed.
Plastid Phylogeny, Gene Loss, and Gene Transfer.
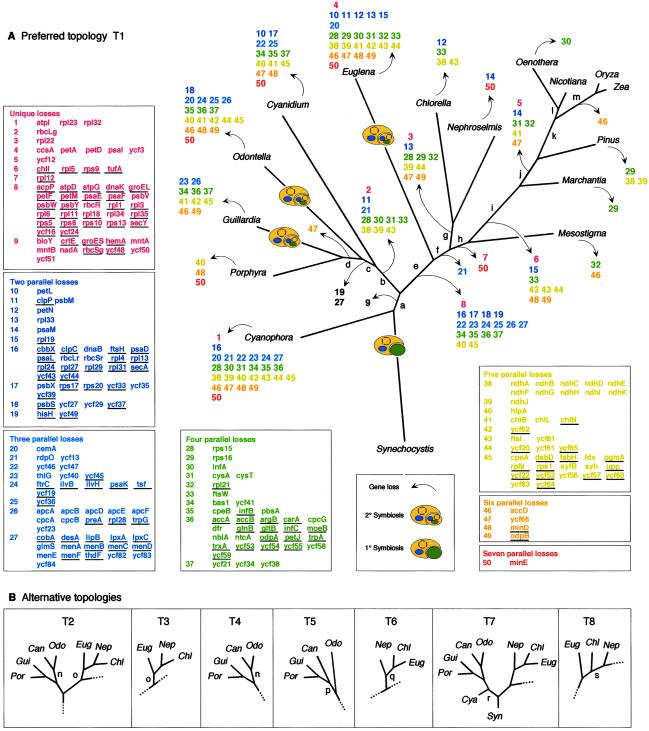
To view the gene transfer process from the standpoint of chloroplast genomes, we examined the 274 protein-coding genes that occur among 16 sequenced plastid genomes. Forty-four of the 274 plastid-encoded proteins are retained in all plastid genomes surveyed, leaving 230 that have been lost from the plastid in at least one lineage, 117 of which were detected as transferred nuclear homologues (Fig. 4 and Table 5, which is published as supporting information on the PNAS web site).
Fig 4.
Phylogeny of chloroplast genomes, gene loss, and gene transfer. (A) Topology preferred by NJ and LD for chloroplast genomes. Branch lengths were estimated with ML using the JTT-F matrix. 1° and 2° endosymbiotic events are indicated. Gene losses inferred at branches are indicated with arrows, designating numbered blocks of genes, which are expanded as gene lists at left and bottom. Genes for which a transferred nuclear homologue was found are underlined. Gene presence matrix and accession numbers are given in Table 5. Numbers of parallel losses are color-coded. Support for branches (lowercase letters), is given in Table 2. (B) Alternative topolgies T2–T8 detected in various subsets of the data and with various methods. Dotted lines indicate that the topology is otherwise identical to T1.
Reconstructing the process of gene migration from plastid genomes to the nucleus requires a plastid phylogeny, which we constructed with concatenated amino acid sequences (41 proteins and 8,308 sites per genome). Biased amino acid composition can dramatically affect the performance of various phylogenetic methods (34, 37). In all data sets investigated, the amino acid composition of the Cyanidium, Chlorella, Euglena, Nephroselmis, and Synechocystis proteins differed at P = 0.05 from the expected frequency distribution (34, 36). Rather than exclude these taxa, which would remove both the root and several important species, we included them and used a variety of methods.
Fig. 4 shows the topology T1 preferred by LD and NJ, in addition to alternative topologies found (T2–T8). Support for branches with different methods is summarized in Tables 2 and 3. Only four genomes, all of which possess significant amino acid composition bias, changed positions in the various analyses: Cyanidium and Euglena frequently, and Synechocystis and Nephroselmis once each. The LD result favoring T1 is important in this respect, because LD can effectively compensate for composition bias (34, 37). NJ also prefered T1, particularly with uncorrected distances (Table 2), whereas MP, quartet puzzling (QP), and ML did not. LD always found T1, except when constant sites were down-weighted by 90% and 100%, where it found T8.
Table 2.
Topologies supported
| Dataset
|
Preferred topology | ||||||
|---|---|---|---|---|---|---|---|
| LD | NJP | NJK | NJD | MP | ML | QP | |
| 8308-site | – | 1 | 1 | 1 | 2 | 2 | 2 |
| 7474-site | 1 | 1 | 1 | 1 | 4 | 2 | 3 |
| 5153-site | – | 1 | 1 | 1 | 2 | 6 | 2 |
| 4139-site | 8 | 1 | 1 | 5 | 4 | 7 | 3 |
T1–T8 shown in Fig. 4, topologies indicate LD, bootstrap consensus topology (NJP, NJK, NJD, MP) or ML result (ML, QP).
LD performed without gapped sites only.
Table 3.
Splits supported
| Split
|
Support for splits | ||||||
|---|---|---|---|---|---|---|---|
| LD | NJP | NJK | NJD | MP | ML | QP | |
| i | 1 | 100 | 100 | 100 | 100 | 100 | 100 |
| m | 2 | 100 | 100 | 100 | 100 | 100 | 100 |
| k | 3 | 100 | 100 | 100 | 100 | 100 | 100 |
| e | 4 | 100 | 100 | 100 | 100 | 100 | 100 |
| j | 5 | 100 | 100 | 100 | 100 | 100 | 100 |
| a | 6 | 100 | 100 | 100 | 83 | – | 98 |
| b | 7 | 100 | 100 | 100 | 100 | 100 | 100 |
| d | 8 | 100 | 100 | 100 | 94 | 98 | 100 |
| l | 9 | 100 | 100 | 100 | 100 | 100 | 100 |
| f | 10 | 100 | 86 | 98 | – | – | – |
| h | 11 | 96 | 99 | 99 | 79 | 89 | 96 |
| c | 12 | 98 | 70 | 83 | – | – | – |
| g | 13 | 93 | 84 | 95 | 81 | – | 66 |
| o | 14 | – | – | – | – | 85 | 65 |
| p | 15 | – | – | – | – | – | – |
| r | 16 | – | – | – | – | – | – |
| n | – | – | – | – | 98 | 99 | – |
| q | – | – | – | – | – | 64 | – |
Splits a–q in Fig. 4. Except columns LD and QP, numbers indicate the average bootstrap proportion across the four data sets with the given method. Average values <50 are indicated with a dash.
Splits listed in order of strength as determined through spectral analysis, e.g., branch “i” was the strongest split detected with LD.
Average of resampling estimated log-likelihood (RELL) bootstrap proportions.
Average of quartet puzzling support values.
The prasinophyte Mesostigma branched basal to land plants but above Chlorella and Nephroselmis in all analyses, in contrast to the position inferred previously by using 53 genes and fewer outgroups (32), but compatible with other recent findings (56). T1–T8 all indicate independent secondary symbioses (plastid origins from eukaryotic symbionts; refs. 2, 3, and 8) each for Euglena, and importantly for Guillardia and Odontella. Thus, we found no support for the chromalveolate concept (18, 57), which posits that the plastids of Odontella (a heterokont) and Guillardia (a cryptomonad) should stem from one and the same secondary endosymbiont (18). Four topologies were not excluded by the Shimodaira–Hasegawa test (39) at P = 0.05: T2, T4, T3, and T1, which are permutations of two positions for Cyanidium and Euglena, both of which possess strong amino acid composition bias. Branch “n” for Cyanidium (T2 in Fig. 4B) conflicts with branch “c” in T1 and had high BP values in ML and MP (Table 3), but not in NJ or in LD, which can effectively compensate for amino acid composition bias (34, 37).
Plotting the presence or absence of genes in chloroplast DNA onto T1 reveals that multiple parallel gene losses in independent lineages far outnumber unique losses. Under the unlikely premise that gene losses occurred in a minimum of events as shown in Fig. 4, the 583 parallel losses outnumber the 54 unique losses >10:1. Because of this abundant homoplasy, Dollo parsimony with the binary gene presence data gave incorrect trees. For example, in two of the four equally shortest trees found (458 losses), Pinus and Euglena were sisters. In Fig. 4, T2 was the shortest by the gene loss criterion (593 losses), T1 required 617 losses.
Conclusion
The present results indicate that the cyanobacterial heritage in plants extends well beyond the plastid and is manifest as ≈18% of the protein-coding genes in the Arabidopsis nuclear genome. The transition of a cyanobacterium into a plastid involved not only inheritance, but also many evolutionary innovations. Among the most important of these was the light-harvesting antenna complex of higher plants. A striking functional homologue of the higher plant antenna was recently discovered in cyanobacteria (58) that surprisingly consists of completely different light harvesting proteins than those in plastids and that was hence reinvented—not inherited—during plastid evolution.
Supplementary Material
Acknowledgments
We thank A. Roger, T. M. Embley, and M. Müller for critical comments, A. Trebst for discussions, and T. Preuten and D. Mainz for help with the chloroplast table. This work was supported by Japan Society for the Promotion of Science and Uehara Foundation (to M.H.), the Marsden Foundation (to D.P.), the Deutsche Forschungsgemeinschaft through SFB-TR/1 (to W.M.).
Abbreviations
ML, maximum likelihood
NJ, neighbor joining
NJP, NJ with uncorrected distances, NJD, NJ with Dayhoff distances
NJK, NJ with Kimura distances
LD, log determinant
MP, maximum parsimony
BP, bootstrap probability
QP, quartet puzzling
See commentary on page 11996.
References
- 1.Goksøyr J. (1967) Nature (London) 214, 1161. [DOI] [PubMed] [Google Scholar]
- 2.Douglas S. E. (1998) Curr. Opin. Gen. Dev. 8, 655-661. [DOI] [PubMed] [Google Scholar]
- 3.Delwiche C. W. (1999) Am. Nat. 154, S164-S177. [DOI] [PubMed] [Google Scholar]
- 4.Tomitani A., Okada, K., Miyashita, H., Matthijs, H. C. P., Ohno, T. & Tanaka, A. (1999) Nature (London) 400, 159-162. [DOI] [PubMed] [Google Scholar]
- 5.Herrmann R. G. (1997) in Eukaryotism and Symbiosis, eds. Schenk, H. E. A., Herrmann, R. G., Jeon, K. W. & Schwemmler, W. (Springer, Heidelberg), pp. 73–118.
- 6.Allen J. F. & Fornsberg, J. (2001) Trends Plant Sci. 6, 317-326. [DOI] [PubMed] [Google Scholar]
- 7.Wolfe G. R., Cunningham, F. X., Durnford, D., Green, B. R. & Gantt, E. (1994) Nature (London) 367, 566-568. [Google Scholar]
- 8.Douglas S., Zauner, S., Fraunholz, M., Beaton, M., Penny, S., Deng, L. T., Wu, X. N., Reith, M., Cavalier-Smith, T. & Maier, U.-G. (2001) Nature (London) 401, 1091-1096. [DOI] [PubMed] [Google Scholar]
- 9.Steiner J. M. & Löffelhardt, W. (2002) Trends Plant Sci. 7, 72-77. [DOI] [PubMed] [Google Scholar]
- 10.Osteryoung K. W. & McAndrew, R. S. (2001) Annu. Rev. Plant Phys. 52, 315-333. [DOI] [PubMed] [Google Scholar]
- 11.Heins L. & Soll, J. (1998) Curr. Biol. 8, R215-R217. [DOI] [PubMed] [Google Scholar]
- 12.Pfannschmidt T., Nilsson, A. & Allen, J. F. (1999) Nature (London) 397, 625-628. [Google Scholar]
- 13.Martin W. F. & Herrmann, R. G. (1998) Plant Physiol. 118, 9-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kubo N., Takano, M., Nishiguchi, M. & Kadowaki, K. (2001) Gene 271, 193-201. [DOI] [PubMed] [Google Scholar]
- 15.Abdallah F., Salamini, F. & Leister, D. (2000) Trends Plant Sci. 5, 141-142. [DOI] [PubMed] [Google Scholar]
- 16.Kaneko T., Sato, S., Kotani, H., Tanaka, A., Asamizu, E., Nakamura, Y., Miyajima, N., Hirosawa, M., Sugiura, M., Sasamoto, S., et al. (1996) DNA Res. 3, 109-136. [DOI] [PubMed] [Google Scholar]
- 17.Rujan T. & Martin, W. (2000) Trends Genet. 17, 113-120. [DOI] [PubMed] [Google Scholar]
- 18.Cavalier-Smith T. (2000) Trends Plant Sci. 5, 174-182. [DOI] [PubMed] [Google Scholar]
- 19.Martin W., Stoebe, B., Goremykin, V., Hansmann, S., Hasegawa, M. & Kowallik, K. V. (1998) Nature (London) 393, 162-165. [DOI] [PubMed] [Google Scholar]
- 20.Millen R. S., Olmstead, R. G., Adams, K. L., Palmer, J. D., Lao, N. T., Heggie, L., Kavanagh, T. A., Hibberd, J. M., Gray, J. C., Morden, C. W., et al. (2001) Plant Cell 13, 645-658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.The Arabidopsis Genome Initiative (2000) Nature (London) 408, 796-815. [DOI] [PubMed] [Google Scholar]
- 22.Meeks J. C., Elhai, J., Thiel, T., Potts, M., Larimer, F., Lamerdin, J., Predki, P. & Atlas, R. (2001) Photosynth. Res. 70, 85-106. [DOI] [PubMed] [Google Scholar]
- 23.Altschul S. F., Madden, T. L., Schaffer, A. A., Zhang, J. H., Zhang, Z., Miller, W. & Lipman, D. J. (1997) Nucleic Acids Res. 25, 3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Thompson J. D., Higgins, D. G. & Gibson, T. J. (1994) Nucl. Acids Res. 22, 4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Adachi J. & Hasegawa, M., (1996) MOLPHY (Institute of Statistical Mathematics, Tokyo), Computer Science Monographs, No. 28, Version 2.3.
- 26.Saitou N. & Nei, M. (1987) Mol. Biol. Evol. 4, 406-425. [DOI] [PubMed] [Google Scholar]
- 27.Emanuelsson O., Nielsen, H., Brunak, S. & von Heijne, G. (2000) J. Mol. Biol. 300, 1005-1016. [DOI] [PubMed] [Google Scholar]
- 28.Douglas S. E. & Penny, S. L. (1999) J. Mol. Evol. 48, 236-244. [DOI] [PubMed] [Google Scholar]
- 29.Glockner G., Rosenthal, A. & Valentin, K. (2000) J. Mol. Evol. 51, 382-390. [DOI] [PubMed] [Google Scholar]
- 30.Wakasugi T., Nagai, T., Kapoor, M., Sugita, M., Ito, M., Ito, S., Tsudzuki, J., Nakashima, K., Tsudzuki, T., Suzuki, Y., et al. (1997) Proc. Natl. Acad. Sci. USA 94, 5967-5972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Turmel M., Otis, C. & Lemieux, C. (1999) Proc. Natl. Acad. Sci. USA 96, 10248-10253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lemieux C., Otis, C. & Turmel, M. (2000) Nature (London) 403, 649-652. [DOI] [PubMed] [Google Scholar]
- 33.Hupfer H., Swiatek, M., Hornung, S., Herrmann, R. G., Maier, R. M., Chiu, W. L. & Sears, B. (2000) Mol. Gen. Genet. 263, 581-585. [DOI] [PubMed] [Google Scholar]
- 34.Lockhart P. J., Howe, C. J., Barbrook, A. C., Larkum, A. W. D. & Penny, D. (1999) Mol. Biol. Evol. 16, 573-576. [Google Scholar]
- 35.Nei M. & Kumar, S., (2000) Molecular Evolution and Phylogenetics (Oxford Univ. Press, Oxford).
- 36.Strimmer K. & von Haeseler, A. (1996) Mol. Biol. Evol. 13, 964-969. [Google Scholar]
- 37.Lockhart P. J., Steel, M. A., Hendy, M. D. & Penny, D. (1994) Mol. Biol. Evol. 11, 605-612. [DOI] [PubMed] [Google Scholar]
- 38.Hendy M. D. & Penny, D. (1993) J. Classif. 10, 5-24. [Google Scholar]
- 39.Shimodaira H. & Hasegawa, M. (1999) Mol. Biol. Evol. 16, 1114-1116. [Google Scholar]
- 40.Nei M. (1996) Annu. Rev. Genet. 30, 371-403. [DOI] [PubMed] [Google Scholar]
- 41.Salzberg S. L., White, O., Peterson, J. & Eisen, J. A. (2001) Science 292, 1903-1906. [DOI] [PubMed] [Google Scholar]
- 42.Stoebe B. & Kowallik, K. V. (1999) Trends Genet. 15, 344-347. [DOI] [PubMed] [Google Scholar]
- 43.Moreira D., Le Guyader, H. & Philippe, H. (2000) Nature (London) 405, 69-72. [DOI] [PubMed] [Google Scholar]
- 44.Woese C. R. (1987) Microbiol. Rev. 51, 221-271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Xiong J., Fischer, W. M., Inoue, K., Nakahara, M. & Bauer, C. E. (2000) Science 289, 1724-1730. [DOI] [PubMed] [Google Scholar]
- 46.Schütz M., Brugna, M., Lebrun, E., Baymann, F., Huber, R., Stetter, K. O., Hauska, G., Toci, R., Lemesle-Meunier, D., Tron, P., et al. (2000) J. Mol. Biol. 300, 663-675. [DOI] [PubMed] [Google Scholar]
- 47.Hansmann S. & Martin, W. (2000) Int. J. Syst. Evol. Microbiol. 50, 1655-1663. [DOI] [PubMed] [Google Scholar]
- 48.Wächtershäuser G. (1998) Syst. Appl. Microbiol. 21, 473-477. [Google Scholar]
- 49.Maeda S.-I. & Omata, T. (1997) J. Biol. Chem. 272, 3036-3041. [DOI] [PubMed] [Google Scholar]
- 50.Martin W. & Schnarrenberger, C. (1997) Curr. Genet. 32, 1-18. [DOI] [PubMed] [Google Scholar]
- 51.Hoiike T., Hamada, K., Kanaya, S. & Shinozawa, T. (2001) Nat. Cell Biol. 3, 210-214. [DOI] [PubMed] [Google Scholar]
- 52.Palenik B. & Haselkorn, R. (1992) Nature (London) 355, 265-267. [DOI] [PubMed] [Google Scholar]
- 53.Urbach E., Robertson, D. L. & Chisolm, S. W. (1992) Nature (London) 355, 267-270. [DOI] [PubMed] [Google Scholar]
- 54.Ochman H., Lawrence, J. G. & Groisman, E. S. (2000) Nature (London) 405, 299-304. [DOI] [PubMed] [Google Scholar]
- 55.Doolittle W. F. (1999) Science 284, 2124-2128. [DOI] [PubMed] [Google Scholar]
- 56.Karol K. G., McCourt, R. M., Cimino, M. T. & Delwiche, C. F. (2001) Science 294, 2351-2353. [DOI] [PubMed] [Google Scholar]
- 57.Fast N. M., Kissinger, J. C., Roos, D. S. & Keeling, P. J. (2001) Mol. Biol. Evol. 18, 418-426. [DOI] [PubMed] [Google Scholar]
- 58.Boekema E. J., Hifney, A., Yakushevska, A. E., Piotrowski, M., Keegstra, W., Berry, S., Michel, K. P., Pistorius, E. K. & Kruip, J. (2001) Nature (London) 412, 745-748. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.