Abstract
The number of genes in the human genome is still a controversial issue. Whereas most of the genes in the human genome are said to have been physically or computationally identified, many short cDNA sequences identified as tags by use of serial analysis of gene expression (SAGE) do not match these genes. By performing experimental verification of more than 1,000 SAGE tags and analyzing 4,285,923 SAGE tags of human origin in the current SAGE database, we examined the nature of the unmatched SAGE tags. Our study shows that most of the unmatched SAGE tags are truly novel SAGE tags that originated from novel transcripts not yet identified in the human genome, including alternatively spliced transcripts from known genes and potential novel genes. Our study indicates that by using novel SAGE tags as probes, we should be able to identify efficiently many novel transcripts/novel genes in the human genome that are difficult to identify by conventional methods.
One of the goals of human genome studies is to identify all of the genes in the human genome for further functional analysis of each gene. However, the correct number of genes in the human genome remains a controversial issue. Among various estimates, the Human Genome Project predicted the presence of 29,691 genes (ensembl Ver. 0.8) (1), and the Celera Human Genome Project estimated around 39,114 genes (2). Comparison between these two sets of data shows, however, that there is little overlap between novel genes predicted by these two studies (3). A recent study of human chromosomes 21 and 22 shows that the number of transcriptional units in the human genome could be an order of magnitude higher than the current estimates (4). We believe that the definitive determination of the correct number of genes in the human genome depends on the physical identification of all of the genes.
One of the major ways for physical gene identification is to analyze the expressed transcripts by using the expressed sequence tag (EST) approach (5–7). Data collected by large-scale EST projects over the past decades provide a significant contribution toward this goal (www.ncbi.nlm.nih.gov/GenBank/). Analysis of the EST data shows an inverted relationship over time between the total collected sequences and the proportion of novel sequences identified from them. That is, as the number of collected sequences increases, the rate of novel sequences identified from these sequences decreases. For example, 10.4% of ESTs collected in 1996 were novel sequences, whereas only 2.7% of ESTs collected in 1998 were novel sequences (8). The novel sequences in the 2,512,344 ESTs collected between 1998 and 2001 are about 1.6% (41,417) (http://www.ncbi.nlm.nih.gov/ncigap/lib_report.html). There are at least two possible explanations for these results: (i) most of the transcripts expressed in the human genome have been identified by use of the EST approach, or (ii) the identification of novel transcripts in the human genome has nearly reached the technical limitation of the EST approach, leaving many novel transcripts unidentified.
Recently, millions of short cDNA sequences named serial analysis of gene expression (SAGE) tags have been collected from human tissues by use of the SAGE method (refs. 9–12; http://www.ncbi.nlm.nih.gov/SAGE/). When the SAGE data are analyzed, a frequent observation is that a large number of SAGE tags do not match the existing expressed sequences (10, 13, 14).
Given the uncertainty of gene numbers, the saturation of novel EST identification, and the presence of a large number of unmatched SAGE tags in the human genome, we wondered whether it was possible that the unmatched SAGE tags originated from potentially novel transcripts or novel genes that were unidentified in the human genome. If this were the case, it would imply that a significant number of novel transcripts or genes have not been identified in the human genome. We performed experiments and analyzed the current SAGE tag database to test systematically this hypothesis. Data from these studies provide evidence that most of the unmatched SAGE tags have indeed originated from novel transcripts and potentially represent many novel genes. We present our data in this report.
Materials and Methods
Conversion of SAGE Tags into 3′ cDNA.
A high-throughput GLCI (generation of longer cDNA fragments from SAGE tags for gene identification) procedure was used for simultaneous conversion of a large number of SAGE tags into their corresponding 3′ cDNAs (15, 16). Briefly, the sense primers were designed on the basis of each SAGE tag (GGATCCCATGxxxxxxxxxx, where x represents the sequences of the tag), and the antisense primer used the sequence (ACTATCTAGAGCGGCCGCTT) in the 3′ end of all 3′ cDNAs incorporated from reverse transcription primers. PCR was performed by using the sense primer, antisense primer, and the same 3′ cDNA sample used previously for the SAGE analysis. The amplified products were cloned and sequenced. The sequences were matched to the GenBank Database (nonredundant and ESTs, http://www.ncbi.nlm.nih.gov/blast/). All unmatched sequences were matched to the human genomic sequences for sequence confirmation (http://genome.ucsc.edu/goldenPath/hgTracks.html; December 12, 2000, or April 1, 2001).
Conversion of 3′ cDNA into Full-Length cDNA.
The full-length cDNAs were generated by using a modified 5′ rapid amplification of cDNA ends (RACE) method (17). Briefly, total RNA and mRNA were isolated from human CD15+ bone marrow mononuclear cells with Trizol reagent (Invitrogen) and oligo(dT)25 beads (Dynal, Lake Success, NY). cDNA templates were synthesized with a modified RNA ligase-mediated-5′-RACE method by use of the GeneRacer Kit (Invitrogen) following the manufacturer's instructions, except that the regular oligo(dT) primer was replaced by the 5′ biotinylated, 3′ anchored oligo(dT) primers (5′ biotin-ATCTAGAGCGGCCGC-T16-A, G, CA, CG, and CC) to generate poly dA/dT(−) cDNAs (8). On the basis of the GLGI-amplified 3′ cDNA sequence, two reverse primers were designed for each gene: the primary reverse primer based on the sequence at the 3′ end of the cDNA and the nested reverse primer located upstream of the primary reverse primer. The full-length cDNAs were amplified by use of the GeneRacer 5′ primer (5′-CGACTGGAGCACGAGGACACTGA-3′) and primary reverse primers. If there were multiple products, nested PCR was performed by use of the Generacer 5′ nested primer (5′-GGACACTGACATGGACTGAAGGAGTA-3′) and the nested reverse primer. PCR products were cloned and sequenced. Only the sequences that contained the original SAGE tag and the 3′ cDNA sequences were used for analysis. The resulting cDNA sequences were matched to GenBank (NR and ESTs, http://www.ncbi.nlm.nih.gov/blast/) and the human genomic sequences (http://genome.ucsc.edu/goldenPath/hgTracks.html, December 12, 2000 or April 1, 2000). The putative ORFs and amino acids for the full-length sequences were determined by using the editseq program (DNAstar). blastp (http://www4.ncbi.nlm.nih.gov/blast/) was used to search SwissProt to identify potential domains in the putative ORFs.
Strand-Specific RT-PCR.
The strand-specific RT-PCR (18) was used for confirmation that the 3′ cDNAs and full-length cDNAs generated by GLGI and 5′ RACE were derived from targeted mRNA rather than from genomic DNA contamination. Briefly, two gene-specific primers located on both ends of a 3′ cDNA or full-length cDNA were designed. The first-strand cDNA was synthesized by use of strand-specific antisense primer. The specific cDNA was then amplified with the paired sense and antisense primers. The amplified products were confirmed by sequencing.
Determination of the Error Rate of SAGE Tags During the SAGE Process.
Two SAGE tag sequences were used for the analysis. SAGE tag A (GTGCACTGAG) was derived from the HLA-C gene (X58536), and SAGE tag B (TACCTGCAGA) was derived from the S100A8 gene (NM_002964). Four oligos were synthesized (Integrated DNA Technologies, Coralville, IA): A1 (5′-TTTGGATTTGCTGGTGCAGTACAACTAGGCTTAATAGGGACATGGTGCACTGAG-3′), A2 (5′-CTCAGTGCACCATGTCCCTATTAAGCCTAGTTGTACTGCACCAGCAAATCC-3′), B1 (TTTCTGCTCGAATTCAAGCTTCTAACGATGTACGGGGACATGTACCTGCAGA-3′) and B2 (TCTGCAGGTCATGTCCCCGTACATCGTTAGAAGCTTGAATTCGAGCAG-3′). Oligos A1 and A2 were annealed to form dimer A, and oligos B1 and B2 were annealed to form dimer B. Dimers A and B resemble the fragments containing SAGE tag A and SAGE tag B released from 3′ cDNA by BsmFI digestion in the beginning of the SAGE process. The SAGE steps from ditag formation and release, concatamerization, and cloning were performed following the SAGE protocol (19). Sequencing reactions were performed by use of the Big-Dye sequencing reagent (Applied Biosystems), and sequences were collected by use of an ABI 377 automatic sequencer (Applied Biosystems). The error rate was determined by comparison of the experimental SAGE tag A or B sequences with the original SAGE tag A or B sequences.
Analyses of SAGE Data.
SAGE tags from 101 human SAGE libraries were used for the analysis (ftp://ftp.ncbi.nih.gov/pub/sage/extr/, September 4, 2001; Table 6, which is published as supporting information on the PNAS web site, www.pnas.org). A series of java and c programs were designed for this analyses. The analyses included the following: (i) identification of unique SAGE tags from the total SAGE tags; (ii) determination of the relationship between SAGE tag collection and unique SAGE tag identification from different numbers of SAGE libraries (1, 1–10, 1–20, 1–40, 1–60, 1–80, and 1–101); (iii) determination of the frequency distribution of unique SAGE tags in different numbers of SAGE libraries (1, 1–10 and 1–101); (iv) determination of the origin of unique SAGE tags through matching of the human SAGE tag reference database SAGEmap_tag_ug-full-Nla3-Hs (ftp://ftp.ncbi.nih.gov/pub/sage/map/Hs/NlaIII/); (v) determination of the ratio between unique SAGE tags and the UniGene clusters; (vi) comparison of the proportion of mismatches of the low-frequency unmatched or matched SAGE tags to the high-frequency SAGE tags; (vii) analysis of the lower-frequency SAGE tags with single-base mismatch to the top 10 high-frequency SAGE tags in the SAGE library SAGE-Duke-H1126.
Results and Discussion
Classification of SAGE Tags in the SAGE Database.
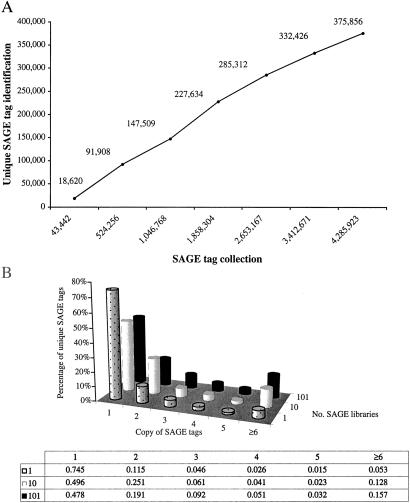
A total of 375,856 unique SAGE tags were identified from 4,285,923 SAGE tags collected from the 101 SAGE libraries of human origin. Comparison of the number of unique SAGE tags and the total number of SAGE tags shows that the number of unique SAGE tags continues to rise with an increase in total SAGE tags, suggesting that more unique SAGE tags would be identified if more SAGE tags were collected (Fig. 1A). Matching the 375,856 unique SAGE tags to existing expressed sequences in the UniGene database yields two subgroups: 141,599 unique SAGE tags match to sequences in 62,946 UniGene clusters (Unique SAGE tag: UniGene cluster = 2.25:1, because of alternative splicing and heterogeneity of polyadenylation sites, etc.; refs. 20, 21), and 234,257 unique SAGE tags do not have any match. The matched SAGE tags tend to be present in higher-copy numbers, whereas the unmatched tags tend to have lower-copy numbers. The origins of the unmatched SAGE tags are uncertain.
Fig 1.
Analyses of the SAGE tags collected from the 101 human SAGE libraries. (A) Relationship between SAGE tag collection and unique SAGE tag identification. The total SAGE tags and unique SAGE tags were extracted from 1, 10, 20, 40, 60, 80, and 101 human SAGE libraries and used for comparison. (B) Changes in frequency of unique SAGE tags with increasing SAGE tag numbers. Unique SAGE tags from 1, 10, and 101 SAGE libraries were divided into groups based on copy number; the rate of unique SAGE tags in each group is illustrated in the bar graph.
About 70% of the Unmatched SAGE Tags Are Derived from Novel Transcripts.
To determine whether the unmatched SAGE tags may be derived from novel transcripts from the human genome, we converted 1,183 human SAGE tags (699 matched and 484 unmatched) to their corresponding 3′ cDNAs experimentally by using a high-throughput GLGI method. Comparison of the 699 3′ cDNAs with their matched known expressed sequences confirmed that 624 (89%) of these 3′ cDNAs are correct. A database search for the 484 3′ cDNAs converted from unmatched SAGE tags showed that 324 (67%) of these 3′ cDNAs still had no match to existing expressed sequences (Table 1); 283 of 324 (87%) of these unmatched 3′ cDNAs were mapped to human genome sequences, of which only 42 and 49 had partial overlap with existing ESTs and predicted expressed sequences, respectively (Tables 7 and 8, which are published as supporting information on the PNAS web site). Not every 3′ cDNA could be amplified by the GLGI method, because a sense primer for the GLGI reaction is based solely on a SAGE tag that does not always provide ideal sequences as the primer for efficient amplification. Therefore, the actual rate of correct 3′ cDNA templates corresponding to the SAGE tags in the cDNA sample should be higher than 67% confirmed by the GLGI reaction.
Table 1.
Classification of GLGI-converted 3′ cDNAs from 484 unmatched SAGE tags
| Classification | Number of GLGI-amplified 3′ cDNAs | Percentage |
|---|---|---|
| Novel 3′ cDNAs | 324 | 66.9 |
| Match to known sequences | 76 | 15.7 |
| With internal CATGs | 14 | 2.9 |
| Single-base mismatch | 18 | 3.7 |
| Artifacts or no amplifications | 52 | 10.7 |
| Total | 484 | 100 |
These are the novel isoforms of known expressed sequences.
These cDNAs are generated by incomplete NlaIII digestion during SAGE library construction.
These 3′ cDNAs matched known expressed sequences, but there are single-base mismatches between their SAGE tag sequences.
Many Unmatched SAGE Tags Represent Novel Genes Not Yet Identified in the Human Genome.
To investigate further whether the 3′ cDNAs generated from novel SAGE tags were originated from novel genes, we converted 17 3′ cDNAs into 22 full-length cDNAs (Table 2). By use of strand-specific RT-PCR, we confirmed that all of these full-length cDNAs were derived from transcripts rather than genomic DNA contamination. Analysis of these full-length cDNA sequences showed that:
Table 2.
Twenty-two full-length cDNAs converted from unmatched SAGE tags
| SAGE tag
|
Copy
|
Full-length cDNA, bp
|
Genomic location (strand)
|
GenBank accession no.
|
Antisense to
|
Putative ORF, aa
|
Domain
|
No. exon
|
Partial overlap | |
|---|---|---|---|---|---|---|---|---|---|---|
| EST | Prediction | |||||||||
| ACCCCAAAGG | 1 | 1090 | 1q23.1:176093307-176144968 (+) | BM285379 | SSR2 | 268–555 (95) | No | 6 | No | No |
| ATGGCGCCTC | 7 | 649 | 19q13.32:58623673-58627513 (−) | BM285381 | RPL18 | 155–517 (120) | No | 7 | No | No |
| ATGGTTATGG | 2 | 1216 | 2q12.1:103454832-103456192 (+) | BM285382 | 381–518 (45) | No | 2 | No | No | |
| CAGATAACTA | 1 | 975 | 1q23.2:180430823-180431959 (−) | BM285383 | 584–841 (85) | No | 2 | No | No | |
| CCTTGAGCCA | 1 | 1048 | 1q23.1:176093570-176144976 (+) | BM285384 | SSR2 | 320–673 (117) | No | 6 | No | No |
| CTCTGTGGCA | 3 | 1040 | 1p36.13:18621891-18622912 (+) | BM285385 | 180–512 (110) | No | 1 | No | No | |
| GCCAACAGTG | 2 | 1359 | 2q12.1:103459588-103461351 (+) | BM285387 | 282–491 (69) | No | 3 | No | No | |
| GTGAAGATTC | 7 | 1091 | 19q13.32-33:60195829-60200846 (−) | BM285388 | CD37 | 640–972 (110) | No | 8 | No | No |
| GTTCACACGG | 3 | 949 | 15q15.3:40860800-40867421 (−) | BM285389 | B2M | 55–192 (45) | No | 4 | No | No |
| ACAGCTATGA | 1 | 819 | 4p16.1:6729161-6822034 (+) | BM285378 | 461–598 (45) | No | 3 | No | Yes | |
| GTTGAATGCT | 3 | 1679 | 12q12:43047796-43145951 (+) | BM285390 | 188–1288 (366) | No | 13 | No | Yes | |
| TTCTTCCTGT | 1 | 1329 | 11p13:36556073-36557467 (−) | BM285393 | 866–1057 (63) | No | 2 | No | Yes | |
| TTTTAGGTGG | 3 | 2014 | 20p13:1439442-1460217 (−) | BM285394 | 231–764 (177) | IG | 8 | No | Yes | |
| ACTAAGATTA | 2 | 1268 | 2p22.2:38076599-38088752 (+) | BM285380 | 68–925 (285) | G_patch | 9 | Yes | Yes | |
| TAACTGCATC | 2 | 1754 | 19q13.42:66283838-66312096 (−) | BM285391 | 121–903 (260) | No | 8 | Yes | Yes | |
| Isoform | ||||||||||
| CTTCTTGTAC | 3 | 1195 | 20p13:1516039-1522630 (−) | BM285386 | 540–779 (79) | No | 3 | Yes | Yes | |
| CTTCTTGTAC | 3 | 824 | 20P13:1516039-1523706 (−) | BQ635328 | 380–712 (110) | No | 4 | Yes | Yes | |
| CTTCTTGTAC | 3 | 923 | 20P13:1516039-1522630 (−) | BQ635329 | 268–507 (79) | No | 3 | Yes | Yes | |
| CTTCTTGTAC | 3 | 944 | 20P13:1516039-1522630 (−) | BQ635330 | 289–528 (79) | No | 3 | Yes | Yes | |
| CTTCTTGTAC | 3 | 963 | 20P13:1516039-1522630 (−) | BQ635331 | 308–547 (79) | No | 3 | Yes | Yes | |
| TTCTGGAAGT | 1 | 1570 | 1p34.2:45499099-45515163 (+) | BM285392 | 91–1227 (378) | KRAB | 5 | Yes | Yes | |
| TTCTGGAAGT | 1 | 1347 | 1P34.2:45499099-45515163 (+) | BQ635332 | 204–1004 (266) | No | 5 | Yes | Yes | |
We define an amplified cDNA as a nonmatch one if it does not match any expressed sequences, or it matches the antisense strand of genomic DNA, whose sense-strand are transcripted for known expressed sequences.
The full-length cDNAs were mapped to human genomic sequences through http://genome.ucsc.edu/goldenPath/hgTracks.html (December 12, 2000, or April 1, 2001).
The putative ORFs and amino acid length were determined with the EDITSEQ program (DNASTAR).
Conserved domains were determined using blastp (http://www4.ncbi.nlm.nih.gov/BLAST/) against SWISSPROT.
Yes refers to partial overlap of some exons between the full-length cDNAs and EST/predicted exons on the same strand of genomic DNA.
(i) Each sequence has a putative ORF, with exons and introns and typical or atypical exon–intron boundaries in the matched genomic sequences.
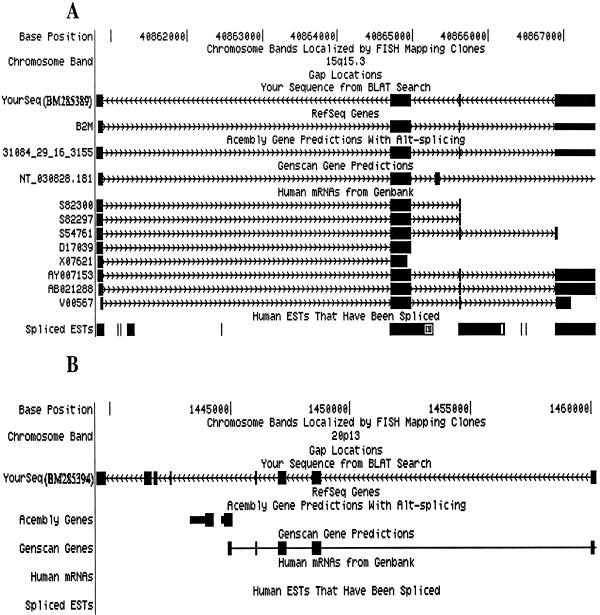
(ii) Thirteen sequences do no match any ESTs, and 9 of these 13 do not match any predicted exons. Among these sequences, two (BM285382 and BM285387) matched introns of the IL18RAP gene (NM_003853). Five sequences (BM285379, BM285381, BM285384, BM285388, and BM285389) matched to the location of known genes (SSR2, RPL18, SSR2 alternative splicing, CD37, and β-2 microglobin), but were transcribed from the antisense strand (Table 2). For example, the full-length cDNA derived from SAGE tag GTTCACACGG present in three copies matches exactly the antisense strand of the β-2 microglobin gene, whereas the SAGE tag GTTGTGGTTA on the sense strand for β-2 microglobin gene was present in 853 copies in the same SAGE library (Fig. 2A). Using semiquantitative strand-specific RT-PCR, we observed that both the sense and antisense transcripts were amplified although the signals from the β-2 microglobin appeared earlier than the antisense signal, reflecting the quantitative differences in the original transcripts. Similar results were found for four other antisense sequences converted from the corresponding SAGE tags. Considering the functional importance of the β-2 microglobin in the regulation of antigen presentation in the immune response, the presence of the in vivo antisense transcript might be related to a regulatory function using the mechanism such as antisense/RNA interference (22). A misconception in gene identification is that the transcripts from both the sense and the antisense strands of the genomic DNA represent the same gene, as reflected in the UniGene database where sense and antisense sequences are grouped into a single UniGene cluster, with only seven UniGene clusters clearly marked as antisense (opposite) in the human UniGene database (http://www.ncbi.nlm.nih.gov/UniGene). We think that the antisense transcript is likely to represent novel genes whose function may or may not be related to regulation of the expression for the genes transcribed from the sense strand.
Fig 2.
Genomic confirmation of novel full-length cDNAs converted from novel SAGE tags. Full-length cDNAs were generated from 3′ cDNAs converted from novel SAGE tags and matched to human genomic sequences. (A) The full-length cDNA originating from novel SAGE tag GTTCACACGG matches exactly the antisense strand of the β-2 microglobin (B2M) gene. (B) Match of a full-length cDNA originating from novel SAGE tag TTTTAGGTGG partially overlapping with predicted exons, but with no matches with known mRNA or EST.
(iii) Four sequences have partial overlap with ESTs, and eight have partial overlap with predicted exons. However, these full-length cDNAs have far more novel exons than these partially overlapped ESTs or predicted genes (Fig. 2B).
(iv) Five sequences are isoforms from two genes, partially overlapped with ESTs and predicted exons.
Most of the Unmatched SAGE Tags Are Not Generated by Experimental Errors.
A regular SAGE tag consists of 10 bases. A single-base difference between two SAGE tags will define these two SAGE tags as being different. A current popular interpretation of the unmatched SAGE tags is that they are largely generated by sequencing errors (6.8–10% per SAGE tag). Because of this concern, the majority of unmatched SAGE tags, especially for the low-frequency SAGE tags, are excluded from further analysis (10, 23, 24). The assumption of the high error rate in SAGE tags is based on estimates using early EST sequences collected before 1996 (25). Since then, the quality of collected sequences has improved significantly because of advances in sequencing technologies (1, 26–29). In fact, most SAGE tag sequences have been collected after 1998 (refs. 10, 12, 23; ftp://ncbi.ftp.nih.gov/pub/sage/extr/).
We consider that the rate of sequencing error in these SAGE tag sequences may be lower than the estimates. To support our hypothesis, we performed experiments to determine the actual error rate covering nearly the whole SAGE process until the sequencing collection. The results of three independent experiments show that the error rate is about 1.67% per SAGE tag (Table 3 Upper), with the erroneous nucleotides being rather randomly distributed along the tag sequences (Table 3 Lower). Although the actual error rate might vary between different SAGE experiments, it is unlikely that the error rates in most SAGE libraries would be up to five times higher than our results. We also observed that all of the erroneous SAGE tags contain only a single-base error (Table 3 Lower), suggesting that the tags containing two or more base errors would be much more rare than those containing a single-base error.
Table 3.
Determination of experimental error rate during SAGE process
| Summary of error rate in three independent SAGE experiments | |||
|---|---|---|---|
| Experiment | Total collected SAGE tags | Erroneous tags | Error rate, % |
| 1 | 1,200 | 20 | 1.67 |
| 2 | 1,086 | 18 | 1.66 |
| 3 | 1,062 | 18 | 1.69 |
| Total | 3,348 | 56 | 1.67 |
| Distribution of error nucleotides | |||
| Wild-type tag A | Detected number | Wild-type tag B | Detected number |
| GTGCACTGAG | 1,638 | TACCTGCAGA | 1,654 |
| Erroneous tag A | Erroneous tag B | ||
| ATGCACTGAG | 2 | CACCTGCAGA | 2 |
| GCGCACTGAG | 4 | TTCCTGCAGA | 2 |
| GTACACTGAG | 2 | TACTTGCAGA | 2 |
| GTGCTCTGAG | 2 | TACCAGCAGA | 2 |
| GTGCAATGAG | 2 | TACCTACAGA | 2 |
| GTGCATTGAG | 12 | TACCTTCAGA | 1 |
| GTGCACCGAG | 2 | TACCTGTAGA | 2 |
| GTGCACTGAA | 6 | TACCTGCTGA | 2 |
| GTGCACTGAC | 2 | TACCTGCAAA | 2 |
| GTGCACTGAT | 2 | TACCTGCAGG | 3 |
We analyzed the SAGE tags in the current SAGE database to determine whether the unmatched SAGE tags were related to sequencing errors. We focused on the analysis of a single-base error for the reason described above. A SAGE tag has 10 bases. If the error rate is M per base, the probability (P) that a single-base-error SAGE tag with a specific nucleotide replacing a wild-type nucleotide at a specific position will follow the formula P = [(1 − M)9M/3], in which M/3 represents the probability of a wild-type nucleotide replaced by one of the other three nucleotides at its position (for example, a wild-type nucleotide A can be replaced by G, C, or T), whereas (1 − M)9 represents the probability that the other nine bases in this tag would be error-free. If the average sequencing error rate is about 2% per SAGE tag as our results indicated, in which M = 0.2%, then P = (1–0.002)9 × 0.002/3 = 6.55 × 10−4 = 1/1,527. Even if the estimated sequencing error rate of about 10% per SAGE tag (23, 24) is used for the calculation, in which M = 1%, then P = (1–0.01)9 × 0.01/3 = 3.04 × 10−3 = 1/329. These probabilities mean that a collection of 1,527-fold or at least 329-fold more tags would be needed for detecting another erroneous tag with the same error derived from the same wild-type SAGE tag. We observed that the number of SAGE tags increased only 12.1-fold from the first SAGE library to the 10 SAGE libraries and 8.2-fold from the 10 to the 101 SAGE libraries (524,256/43,442 and 4,285,923/524,256, respectively; Fig. 1A), far less than 329- or 1,527-fold. If a high error does exist and the single-copy SAGE tags in the 1 or 10 SAGE libraries were largely generated from sequencing errors, then most of these single-copy error SAGE tags will stay as a singleton in this scale of SAGE tag collection, and the percentage of single-copy tags should increase significantly when the collection of SAGE tags increases from 1 to 10 libraries and from 10 to 101 libraries. Analysis of the SAGE tags collected from the 101 human SAGE libraries shows, however, that the percentage of single-copy unique SAGE tags decreases with increased collection of SAGE tags (Fig. 1B). For example, the rate of single-copy tags is 75% in the first SAGE library, but it is only 50% in the 10 SAGE libraries and only 48% in the 101 SAGE libraries, suggesting that most of the single-copy SAGE tags are not generated from experimental errors.
We further analyzed the rate of mismatches between the low- and higher-frequency tags of matched and unmatched tag sets to investigate whether there was a difference between these two sets of SAGE tags. If the unmatched low-frequency SAGE tags contain more erroneous forms derived from the higher-frequency SAGE tags than do the matched low-frequency SAGE tags, the rate of mismatch between the unmatched low-frequency SAGE tags and the higher-frequency tags should be higher than that between the matched low- and the higher-frequency SAGE tags. However, the results showed that the rates of mismatch were similar between the unmatched and the matched low-frequency SAGE tags, indicating that the unmatched SAGE tags do not have a higher probability of errors than do the matched SAGE tags (Table 4). Although more than 60% of low-frequency SAGE tags have single-base mismatch with higher-frequency SAGE tags (Table 4), this does not mean that these low-frequency SAGE tags originated from higher-frequency SAGE tags because of sequencing errors (12). We randomly selected a SAGE library (SAGE-Duke-H1126) from the 101 libraries and analyzed its top 10 SAGE tags and the low-frequency tags with single-base mismatches to each of the top 10 tags. The result shows that 97 and 83% copies of the mismatched low-frequency tags perfectly match mRNAs and genomic DNA sequences, respectively (Table 9 Upper and Lower, which is published as supporting information on the PNAS web site). Furthermore, it is possible that SAGE tags without matches could be due to the presence of single-nucleotide polymorphism sites or the incomplete gaps in human genomic DNA sequences where the gene contributing these SAGE tag may be located, rather than errors. An example is the RPL38 (Hs.2017) identified by SAGE tag GCGACGAGGC with 279 copies. This gene has been well studied, and its mRNA sequence is included in the RefSeq database (www.ncbi.nlm.nih.gov/LocusLink/refseq.html). However, this mRNA sequence does not match the human genomic sequence. Further analysis of 1,500 unmatched single-copy SAGE tags randomly selected from the first 10 libraries reveals that 75% of these single-copy SAGE tags become higher-copy SAGE tags in the 101 SAGE libraries (Table 5). These analyses support the concept that experimental errors contribute a much lower fraction in the unmatched SAGE tags than currently estimated.
Table 4.
Single-base mismatch between unmatched/matched low-frequency tags and high-frequency tags
| SAGE tags | LF tags/HF tags | Mismatched LF SAGE tags (%) | Rate of HF tags with mismatches/mismatched LF tags |
|---|---|---|---|
| 1–2 copies/>2 copies | |||
| Unmatched SAGE tags | 31,812/23,269 | 19,615 (62) | 1.7 (33,560/19,615) |
| Matched SAGE tags | 36,827/23,269 | 24,055 (65) | 1.9 (45,322/24,055) |
| 3–4 copies/>4 copies | |||
| Unmatched SAGE tags | 1,802/13,841 | 1,150 (64) | 1.6 (1,829/1,150) |
| Matched SAGE tags | 7,626/13,841 | 4,548 (60) | 1.9 (8,636/4548) |
All unique SAGE tags were extracted from the first 10 libraries and matched to SAGEmap database.
LF: low-frequency; HF: high-frequency. The low-frequency SAGE tags were divided into unmatched SAGE tags and matched SAGE tags; the high-frequency SAGE tags included both unmatched and matched SAGE tags.
Table 5.
Distribution of 1,500 unmatched single-copy tags from 1 to 101 SAGE libraries
| Tag copy
|
SAGE libraries | ||||||
|---|---|---|---|---|---|---|---|
| 1 | 10 | 20 | 40 | 60 | 80 | 101 | |
| 1 | 1,500 (100) | 797 (53) | 679 (45) | 556 (37) | 479 (32) | 416 (27) | 374 (25) |
| 2 | 0 | 467 (31) | 461 (31) | 437 (29) | 408 (27) | 402 (27) | 361 (24) |
| 3 | 0 | 123 (8) | 136 (9) | 178 (12) | 182 (12) | 174 (12) | 192 (13) |
| 4 | 0 | 44 (3) | 77 (5) | 78 (5) | 104 (7) | 116 (8) | 117 (8) |
| 5 | 0 | 20 (1) | 29 (2) | 49 (3) | 68 (5) | 71 (5) | 88 (6) |
| 6 to 10 | 0 | 23 (2) | 67 (5) | 109 (7) | 114 (8) | 131 (9) | 146 (10) |
| 11 to 50 | 0 | 23 (2) | 40 (3) | 74 (5) | 120 (8) | 156 (10) | 180 (12) |
| over 50 | 0 | 3 (0) | 11 (1) | 19 (1) | 25 (2) | 34 (2) | 42 (3) |
The 1,500 unique SAGE tags were randomly selected from unmatched single-copy tags in the first 10 SAGE libraries.
The numbers in parentheses are the percentage of the SAGE tags in the 1,500 SAGE tags and are rounded to 1.
On the basis of these analyses, we conclude that most of the unmatched SAGE tags are novel SAGE tags derived from novel transcripts. These novel transcripts may originate from the alternatively spliced transcripts (20, 21); they may also belong to the noncoding transcripts that have multiple regulatory functions (4, 30, 31). A significant number of these novel transcripts may represent novel genes not yet identified in the human genome. Recent experimental data using different approaches also show similar results. For example, the number of genes estimated by using the longSAGE approach suggests that the number of genes in the human genome could be doubled from current estimates (32, 33). The analysis of transcriptional units in human chromosomes 21 and 22 using oligo-microarray method shows that the number of transcribed sequences in the human genome could be an order of magnitude greater than current estimates (4), although full-length sequences will be needed to provide the final proof.
Why Such a Large Number of Novel Transcripts Have Not Been Identified.
It is interesting that we still identify a large number of novel transcripts in the human cells, despite the decade-long effort in identifying the genes in the human genome. There are basically two ways to identify genes: computational prediction based on genomic sequences, etc., and experimental identification through analysis of the expressed transcripts. It has been shown that the current computational tools are inadequate for gene prediction, particularly for complex genomes such as the human genome (34), due largely to the high signal-to-noise ratio between coding and noncoding sequences (1). For experimental identification, a major barrier is the issue of redundancy of gene expression. Unlike the genomic sequencing in which the DNA sequences are rather evenly distributed in the genome and can be identified through increased sequencing coverage several-fold, the identification of genes through analysis of the expressed transcripts has to face the issue of redundant expression, in which a few genes express at higher levels contributing a large portion of the total transcripts, whereas most of the genes express at lower levels and account for only a small portion of the total transcripts. Although the approach of subtraction/normalization can certainly decrease the redundancy by reducing the highly expressed transcripts, many of the lower-expressed transcripts could be lost because of factors such as cross-hybridization between unrelated transcripts (8). In contrast, SAGE collects a short tag from a transcript and forms a concatemer of multiple tags from many transcripts for a single sequencing reaction, leading to a significant decrease in the sequencing scale. Such an approach overcomes the obstacle of redundancy and makes it possible to identify transcripts expressed from high to low levels without exposing the samples to subtraction/normalization. By use of methods such as GLGI and 5′ rapid amplification of cDNA ends, novel SAGE tags can be converted back to their corresponding 3′ cDNAs and full-length cDNAs. Applying the approach of converting novel SAGE tag to longer sequence should significantly accelerate the rate of discovery of novel transcripts/novel genes in the human genome. The same approach should also be applicable to gene identification in other eukaryotic genomes.
Supplementary Material
Acknowledgments
We thank G. Riggins, C.-I. Wu, R. Watertson, E. Green, and E. H. Margulies for critical reading of and valuable comments on the manuscript. This work was supported by National Institutes of Health Grant CA84405 (J.D.R.), This work was supported by National Institutes of Health Grant CA84405 (J.D.R.), a Fellowship from the University of Chicago Committee on Cancer Biology (J.C.), and the G. Harold and Lelia Y. Mathers Charitable Foundation (S.M.W.).
Abbreviations
EST, expressed sequence tag
SAGE, serial analysis of gene expression
GLGI, generation of longer cDNA fragments from SAGE tags for gene identification
References
- 1.Lander E. S., Linton, L. M., Birren, B., Nusbaum, C., Zody, M. C., Baldwin, J., Devon, K., Dewar, K., Doyle, M., FitzHugh, W., et al. (2001) Nature (London) 409, 860-921. [DOI] [PubMed] [Google Scholar]
- 2.Venter J. C., Adams, M. D., Myers, E. W., Li, P. W., Mural, R. J., Sutton, G. G., Smith, H. O., Yandell, M., Evans, C. A., Holt, R. A., et al. (2001) Science 291, 1304-1351. [DOI] [PubMed] [Google Scholar]
- 3.Hogenesch J. B., Ching, K. A., Batalov, S., Su, A. I., Walker, J. R., Zhou, Y., Kay, S. A., Schultz, P. G. & Cooke, M. P. (2001) Cell 106, 413-415. [DOI] [PubMed] [Google Scholar]
- 4.Kapranov P., Cawley, , Simon, E., Drenkow, J., Bekiranov, S., Strausberg, R. L., Fodor, S. P. A. & Gingeras, T. R. (2002) Science 296, 916-919. [DOI] [PubMed] [Google Scholar]
- 5.Adams M. D., Kelley, J. M., Gocayne, J. D., Dubnick, M., Polymeropoulos, M. H., Xiao, H., Merril, C. R., Wu, A., Olde, B., Moreno, R. F., et al. (1991) Science 252, 1651-1656. [DOI] [PubMed] [Google Scholar]
- 6.Bonaldo M. F., Lennon, G. & Soares, M. B. (1996) Genome Res. 6, 791-806. [DOI] [PubMed] [Google Scholar]
- 7.Strausberg R. L., Dahl, C. A. & Klausner, R. D. (1997) Nat. Genet. 15, 415-416. [DOI] [PubMed] [Google Scholar]
- 8.Wang S. M., Fears, S. C., Zhang, L., Chen, J. J. & Rowley, J. D. (2000) Proc. Natl. Acad. Sci. USA 97, 4162-4167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Velculescu V. E., Zhang, L., Vogelstein, B. & Kinzler, K. W. (1995) Science 270, 484-487. [DOI] [PubMed] [Google Scholar]
- 10.Velculescu V. E., Madden, S. L., Zhang, L., Lash, A. E., Yu, J., Rago, C., Lal, A., Wang, C. J., Beaudry, G. A., Ciriello, K. M., et al. (1999) Nat. Genet. 23, 387-388. [DOI] [PubMed] [Google Scholar]
- 11.Lal A., Lash, A. E., Altschul, S. F., Velculescu, V., Zhang, L., McLendon, R. E., Marra, M. A., Prange, C., Morin, P. J., Polyak, K., et al. (1999) Cancer Res. 59, 5403-5407. [PubMed] [Google Scholar]
- 12.Boon K., Osorio, E. C., Greenhut, S. F., Schaefer, C. F., Shoemaker, J., Polyak, K., Morin, P. J., Buetow, K. H., Strausberg, R. L., De Souza, S. J. & Riggins, G. J. (2002) Proc. Natl. Acad. Sci. USA 99, 11287-11292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee S., Zhou, G., Clark, T., Chen, J., Rowley, J. D. & Wang, S. M. (2001) Proc. Natl. Acad. Sci. USA 98, 3340-3345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhou G., Chen, J., Lee, S., Clark, T., Rowley, J. D. & Wang, S. M. (2001) Proc. Natl. Acad. Sci. USA 98, 13966-13971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen J., Rowley, J. D. & Wang, S. M. (2000) Proc. Natl. Acad. Sci. USA 97, 349-353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen J., Lee, S., Zhou, G. & Wang, S. M. (2002) Genes Chromosomes Cancer 33, 252-261. [DOI] [PubMed] [Google Scholar]
- 17.Maruyama K. & Sugano, S. (1994) Gene 138, 171-174. [DOI] [PubMed] [Google Scholar]
- 18.Lee J. T., Davidow, L. S. & Warshawsky, D. (1999) Nat. Genet. 21, 400-404. [DOI] [PubMed] [Google Scholar]
- 19.Lee S., Chen, J., Zhou, G. & Wang, S. M. (2001) BioTechniques 31, 348-354. [DOI] [PubMed] [Google Scholar]
- 20.Mironov A. A., Fickett, J. W. & Gelfand, M. S. (1999) Genome Res. 9, 1288-1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pauws E., van Kampen, A. H., van de Graaf, S. A., de Vijlder, J. J. & Ris-Stalpers, C. (2001) Nucleic Acids Res. 29, 1690-1694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brantl S. (2002) Biochim. Biophys. Acta 1575, 15-25. [DOI] [PubMed] [Google Scholar]
- 23.Lash A. E., Tolstoshev, C. M., Wagner, L., Schuler, G. D., Strausberg, R. L., Riggins, G. J. & Altschul, S. F. (2000) Genome Res. 10, 1051-1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stollberg J., Urschitz, J., Urban, Z. & Boyd, C. D. (2000) Genome Res. 10, 1241-1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hillier L. D., Lennon, G., Becker, M., Bonaldo, M. F., Chiapelli, B., Chissoe, S., Dietrich, N., DuBuque, T., Favello, A., Gish, W., et al. (1996) Genome Res. 6, 807-828. [DOI] [PubMed] [Google Scholar]
- 26.Lee L. G., Spurgeon, S. L., Heiner, C. R., Benson, S. C., Rosenblum, B. B., Menchen, S. M., Graham, R. J., Constantinescu, A., Upadhya, K. G. & Cassel, J. M. (1997) Nucleic Acids Res. 25, 2816-2822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rosenblum B. B., Lee, L. G., Spurgeon, S. L., Khan, S. H., Menchen, S. M., Heiner, C. R. & Chen, S. M. (1997) Nucleic Acids Res. 25, 4500-4504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ewing B. & Green, P. (1998) Genome Res. 8, 186-194. [PubMed] [Google Scholar]
- 29.Ewing B., Hillier, L., Wendl, M. C. & Green, P. (1998) Genome Res. 8, 175-185. [DOI] [PubMed] [Google Scholar]
- 30.Erdmann V. A., Barciszewska, M. Z., Hochberg, A., Groot, N. & Barciszewski, J. (2001) Cell Mol. Life Sci. 58, 960-977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kelley R. L. & Kuroda, M. I. (2000) Cell 103, 9-12. [DOI] [PubMed] [Google Scholar]
- 32.Saha S., Sparks, A. B., Rago, C., Akmaev, V., Wang, C. J., Vogelstein, B., Kinzler, K. W. & Velculescu, V. E. (2002) Nat. Biotechnol. 20, 508-512. [DOI] [PubMed] [Google Scholar]
- 33.Shoues B. (2002) Science 295, 1457. [Google Scholar]
- 34.Guigo R., Agarwal, P., Abril, J. F., Burset, M. & Fickett, J. W. (2000) Genome Res. 10, 1631-1642. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.