Abstract
T helper 1 (TH1) differentiation and IFN-γ production are crucial in cell-mediated immune responses. IL-12 is an important regulator of this process and mediates its effects through signal transducer and activator of transcription 4 (STAT4). IFN-γ production is also regulated by the p38 mitogen-activated kinase pathway, although the mechanisms are ill-defined. We show here that GADD45-β and GADD45-γ can induce STAT4 S721 phosphorylation via the MKK6/p38 pathway. Thus, STAT4 could be a target that accounts for the defects in cell-mediated immunity associated with perturbations in the p38 pathway. To investigate the biological significance of STAT4 S721 phosphorylation, we reconstituted primary spleen cells from STAT4-deficient mice with wild-type and mutated STAT4, by using a retroviral gene transduction. We demonstrated that expression of wild-type STAT4, but not the S721A mutant, restored normal TH1 differentiation and IFN-γ synthesis. The inability of STAT4 S721 to restore IFN-γ production was not caused by decreased IL-12R expression because the STAT4 S721 mutant also failed to restore IFN-γ production in STAT4-deficient IL-12Rβ2 transgenic cells. Importantly, STAT4 S721A-transduced cells showed normal proliferative response to IL-12, illustrating that serine phosphorylation is not required for IL-12-induced proliferation. Additionally, the results imply the existence of STAT4 serine phosphorylation-dependent and -independent target genes. We conclude that phosphorylation of STAT4 on both tyrosine and serine residues is important in promoting normal TH1 differentiation and IFN-γ secretion.
Interferon-γ (IFN-γ) is critical for host defense against many pathogens and contributes to the pathogenesis of autoimmune disease (1). Because of its importance in health and disease, the regulation of IFN-γ production in T lymphocytes and other cells has been intensively studied. Although the transcriptional control of the IFN-γ gene is still incompletely understood, it is nonetheless clear that a variety of cytokines, signaling molecules, and transcription factors contribute to IFN-γ production and T helper 1 (TH1) development (2–7). For instance, multiple lines of evidence have implicated the p38 mitogen-activated protein kinase (MAPK) pathway in this process (8). That is, a selective p38 inhibitor blocks IFN-γ production, and transgenic mice expressing dominant negative p38 show impaired TH1 differentiation (9). Upstream of p38 are the kinases MKK6 and MKK3, and MKK3-deficient mice have defective cell-mediated immunity (10). How the MKK3/6/p38 pathway is activated is incompletely understood but the small guanine nucleotide binding protein Rac2 is thought to be one intermediate. Accordingly, mice lacking Rac2 also have defective TH1 development and IFN-γ gene expression (11). GADD45 members have also been reported to be upstream activators of p38 MAPK, and overexpression of GADD45-β augments IFN-γ production (12), whereas GADD45-γ−/− mice have impaired IFN-γ production (13). However, despite the abundance of evidence supporting the role of the p38 MAPK pathway in controlling IFN-γ production, candidate p38 substrates that explain this regulation are lacking.
Cytokines are also important contributors to IFN-γ regulation, IL-12 being the key cytokine that promotes TH1 differentiation (14, 15). The importance of this cytokine is most clearly evidenced in IL-12- and IL-12 receptor (IL-12R)-deficient mice (16–18) and humans (19–21), which exhibit increased susceptibility to intracellular pathogens. IL-12 stimulation results in the activation of the Janus kinases Jak2 and Tyk2, which in turn phosphorylate IL-12R, providing docking sites for the transcription factor STAT4 (signal transducers and activators of transcription 4) (22, 23). The critical functions of STAT4 are illustrated by the fact that most IL-12 functions are disrupted in STAT4−/− mice, their phenotype being concordant with IL-12 and IL-12R deficiency in mice and humans; these mice have markedly impaired IFN-γ production and TH1 differentiation (24, 25). Receptor-bound STAT4 is phosphorylated on tyrosine 693 by the Jaks, promoting STAT dimerization, translocation to the nucleus, and regulation of gene expression (26, 27). Whether STAT4 contributes directly to IFN-γ gene regulation remains controversial, although potential STAT4 binding sites have been reported in the first intron and promoter of the IFN-γ gene (28, 29).
Because of the evidence pointing to the importance of the p38 pathway and STAT4 in TH1 differentiation and IFN-γ production, we have looked for a link between p38 and STAT4. Recently, we and others have reported that IL-12 activates p38 (30, 31). Moreover, we showed that serine 721 of STAT4 is also phosphorylated upon IL-12 stimulation in a MKK6/p38 MAPK-dependent manner. Furthermore, mutation of S721 or inhibition of p38 MAPK diminished STAT4-mediated transactivation of a reporter construct (30). Together these data suggested that STAT4 is a candidate p38 MAPK target that could regulate IFN-γ production. However, it was not known whether STAT4 serine phosphorylation was relevant for IL-12-dependent TH1 differentiation and IFN-γ production. Therefore, we set out to clarify the biological roles of STAT4 serine phosphorylation. Herein, we show that STAT4 S721 phosphorylation is important for optimal IFN-γ production and TH1 differentiation, and that Rac and GADD45-β and GADD45-γ, intermediates upstream of p38 MAPK involved in TH1 differentiation, can induce STAT4 serine phosphorylation. Thus, STAT4 S721 phosphorylation may be one important mechanism that links the p38 pathway with IFN-γ gene regulation.
Materials and Methods
Reagents.
Mouse IL-12 and IL-18 were purchased from R & D Systems. Human IL-2 was from Craig Reynolds (National Cancer Institute, Frederick, MD). Anti-STAT4 and anti-phosphoserine STAT3 antibodies have been described (30). Anti-phospho STAT4 antibody was from Zymed, anti-phospho p38 and phospho MKK3/6 antibodies were from Cell Signaling Technology (Beverly, MA), and anti-p38 antibody was from Transduction Laboratories (Lexington, KY). Antibodies to CD3, CD28, FITC-CD4, PE-CD62L, IFN-γ, and isotype-matched antibody were purchased from PharMingen. Phorbol myristate acetate and ionomycin were from Sigma.
Cell Preparation.
STAT4-deficient mice, BALB/c mice, and C57BL/6 mice were purchased from The Jackson Laboratory. IL-12Rβ2 transgenic mice and IL-12Rβ2 transgenic/STAT4-deficient mice were generated as described (32). Naive CD4+ T cells were purified by fluorescence-activated cell sorting for CD4+CD62L+ populations, as described (32). Purity of CD4+CD62L+ cells was >99%.
Transfection, Immunoprecipitation, Immunoblotting, and Electrophoretic Mobility-Shift Assay (EMSA).
293T cells were transfected by using Fugene (Roche Diagnostics) and harvested after 24 h for analysis. PCDNA3-STAT4, pCDNA3-STAT4-S721A, pCEFL-MKK6, pCEFL-MKK6KR, PCDNA3-GADD45-β, and pCDNA3-GADD45-γ have been described (30, 33, 34). Immunoprecipitation, immunoblotting, and EMSA were carried out as described (26, 35).
Retroviral Vectors and Transduction.
S721A and Y693F mutants of STAT4 have been described (30). Wild type, mutated STAT4, GADD45-β, and GADD45-γ were PCR-amplified with Pfu Turbo DNA Polymerase (Stratagene). Gel-purified PCR products were subcloned into PCR4-Topo vector (Invitrogen), and then subcloned into the PBMN-I-GFP retroviral vector, provided by Garry Nolan (Stanford University, Stanford, CA). The integrity of the constructs was confirmed by sequencing.
The Phoenix-Eco cell line was transfected with retroviral vectors by using calcium phosphate precipitation and was further cultured at 32°C for 48–72 h to generate the retroviral supernatant. Mouse spleen cells or naive CD4+ T cells were stimulated with plate-coated anti-CD3 and anti-CD28 antibodies in the presence of 100 units/ml IL-2. The cells were cultured in the retroviral supernatant on days 1 and 2 and harvested on days 4 or 5. The percentage of GFP-positive cells was typically 35–45% on day 4 and 25–35% on day 5, and there was no significant difference among the three STAT4 retrovirus constructs. For TH1 differentiation, IL-12 (5 ng/ml) was added throughout the culture period.
Real-Time PCR.
Total RNA was extracted with Trizol (Life Technologies, Gaithersburg, MD) and reverse-transcribed with a first-strand cDNA synthesis kit (Roche Diagnostics). TaqMan PCR was performed as described (36). Reagents to amplify ribosomal 18S and mouse IFN-γ were purchased from Perkin–Elmer. IFN-γ mRNA levels were standardized by ribosomal 18S RNA levels and expressed as relative ratio to those of nonstimulated cells transfected with RV (retroviral vector)-GFP.
ELISA and Fluorescence-Activated Cell Sorting.
Cells were incubated at 1 × 106 per ml in the presence or absence of 10 ng/ml IL-12 and/or 10 ng/ml IL-18 for 44 h. IFN-γ levels in the supernatant were measured by using a Quantikine mouse IFN-γ kit (R & D Systems). For intracellular IFN-γ staining, cells were stimulated with 50 ng/ml phorbol myristate acetate plus 5 μg/ml ionomycin for 4 h with the last 2 h pulse of GolgiStop (PharMingen). The cells were fixed and permeabilized with Cytofix/Cytoperm (PharMingen), stained with anti-IFN-γ-phycoerythrin antibody or isotype-matched control antibody (PharMingen), and analyzed on a FACSCalibur (Becton Dickinson).
Proliferation Assay.
Cells were cultured in 96-well plates at 1 × 106/ml for 24 h in the presence of indicated doses of IL-12. [3H]thymidine (1 μCi; 1 Ci = 37 GBq) was pulsed for the last 4 h of the culture, and thymidine incorporation was measured as described (32).
Results
GADD45 Induces STAT4 S721 Phosphorylation.
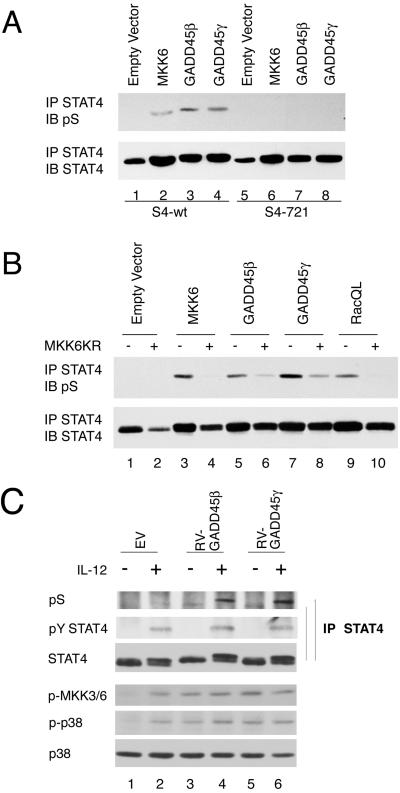
GADD45 members are recently described upstream activators of the p38 MAPK pathway (37), and GADD45-γ-deficient mice have defective TH1 differentiation and IFN-γ production (13). Furthermore, IL-12 and IL-18 induce GADD45-β and GADD45-γ expression, and overexpression of GADD45-β augments IL-12 + IL-18-induced IFN-γ production (12). Having previously shown that IL-12 activates p38 and that MKK6 and p38 can mediate STAT4 S721 phosphorylation, in the present study we first examined whether overexpression of GADD45-β and GADD45-γ could induce STAT4 S721 phosphorylation. Consistent with previous findings (30), STAT4 S721 was not constitutively phosphorylated (Fig. 1A, lane 1) but was inducibly phosphorylated upon overexpression of MKK6, the kinase upstream of p38 (Fig. 1A, lane 2). In addition, overexpression of GADD45-β and GADD45-γ induced S721 phosphorylation (Fig. 1A, lanes 3 and 4), whereas this modification was not detected in a STAT4 S721 mutant (Fig. 1A, lanes 5–8). To provide further evidence of a connection between GADD45 and MKK6, we next expressed the former with a dominant negative version of the latter to see whether the dominant negative MKK6 interfered with the induction of S721 phosphorylation. MKK6 (Fig. 1B, lanes 3 and 4) and a gain-of-function Rac1 mutant, RacQL (Fig. 1B, lanes 9 and 10), were also expressed as controls in the absence and presence of a dominant negative MKK6 allele (Fig. 1B, lanes 4, 6, 8, and 10). GADD45-β and GADD45-γ induced S721 phosphorylation, and this modification was inhibited by expression of dominant negative MKK6 (Fig. 1B, lanes 6 and 8). Next, we sought to confirm the effects of GADD45-β and GADD45-γ in CD4+ T cells. As shown in Fig. 1C, IL-12 induced phosphorylation of MKK6 and p38 (lanes 1, 3, and 5). Expression of GADD45-β and GADD45-γ α also induced MKK6 and p38 activation, which was modestly affected by IL-12 (Fig. 1C, lanes 4 and 6). Accordingly, serine phosphorylation of STAT4 was markedly enhanced by GADD45-β and GADD45-γ expression but, interestingly, depended on IL-12 stimulation. Tyrosine phosphorylation of STAT4 was not affected by overexpression of GADD45-β and GADD45-γ (Fig. 1C, lanes 1, 3, and 5).
Fig 1.
Overexpression of GADD45-β and GADD45-γ induces STAT4 S721 phosphorylation in an MKK6-p38-dependent manner. (A) 293T cells were transfected with vectors encoding 1 μg of wild-type (wt) STAT4 (lanes 1–4) or S721A STAT4 (lanes 5–8) alone (lanes 1 and 5) or with 5 μg of MKK6 (lanes 2 and 6), GADD45-β (lanes 3 and 7), and GADD45-γ (lanes 4 and 8). The cells were lysed and immunoprecipitated with anti-STAT4, and immunoblotted with an antibody that specifically recognizes phosphorylated S721 (pS). (B) 293T cells were transfected with STAT4 with 1 μg of MKK6 (lanes 3 and 4), GADD45-β (lanes 5 and 6), and GADD45-γ (lanes 7 and 8) in the absence or presence of 8 μg MKK6KR. (C) C57BL/6 spleen CD4+ T cells were stimulated with immobilized anti-CD3 plus CD28 in TH1 conditions, and retrovirally transfected with GADD45-β and GADD45-γ. Cells were rested and stimulated with IL-12 for pY and pS STAT4, MKK6, and p38 activation. IP, immunoprecipitation; IB, immunoblotting.
STAT4 Serine 721 Phosphorylation Does Not Abrogate Tyrosine Phosphorylation or IL-12-Induced Cell Proliferation.
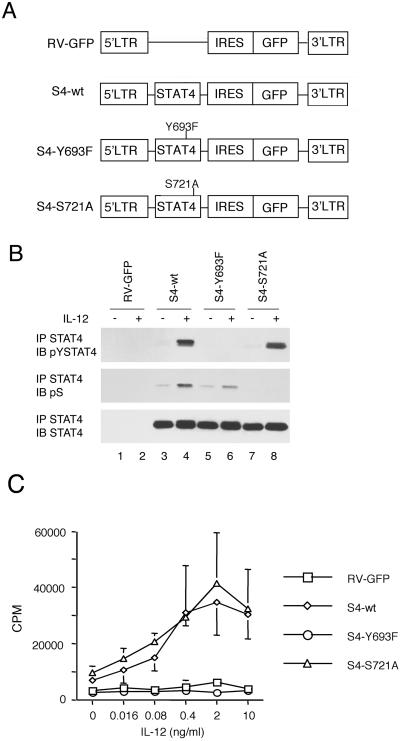
Although we previously showed that mutation of S721 led to impaired STAT4-mediated transactivation of a reporter construct, the functional relevance of STAT4 S721 phosphorylation to IFN-γ production and TH1 differentiation had not been analyzed. Therefore we next sought to address this issue. We reconstituted spleen cells from STAT4−/− mice with wild-type or mutant versions of STAT4 by using retroviral gene transduction (Fig. 2A). Transduction efficiency, as measured by GFP expression, was not different among three STAT4 constructs. In addition, the levels of expression of the wild-type and mutant versions of STAT4 were equivalent (Fig. 2B Bottom, lanes 3–8). We next assessed whether IL-12-dependent signaling could be restored in the STAT4-deficient cells. As expected, IL-12 induced STAT4 phosphorylation in cells transduced with wild-type STAT4 (Fig. 2B Top, lane 4) but not in cells transduced with the Y693F STAT4 mutant (Fig. 2B Top, lane 6) or in cells transduced with GFP without STAT4 (Fig. 2B Top, lane 2). Furthermore, IL-12 induced phosphorylation of S721 in wild-type STAT4 and, to a lesser extent, in Y693F STAT4 (Fig. 2B Middle, lanes 4 and 6). By contrast, S721A STAT4 showed normal IL-12-dependent tyrosine phosphorylation (Fig. 2B Top, lane 8) but did not undergo IL-12-induced serine phosphorylation (Fig. 2B Middle, lane 8). The results indicate that serine phosphorylation partially depends on tyrosine phosphorylation but tyrosine phosphorylation takes place in the absence of serine phosphorylation. The former fact, together with the finding that GADD45-induced serine phosphorylation is IL-12 dependent (Fig. 1C), raise the possibility that STAT4 serine phosphorylation takes place at least in part in the nucleus.
Fig 2.
IL-12 signaling is restored in STAT4-deficient splenocytes transduced with wild-type and mutant versions of STAT4. (A) Retroviral constructs encoding wild-type (wt) and Y693F and S721A STAT4 mutants. LTR, long terminal repeat. IRES, internal ribosomal entry site. (B) Tyrosine and serine phosphorylation of wild-type and mutant versions of STAT4 in reconstituted STAT4−/− spleen cells. Reconstituted spleen cells were serum-starved and stimulated with IL-12 for 45 min. The cells were lysed and immunoprecipitated with anti-STAT4. The immunoprecipitates were electrophoresed and immunoblotted with an antibody that recognizes phosphorylated Y693 (pYSTAT4), pS, and anti-STAT4. IP, immunoprecipitation; IB, immunoblotting. (C) IL-12-dependent proliferation of STAT4−/− spleen cells reconstituted with wild-type (wt) or mutant STAT4. Transduced spleen cells were stimulated with indicated doses of IL-12 for 24 h, and proliferative responses were determined.
We next analyzed DNA binding activity of wild-type and mutant STATs in transduced splenocytes by using an electrophoretic mobility-shift assay (EMSA). The DNA binding activity of S721A STAT4 mutant was comparable to that of wild-type STAT4, whereas Y693F STAT4 did not bind DNA (data not shown). In separate experiments we also examined nuclear localization of wild-type and mutant versions of STAT4 by using confocal microscopy, and our results were consistent with the EMSA experiments (Fig. 5, which is published as supporting information on the PNAS web site, www.pnas.org). Thus, we concluded that serine phosphorylation of STAT4 is not required for nuclear translocation and DNA binding of STAT4. It might be predicted therefore that the S721 STAT4 mutant should be capable of supporting some IL-12-dependent responses.
STAT4 has been shown to be required for IL-12-induced proliferation as evidenced by the defective response in STAT4-deficient mice (24, 25). The experiments in Fig. 2 argued that IL-12 signaling could be restored in the transduced STAT4-deficient cells, but we next sought to confirm that this reconstitution provided a functionally relevant signal and to determine whether S721 phosphorylation was important for this IL-12 response. As shown in Fig. 2D, IL-12-induced proliferation is STAT4 dependent because STAT4-deficient and Y693F-reconstituted cells failed to proliferate in response to IL-12. Interestingly, though, S721A STAT4 lymphocytes proliferated upon IL-12 stimulation to the same extent as wild-type STAT4 lymphocytes, confirming that transduced S721A STAT4 can mediate IL-12 signaling. The results indicate that serine phosphorylation of STAT4 is not required for IL-12-induced cell proliferation.
Serine 721 Phosphorylation of STAT4 Is Required for IFN-γ Production in Response to IL-12.
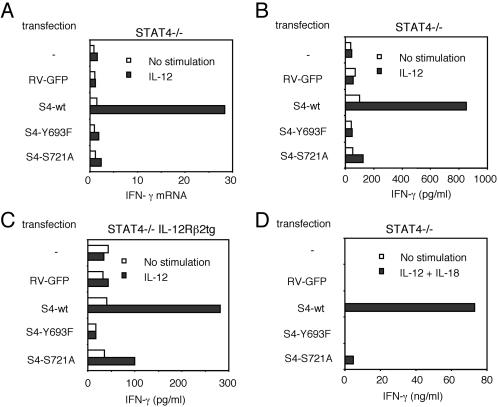
Because the S721A mutant became tyrosine-phosphorylated, translocated to the nucleus, bound DNA normally, and supported IL-12-induced proliferation, we next sought to determine whether this modification had a functional role in IL-12-mediated IFN-γ induction. As shown in Fig. 3A, STAT4-deficient cells fail to up-regulate IFN-γ mRNA in response to IL-12, whereas wild-type STAT4 reconstituted cells up-regulated IFN-γ mRNA; in fact, the level seen in wild-type STAT4 reconstituted cells was comparable to that seen in normal STAT4-expressing splenocytes. As expected, Y693F STAT4 did not support responsiveness to IL-12. Importantly, S721A STAT4-transduced cells also did not show IFN-γ mRNA up-regulation upon IL-12 stimulation, indicating that serine phosphorylation is critical for IL-12 induction of IFN-γ mRNA expression.
Fig 3.
STAT4 tyrosine and serine phosphorylation are required for normal IL-12-induced IFN-γ production. (A) IL-12 induced IFN-γ mRNA expression in STAT4−/− spleen cells reconstituted with wild-type (wt) or mutant versions of STAT4. Transduced cells were serum starved and stimulated with IL-12 for 6 h. Levels of IFN-γ mRNA expression were determined by real-time PCR, as described in Materials and Methods. IL-12-induced IFN-γ mRNA expression in STAT4+/+ spleen cells was 37.4. (B) IL-12 induced IFN-γ production in STAT4−/− spleen cells reconstituted with wild-type or mutant versions of STAT4. (C) IL-12 induced IFN-γ production in STAT4−/− IL-12Rβ2tg spleen cells reconstituted with wild-type or mutant STAT4 constructs. (D) IL-12 + IL-18 induced IFN-γ production in reconstituted STAT4−/− spleen cells. tg, transgenic.
To confirm these results, we next examined IFN-γ production in response to IL-12 by ELISA. As shown in Fig. 3B, IL-12-induced IFN-γ production was completely STAT4 dependent because neither STAT4-deficient nor Y693F STAT4-transduced cells produced IFN-γ in response to IL-12. Importantly, IFN-γ production from the cells transduced with S721A STAT4 mutant was reduced to 10% of that from the cells transfected with wild-type STAT4.
One potential criticism of the aforementioned experiments could be that STAT4 deficiency is associated with reduced IL-12Rβ2 expression (38). Our experiments clearly indicated that IL-12 signaling was restored, both biochemically and functionally, in cells transduced with wild-type STAT4. Nonetheless, it was important to establish that impaired receptor expression was not the major mechanism underlying impaired IFN-γ production. We therefore examined cells from STAT4−/− IL-12Rβ2 transgenic mice in which expression of the IL-12Rβ2 subunit was under the control of the CD2 promoter (32). As shown in Fig. 3C, IFN-γ production depends on STAT4 expression in cells from the receptor transgenic mice. The response to IL-12 was reduced in S721A versus wild-type STAT4 reconstituted cells, although the reduction was not as profound as that seen in the nonreceptor transgenic mice (Fig. 3B). Thus it is possible that the reduced receptor expression might exacerbate the defect seen in S721A-transduced cells. Nonetheless, it is also very clear that IFN-γ production is substantially impaired in S721A-expressing cells, arguing for a direct contribution of serine phosphorylation of STAT4 in IL-12-induced IFN-γ production.
We also determined the effect of the STAT4 mutants on IFN-γ production in response to IL-12 and IL-18, as this stimulation is a more potent means of induction (39). As shown in Fig. 3D, expression of wild-type STAT4 permitted high levels of IFN-γ production in response to IL-12 and IL-18, but it is clearly STAT4 dependent, because no induction was seen in splenocytes expressing GFP alone or in cells transduced with the Y693F STAT4 mutant. Importantly, the production of IFN-γ was markedly impaired in S721A STAT4-expressing splenocytes. This impaired response to IL-12 and IL-18 was also evident in S721A reconstituted cells from STAT4−/−, IL-12Rβ2 transgenic mice (data not shown). Taken together, the data argue that tyrosine phosphorylation is prerequisite for all STAT4 functions and serine phosphorylation is critical for some, e.g., IFN-γ induction, but not others, e.g., proliferation.
Serine 721 Phosphorylation of STAT4 Is Required for TH1 Differentiation.
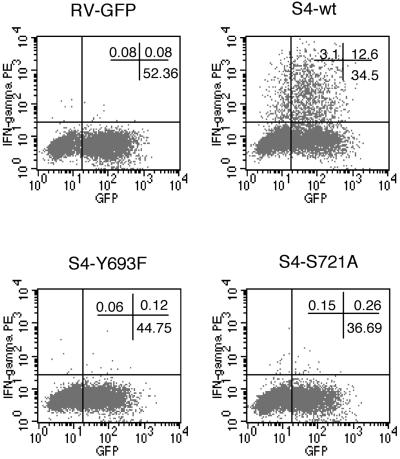
We examined whether serine phosphorylation of STAT4 is required for TH1 differentiation. Purified naive CD4+ T cells were stimulated with anti-CD3, anti-CD28, and IL-12 (10 ng/ml) for 3 days followed by an additional day of culture in IL-2 and IL-12. During this culture period the cells were transduced with wild-type and mutant versions of STAT4 constructs, as in the previous experiments. On day 4, cells were stimulated with phorbol myristate acetate and ionomycin and stained for intracellular IFN-γ. As shown in Fig. 4, transduction with wild-type STAT4 readily permitted the development of IFN-γ-producing cells, compared with Y693F STAT4 and GFP-only transduced cells. Of note, cells transduced with S721A STAT4 had very poor IFN-γ production, indicating STAT4 serine phosphorylation is critical for IL-12-mediated TH1 development of naive CD4+ T cells.
Fig 4.
STAT4 tyrosine and serine phosphorylation are required for normal TH1 differentiation. Purified naive CD4 T cells from STAT4−/− spleen cells were stimulated in the presence of IL-12 and reconstituted with wild-type (wt) and mutant STAT4. The cells were harvested and restimulated with phorbol myristate acetate and ionomycin for intracellular IFN-γ staining.
Discussion
Here, we show that serine phosphorylation of STAT4 is critical for optimal IFN-γ production and TH1 differentiation. Because the S721A STAT4 mutant exhibited normal tyrosine phosphorylation, nuclear translocation, and DNA binding, serine phosphorylation of STAT4 itself appears to play an important role in the transcriptional regulation of target genes. However, it is equally notable that serine phosphorylation was not required for all IL-12-mediated events; IL-12 proliferation, although clearly STAT4 dependent, was not impaired in cells expressing the S721A mutant. This finding indicates that STAT4 selectively regulates gene expression in serine phosphorylation-dependent and -independent manners, depending on the exact gene(s) that are regulated. This finding is also of interest as it provides a mechanism for linking two players in TH1 differentiation: STAT4 and the p38 MAPK.
Serine phosphorylation of a MAPK consensus site within the transcriptional STAT1 and STAT3 has been documented, and it has been reported to have important, but selective, effects on gene regulation (40, 41). The mechanisms through which STAT serine phosphorylation in this region regulates transcriptional activity have not been defined, however. The simplest explanation would be this modification might control the binding of STAT proteins to other nuclear proteins including coactivators or other transcription factors like CBP/p300 (42, 43), MCM5 (44), and AP-1 (45, 46). At present the molecular basis for this mode of control of STAT transcriptional activity is unknown. Defining the mechanism underlying STAT4 serine phosphorylation in control of IFN-γ mRNA expression is an important issue that will need to be resolved.
For STAT1 and STAT3 it appears that both Erk1/2 and p38 can mediate serine phosphorylation (40). For STAT4, however, there is no evidence that a MAPK other than p38 mediates serine phosphorylation. IL-12 does not activate Erk1/2 or JNK, and these kinases do not affect IL-12-induced STAT4 transcriptional activity (30). How, then, is the p38 linked to the IL-12R? Interestingly, Rac2 (11), GADD45-β (12), and GADD45-γ (13) are induced in TH1 conditions and activate p38. We show here that they can promote STAT4 S721 phosphorylation. Although these molecules are attractive candidates linking IL-12 stimulation to p38 activation, it is completely unknown how they might be linked to a cytokine receptor. It should be emphasized, though, that STAT4 is likely not the only p38 substrate relevant to IFN-γ gene regulation (31). IL-12 may activate other p38 substrates such as ATF2 (47), Elk-1 (48), SAP-1 (48), or MEF2C (49). If and how these transcription factors are involved in IL-12-mediated IFN-γ production needs to be clarified, particularly because transcription factors typically function coordinately in gene regulation.
Although the importance of both STAT4 and the p38 pathway in TH1 differentiation and IFN-γ production is clear, it is equally important to emphasize that IFN-γ production can occur independently of IL-12 and STAT4 (50). It should also be emphasized that it has not been established that STAT4 is directly involved in IFN-γ gene regulation, despite the finding by one group that a STAT4 footprint is present in the human IFN-γ gene (28, 29). Additionally, crosslinking the T cell receptor induces IFN-γ and can activate the p38 MAPK. Thus genetic lesions in the p38 pathway would be expected not only to impair IL-12 signaling but other modes of IFN-γ induction. Moreover, other regulators like the transcription factor T-bet are also involved in TH1 differentiation (51, 52). T-bet is regulated by STAT1 but whether the p38 pathway contributes to T-bet regulation is not known (36). At present, it seems that the complexity of the control of IFN-γ gene expression is growing, but perhaps this is not surprising given its importance in the immune response. For all of these reasons, it is not surprising that mutation of STAT4 S721 or deficiency of STAT4 itself has partial and not absolute effects on IFN-γ gene regulation. On the other hand, IL-12 stimulation can induce IFN-γ production in the absence of T cell receptor ligation (12), and the present study argues that tyrosine and serine phosphorylation of STAT4 are important events for normal cytokine control of IFN-γ production.
In conclusion, serine phosphorylation of STAT4 appears to be mediated by p38 MAPK and can be induced by Rac and GADD45-β and GADD45-γ, intermediates that activate p38. The lack of these intermediates and other elements leading to p38 activation is associated with deficits in cell-mediated immunity. Therefore we suggest that STAT4 is one important target of this pathway, and failure to fully activate STAT4 is an important, but presumably not the sole, contributor to the phenotype associated with perturbations in this pathway. Defining exactly how the p38 pathway is linked to the IL-12R and dissecting the mechanisms by which serine phosphorylation regulates STAT-mediated transcription should prove to be interesting areas of investigation. Pragmatically, though, p38 inhibitors are being developed for use in autoimmune disease (53). It will be important to determine whether inhibition of STAT4-dependent transcription is an important aspect of their immunosuppressive mode of actions.
Supplementary Material
Acknowledgments
We thank Dr. Silvio Gutkind, National Institute of Dental and Craniofacial Research, National Institutes of Health, and Dr. Kendall A. Smith, Cornell University (Ithaca, NY), for providing us with plasmids.
Abbreviations
TH1, T helper 1
MAPK, mitogen-activated protein kinase
IL-12R, IL-12 receptor
STAT4, signal transducers and activators of transcription
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Bach E. A., Aguet, M. & Schreiber, R. D. (1997) Annu. Rev. Immunol. 15, 563-591. [DOI] [PubMed] [Google Scholar]
- 2.Seder R. A. & Paul, W. E. (1994) Annu. Rev. Immunol. 12, 635-673. [DOI] [PubMed] [Google Scholar]
- 3.Abbas A. K., Murphy, K. M. & Sher, A. (1996) Nature (London) 383, 787-793. [DOI] [PubMed] [Google Scholar]
- 4.Murphy K. M., Ouyang, W., Farrar, J. D., Yang, J., Ranganath, S., Asnagli, H., Afkarian, M. & Murphy, T. L. (2000) Annu. Rev. Immunol. 18, 451-494. [DOI] [PubMed] [Google Scholar]
- 5.Dong C. & Flavell, R. A. (2001) Curr. Opin. Hematol. 8, 47-51. [DOI] [PubMed] [Google Scholar]
- 6.Glimcher L. H. & Murphy, K. M. (2000) Genes Dev. 14, 1693-1711. [PubMed] [Google Scholar]
- 7.O'Shea J. J. & Paul, W. E. (2002) Nat. Immunol. 3, 506-508. [DOI] [PubMed] [Google Scholar]
- 8.Rincon M., Flavell, R. A. & Davis, R. J. (2001) Oncogene 20, 2490-2497. [DOI] [PubMed] [Google Scholar]
- 9.Rincon M., Enslen, H., Raingeaud, J., Recht, M., Zapton, T., Su, M. S., Penix, L. A., Davis, R. J. & Flavell, R. A. (1998) EMBO J. 17, 2817-2829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lu H. T., Yang, D. D., Wysk, M., Gatti, E., Mellman, I., Davis, R. J. & Flavell, R. A. (1999) EMBO J. 18, 1845-1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li B., Yu, H., Zheng, W., Voll, R., Na, S., Roberts, A. W., Williams, D. A., Davis, R. J., Ghosh, S. & Flavell, R. A. (2000) Science 288, 2219-2222. [DOI] [PubMed] [Google Scholar]
- 12.Yang J., Zhu, H., Murphy, T. L., Ouyang, W. & Murphy, K. M. (2001) Nat. Immunol. 2, 157-164. [DOI] [PubMed] [Google Scholar]
- 13.Lu B., Yu, H., Chow, C., Li, B., Zheng, W., Davis, R. J. & Flavell, R. A. (2001) Immunity 14, 583-590. [DOI] [PubMed] [Google Scholar]
- 14.Gately M. K., Renzetti, L. M., Magram, J., Stern, A. S., Adorini, L., Gubler, U. & Presky, D. H. (1998) Annu. Rev. Immunol. 16, 495-521. [DOI] [PubMed] [Google Scholar]
- 15.Sinigaglia F., D'Ambrosio, D., Panina-Bordignon, P. & Rogge, L. (1999) Immunol. Rev. 170, 65-72. [DOI] [PubMed] [Google Scholar]
- 16.Magram J., Connaughton, S. E., Warrier, R. R., Carvajal, D. M., Wu, C. Y., Ferrante, J., Stewart, C., Sarmiento, U., Faherty, D. A. & Gately, M. K. (1996) Immunity 4, 471-481. [DOI] [PubMed] [Google Scholar]
- 17.Wu C., Ferrante, J., Gately, M. K. & Magram, J. (1997) J. Immunol. 159, 1658-1665. [PubMed] [Google Scholar]
- 18.Wu C., Wang, X., Gadina, M., O'Shea, J. J., Presky, D. H. & Magram, J. (2000) J. Immunol. 165, 6221-6228. [DOI] [PubMed] [Google Scholar]
- 19.Altare F., Lammas, D., Revy, P., Jouanguy, E., Doffinger, R., Lamhamedi, S., Drysdale, P., Scheel-Toellner, D., Girdlestone, J., Darbyshire, P., et al. (1998) J. Clin. Invest. 102, 2035-2040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Altare F., Durandy, A., Lammas, D., Emile, J. F., Lamhamedi, S., Le Deist, F., Drysdale, P., Jouanguy, E., Doffinger, R., Bernaudin, F., et al. (1998) Science 280, 1432-1435. [DOI] [PubMed] [Google Scholar]
- 21.de Jong R., Altare, F., Haagen, I. A., Elferink, D. G., Boer, T., van Breda Vriesman, P. J., Kabel, P. J., Draaisma, J. M., van Dissel, J. T., Kroon, F. P., et al. (1998) Science 280, 1435-1438. [DOI] [PubMed] [Google Scholar]
- 22.Bacon C. M., McVicar, D. W., Ortaldo, J. R., Rees, R. C., O'Shea, J. J. & Johnston, J. A. (1995) J. Exp. Med. 181, 399-404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jacobson N. G., Szabo, S. J., Weber-Nordt, R. M., Zhong, Z., Schreiber, R. D., Darnell, J. E., Jr. & Murphy, K. M. (1995) J. Exp. Med. 181, 1755-1762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Thierfelder W. E., van Deursen, J. M., Yamamoto, K., Tripp, R. A., Sarawar, S. R., Carson, R. T., Sangster, M. Y., Vignali, D. A., Doherty, P. C., Grosveld, G. C. & Ihle, J. N. (1996) Nature (London) 382, 171-174. [DOI] [PubMed] [Google Scholar]
- 25.Kaplan M. H., Sun, Y. L., Hoey, T. & Grusby, M. J. (1996) Nature (London) 382, 174-177. [DOI] [PubMed] [Google Scholar]
- 26.Bacon C. M., Petricoin, E. F., III, Ortaldo, J. R., Rees, R. C., Larner, A. C., Johnston, J. A. & O'Shea, J. J. (1995) Proc. Natl. Acad. Sci. USA 92, 7307-7311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cho S. S., Bacon, C. M., Sudarshan, C., Rees, R. C., Finbloom, D., Pine, R. & O'Shea, J. J. (1996) J. Immunol. 157, 4781-4789. [PubMed] [Google Scholar]
- 28.Xu X., Sun, Y. L. & Hoey, T. (1996) Science 273, 794-797. [DOI] [PubMed] [Google Scholar]
- 29.Barbulescu K., Becker, C., Schlaak, J. F., Schmitt, E., Meyer zum Buschenfelde, K. H. & Neurath, M. F. (1998) J. Immunol. 160, 3642-3647. [PubMed] [Google Scholar]
- 30.Visconti R., Gadina, M., Chiariello, M., Chen, E. H., Stancato, L. F., Gutkind, J. S. & O'Shea, J. J. (2000) Blood 96, 1844-1852. [PubMed] [Google Scholar]
- 31.Zhang S. & Kaplan, M. H. (2000) J. Immunol. 165, 1374-1380. [DOI] [PubMed] [Google Scholar]
- 32.Nishikomori R., Ehrhardt, R. O. & Strober, W. (2000) J. Exp. Med. 191, 847-858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kovalsky O., Lung, F. D., Roller, P. P. & Fornace, J. (2001) J. Biol. Chem. 276, 39330-39339. [DOI] [PubMed] [Google Scholar]
- 34.Fan W., Richter, G., Cereseto, A., Beadling, C. & Smith, K. A. (1999) Oncogene 18, 6573-6582. [DOI] [PubMed] [Google Scholar]
- 35.Wilson K. C. & Finbloom, D. S. (1992) Proc. Natl. Acad. Sci. USA 89, 11964-11968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lighvani A. A., Frucht, D. M., Jankovic, D., Yamane, H., Aliberti, J., Hissong, B. D., Nguyen, B. V., Gadina, M., Sher, A., Paul, W. E. & O'Shea, J. J. (2001) Proc. Natl. Acad. Sci. USA 98, 15137-15142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Takekawa M. & Saito, H. (1998) Cell 95, 521-530. [DOI] [PubMed] [Google Scholar]
- 38.Lawless V. A., Zhang, S., Ozes, O. N., Bruns, H. A., Oldham, I., Hoey, T., Grusby, M. J. & Kaplan, M. H. (2000) J. Immunol. 165, 6803-6808. [DOI] [PubMed] [Google Scholar]
- 39.Yoshimoto T., Takeda, K., Tanaka, T., Ohkusu, K., Kashiwamura, S., Okamura, H., Akira, S. & Nakanishi, K. (1998) J. Immunol. 161, 3400-3407. [PubMed] [Google Scholar]
- 40.Decker T. & Kovarik, P. (2000) Oncogene 19, 2628-2637. [DOI] [PubMed] [Google Scholar]
- 41.Kovarik P., Mangold, M., Ramsauer, K., Heidari, H., Steinborn, R., Zotter, A., Levy, D. E., Muller, M. & Decker, T. (2001) EMBO J. 20, 91-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Horvai A. E., Xu, L., Korzus, E., Brard, G., Kalafus, D., Mullen, T. M., Rose, D. W., Rosenfeld, M. G. & Glass, C. K. (1997) Proc. Natl. Acad. Sci. USA 94, 1074-1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bhattacharya S., Eckner, R., Grossman, S., Oldread, E., Arany, Z., D'Andrea, A. & Livingston, D. M. (1996) Nature (London) 383, 344-347. [DOI] [PubMed] [Google Scholar]
- 44.Zhang J. J., Zhao, Y., Chait, B. T., Lathem, W. W., Ritzi, M., Knippers, R. & Darnell, J. E., Jr. (1998) EMBO J. 17, 6963-6971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schaefer T. S., Sanders, L. K. & Nathans, D. (1995) Proc. Natl. Acad. Sci. USA 92, 9097-9101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang X., Wrzeszczynska, M. H., Horvath, C. M. & Darnell, J. E., Jr. (1999) Mol. Cell. Biol. 19, 7138-7146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Raingeaud J., Gupta, S., Rogers, J. S., Dickens, M., Han, J., Ulevitch, R. J. & Davis, R. J. (1995) J. Biol. Chem. 270, 7420-7426. [DOI] [PubMed] [Google Scholar]
- 48.Whitmarsh A. J., Yang, S. H., Su, M. S., Sharrocks, A. D. & Davis, R. J. (1997) Mol. Cell. Biol. 17, 2360-2371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Han J., Jiang, Y., Li, Z., Kravchenko, V. V. & Ulevitch, R. J. (1997) Nature (London) 386, 296-299. [DOI] [PubMed] [Google Scholar]
- 50.Kaplan M. H., Wurster, A. L. & Grusby, M. J. (1998) J. Exp. Med. 188, 1191-1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mullen A. C., High, F. A., Hutchins, A. S., Lee, H. W., Villarino, A. V., Livingston, D. M., Kung, A. L., Cereb, N., Yao, T. P., Yang, S. Y. & Reiner, S. L. (2001) Science 292, 1907-1910. [DOI] [PubMed] [Google Scholar]
- 52.Szabo S. J., Sullivan, B. M., Stemmann, C., Satoskar, A. R., Sleckman, B. P. & Glimcher, L. H. (2002) Science 295, 338-342. [DOI] [PubMed] [Google Scholar]
- 53.Lee J. C., Kumar, S., Griswold, D. E., Underwood, D. C., Votta, B. J. & Adams, J. L. (2000) Immunopharmacology 47, 185-201. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.