Abstract
Somatic hypermutation (SHM) of the Ig genes is required for affinity maturation of the humoral response to foreign antigens. Activation-induced cytidine deaminase (AID), which is specifically expressed in germinal center centroblasts, is indispensable for this process. Expression of AID is sufficient to activate SHM in hybridomas, non-B cells, and Escherichia coli, suggesting that it initiates the mutational process by deaminating DNA. However, the cis-acting sequences that are responsible for targeting AID activity to the variable (V) region of Ig genes are unknown. Here we show that expression of AID in B cell lines (i.e., Burkitt's lymphoma Ramos and hybridoma P1–5) not only causes hypermutation of Ig sequences, but also of the AID transgene itself. Because it is possible that B cell-specific transacting factors bind to and recruit the “mutator” to the AID transgene, we tested whether the AID transgene can mutate in non-B cells. Indeed, we show that expression of AID in Chinese hamster ovary cells causes SHM of both the Ig and AID transgenes. These data suggest that high transcription rates alone may predispose any gene to mutation by AID but do not preclude that there are specific B cell factors that account for the extremely high rate of V mutation in vivo and its targeting to the V region.
Affinity maturation of the antibody response is mediated by somatic hypermutation (SHM) of the antibody genes with subsequent selection of B cell clones that produce higher affinity antibodies to the immunizing antigen. SHM causes point mutations and occasional deletions and insertions at very high rates of ≈10−3 bp per generation and occurs during the centroblast stage of B cell differentiation. The mutations are restricted to the variable region (V region) that encodes the antibody binding site and to sequences that are ≈1.5 kb downstream from the Ig promoter. Some of these mutations increase the affinity of the antibody so that it can neutralize viruses and toxins and inactivate pathogenic organisms.
Mice and humans with mutations in activation-induced cytidine deaminase (AID) have defects in SHM (1, 2), class-switch recombination (1), and gene conversion (3, 4). Based on the sequence similarity of AID to the RNA-editing enzyme APOBEC-1 (5), it was postulated that AID might function by editing a specific message that results in the production of an altered protein that subsequently causes mutation (6). However, the higher than expected number of transition mutations in V regions (7) raises the possibility that AID might be a DNA-specific cytidine deaminase that converts C to U nucleotides directly in the DNA of the V region. In fact, we recently showed that AID induced the P1–5 hybridoma to exclusively mutate G⋅C bp, and most of these mutations were transition mutations (8). With the finding that AID can induce SHM in non-B cells (9) and Escherichia coli (4), it is more likely that AID is a DNA-specific cytidine deaminase.
The mechanism for targeting of SHM to the V region of Ig genes is not known. Interestingly, SHM has also been observed to occur in other non-Ig genes. The genes for bcl-6 (10–12) and FasL (13) undergo SHM with similar characteristics to those observed in the V region of Ig genes, albeit at a lower rate. It is possible that these genes share cis-acting sequences with the Ig locus that are responsible for recruiting AID activity. Another possibility is that B cell-specific cis-acting sequences do not exist, and that targeting of SHM to certain genes is caused by high transcription rates and possibly other nonspecific factors (14). In this regard, because the Ig gene is one of the most highly transcribed genes in B cells, it will be mutated at higher rates than other genes. However, other transcribed genes have not been found to mutate above the PCR error rate in germinal center B cells (11, 15, 16). Thus, the issue of whether SHM is targeted by B cell-specific cis-acting sequences is not resolved. In this article, we examine this issue by looking for SHM of other transcribed genes in B cells and non-B cells activated for SHM by expression of AID.
Materials and Methods
Constructs.
Full-length human AID (hAID) cloned into the pCEP4 vector (Invitrogen) has been described (8). The vector was digested with Nru1 and EcoRV before transfection. The Vn/ECMV γ2a and Lκ constructs have been described (17, 18).
Cell Lines, Cell Culture, and Transfection Conditions.
Ramos and P1–5 hybridomas were grown as described (8). Chinese hamster ovary (CHO) Pro-5 cells (obtained from P. Stanley, Albert Einstein College of Medicine) were grown in DMEM supplemented with 10% FCS. CHO cells were first electroporated with 10 μg of the Lκ construct. One transfectant that secreted high levels of light chain (CHO-LC18) was then transfected with 10 μg of the mouse Vn/ECMV γ2a construct. One of these transfectants (CHO-LC18-Vn/ECMV clone 8) was then transfected with 10 μg of the linearized hAID or the empty vectors. CHO cells were transfected in DMEM at 400 V, 960 μF, plated into 96-well plates, and selected with 1.5 mg/ml G418 for the Vn/ECMV γ2a construct and 0.6 mg/ml hygromycin B for the hAID and empty constructs. The ELISA spot assay was performed as reported (19). Briefly, each drug-resistant colony was expanded to ≈1–5 × 106 cells and plated onto 96-well plates that were precoated with anti-mouse IgG2a antibody. After 20 h, the plates were developed for secreted IgG2a.
PCR Amplification, Cloning, and Sequencing V Regions and AID Transgene.
Genomic DNA was prepared as reported (19). V regions from the various B cell lines were amplified with Pfu polymerase (Stratagene) from genomic DNA by using 30 cycles of 95°C/15 sec, 56°C/15 sec, 72°C/30 sec. Primers used were: for AID, 5′ primer, 5′-GAGGCAAGAAGACACTCTGG-3′, 3′ primer, 5′-GTGACATTCCTGGAAGTTGC-3′; for bcl-6, 5′ primer, CCGCTCTTGCCAAATGCTTTG, 3′ primer, CACGATACTTCAT-CTCATCTGG; and for c-myc, 5′ primer, AGAAAATGGTAGGCGCGCGTA, 3′ primer, TCGACTCATCTCAGCATTAAAG. PCR products were cloned and sequenced as reported (19). Stratagene reports that PFU polymerase has an error rate of ≈1/650,000 bp per duplication. Therefore, in a 30-cycle amplification, we expect ≈1 mutation in 20,000 nt to be attributed to PCR error. GenBank accession nos. for mutated sequences of the AID gene for Ramos A.2 and A.5 are AF529815–AF529827, for hybridoma P1–5 A.1 and A.2 they are AF529828–AF529840, and for CHO A.3 and A.9 they are AF529841–AF529856.
Extraction of RNA and Northern Blots.
About 5 × 106 cells were lysed with 1 ml Trizol reagent (GIBCO/BRL), and RNA was extracted according to the manufacturer's instructions. About 1 μg of total RNA was run on formaldehyde gels for Northern blots.
Statistics.
Statistics for primary reversion data (see Fig. 2B) was calculated by the independent-samples t test with equal variances assumed (Version 10, SPSS, Chicago).
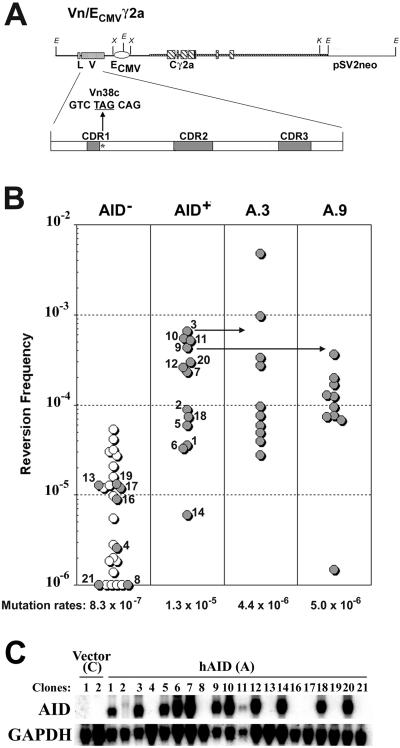
Fig 2.
AID induces SHM in CHO cells. (A) Murine Vn/ECMV γ2a construct transfected into CHO cells to study SHM. As described in ref. 17, this heavy chain Ig construct has replaced the intronic μ enhancer with a CMV enhancer and contains a TAG nonsense codon within an RGYW hotspot motif at codon 38. (B Left) CHO clone CHO-LC18 (see Materials and Methods) stably transfected with heavy and light chain Ig genes was transfected with empty vector (○) or the hAID construct (•). Depending on expression of AID (see below), data were distributed into AID-negative (AID−) and AID-positive (AID+) columns. Frequency of nonsense revertants, as detected with the ELISA spot assay, is plotted. (Right) Ten subclones of CHO clones A.3 and A.9 were further analyzed by using the ELISA spot assay to calculate mutation rates by fluctuation analysis. (C) Northern blots for hAID and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) in CHO transfectants. Clone numbers correspond to numbers in B Left.
Results
We previously showed that the Burkitt's lymphoma Ramos clone 1, which does not undergo SHM and expresses low levels of AID, could be activated for V region SHM by overexpressing AID (8). Specifically, the V region accumulates many mutations in Ramos clones A.2 and A.5 (Table 1) that have ≈25-fold higher levels of AID mRNA than Ramos clones C.1 and A.1 (8). Bcl-6 and the c-myc allele that has translocated into the switch region also have been shown to undergo SHM in B cell lines (20, 21). To determine whether overexpression of AID in Ramos cells also activated mutation in these genes, bcl-6 and c-myc were sequenced from 2-month-old cultures of Ramos clones that overexpressed AID. However, only a few mutations were found in the c-myc and bcl-6 genes in Ramos clones A.2 and A.5 (data not shown). Reverse transcription–PCR analysis revealed that these genes were indeed being transcribed (data not shown). Because SHM correlates with transcription rates (22), it is possible that the rates of transcription of these genes might be too low for SHM to occur at the level seen in the V region.
Table 1.
SHM of the V region and of the AID transgene in Ramos, hybridoma P1-5, and CHO cells
| Clone (months cultured) | Level of AID expression (vector used) | Mutations | Total bp sequenced | Frequency (mutation/bp) ×10−4 | Mutated sequences/total | Mutation rates (mutation/bp per generation) ×10−6 |
|---|---|---|---|---|---|---|
| Ramos C.1 (2) | AIDlow (empty vector) | 1 (V) | 12,900 | 0.78 | 1/30 | 1.1 |
| Ramos A.1 (2) | AIDlow (sense hAID) | 1 (V) | 11,600 | 0.86 | 1/27 | 1.2 |
| Ramos A.2 (2) | AIDhi (sense hAID) | 7 (V) | 12,500 | 5.6 | 7/29 | 7.7 |
| 7 (A) | 17,400 | 4.0 | 6/29 | 5.6 | ||
| Ramos A.5 (2) | AIDhi (sense hAID) | 13 (V) | 15,300 | 8.5 | 9/31 | 11.8 |
| 8 (A) | 15,600 | 5.1 | 7/26 | 6.9 | ||
| Ramos αA.1 (1) | AIDlow (antisense hAID) | 2 (A) | 15,020 | 1.3 | 2/29 | 3.6 |
| Ramos αA.2 (1) | AIDlow (antisense hAID) | I (A) | 8,200 | 1.2 | 1/14 | 3.3 |
| P1-5 A.1 (2) | AIDhi (sense hAID) | 28 (V) | 28,900 | 9.3 | 22/85 | 15.7 |
| 10 (A) | 14,400 | 6.9 | 9/24 | 11.6 | ||
| P1-5 A.2 (2) | AIDhi (sense hAID) | 6 (V) | 5,780 | 10.4 | 6/17 | 17.3 |
| 7 (A) | 6,600 | 10.6 | 5/11 | 17.7 | ||
| CHO A.3 (2) | AIDhi (sense hAID) | 6 (A) | 12,600 | 4.8 | 6/21 | 8.0 |
| CHO A.9 (2) | AIDhi (sense hAID) | 11 (A) | 11,400 | 9.6 | 11/19 | 16.0 |
| CHO αA.1 (1) | AIDneg (antisense hAID) | 1 (A) | 10,800 | 0.9 | 1/18 | 3.0 |
| CHO αA.5 (1) | AIDneg (antisense hAID) | 1 (A) | 9,490 | 1.1 | 1/16 | 3.7 |
Expression levels of AID for Ramos clones have been published (8). Expression of AID was negative (AIDneg), low (AIDlow), or high (AIDhi).
Mutations identified in the V region (V) = 430 bp, AID transgene (A) = 600 bp.
Mutations rates were calculated by using a 20-, a 24-, and a 24-h generation time for Ramos, P1-5, and CHO cells, respectively.
Data published (8) from the identical clones and DNA samples used to analyze AID mutations.
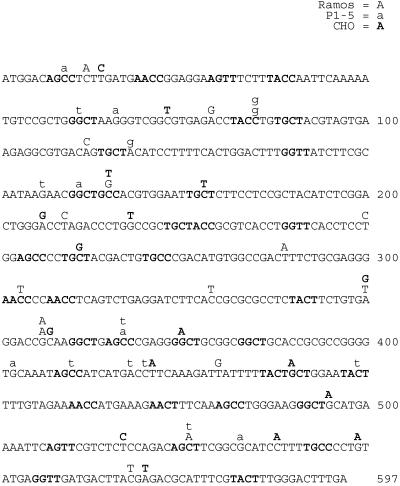
To test whether a highly transcribed non-Ig gene was undergoing SHM in the same Ramos clones, we sequenced the highly transcribed AID transgene that is regulated by the cytomegalovirus (CMV) promoter. Indeed, many mutations were identified within the AID transgene in Ramos clones A.2 and A.5 (Fig. 1 and Table 1), and the calculated rates of mutation were only slightly lower than that of the V region (Tables 1 and 2). In addition, the characteristics of the mutations in the AID transgene were similar to those in the V region (Table 2). In particular, 20% of all mutations occurred at G⋅C base pairs within RGYW or WRCY hotspot motifs, even though only 7% of G/C nucleotides within the AID transgene occur at these sequences (Table 2). RGYW and WRCY hotspot motifs are frequently mutated during SHM in vitro and in vivo (23). To confirm that mutations observed in the AID transgene were caused by the AID protein, the nonmutating Ramos clone 1 was transfected with a construct in which the AID transgene is in the antisense orientation to transcription. In this case, only a few mutations were found within the AID transgene (Ramos clones αA.1 and αA.2; Table 1). To confirm that the hAID construct that was used for transfection did not contain mutations, AID was amplified from the hAID plasmid with PFU polymerase, cloned, and sequenced. Only one mutation was found at an A⋅T base pair in 10 clones (6,000 nt) sequenced.
Fig 1.
Mutations in the AID transgene from Ramos, hybridoma P1–5, and CHO are shown. Mutations located within the AID transgene from the Burkitt's lymphoma Ramos (uppercase), hybridoma P1–5 (lowercase), and CHO cells (uppercase bolded) are shown. Duplicate mutations from each clone were counted once in Table 1. Hotspot motifs (RGYW and WRCY) are bolded.
Table 2.
Characteristics of mutations observed in the V region and the AID transgene in Ramos, P1-5, and CHO cells
| Characteristic
|
Ramos | P1-5 | CHO | ||
|---|---|---|---|---|---|
| V region A.2 and A.5 | AID transgene A.2 and A.5 | V region A.1 and A.2 | AID transgene A.1 and A.2 | AID transgene A.3 and A.9 | |
| Mutation rates | 9.8 | 6.3 | 16.5 | 14.7 | 12.0 |
| GC mutations/total | 25/31 (81%) | 8/15 (53%) | 34/34 (100%) | 15/17 (88%) | 11/17 (65%) |
| Ts mutations‡/total | 13/31 (42%) | 8/15 (53%) | 24/34 (71%) | 10/17 (59%) | 11/17 (65%) |
| Hotspot/total | 10/31 (32%) | 3/15 (20%) | 21/34 (62%) | 8/17 (47%) | 4/17 (24%) |
Published data (8).
Mutation/bp per generation.
Transition mutation (i.e., C to T, T to C, G to A, and A to G).
G⋅C base pairs within RGYW/WRCY motifs are designated as hotspot nucleotides. A total of 39/597 (7%) of nucleotides in AID transgene, and 7% (36/550) and 9% (32/340) of nucleotides in the V region of Ramos and P1-5, respectively, are hotspot nucleotides.
To determine whether SHM of the AID transgene can also occur in other B cell lines, we tested whether the AID transgene was being mutated in the P1–5 hybridoma. This hybridoma is unique in that AID expression induces mutations exclusively at G⋅C base pairs in the endogenous V region that are mostly within RGYW/WRCY hotspot motifs (P1–5 clones A.1 and A.2; Table 2) (8). This finding suggests that this cell line is missing a factor(s) responsible for the A⋅T mutations that are believed to occur during the second phase of SHM that is mismatch repair- and polymerase η-dependent (24–26). Sequencing of the AID transgene in 2-month-old cultures revealed many mutations (Fig. 1 and Table 1). The calculated rates and frequencies of mutation in the endogenous V region and the ectopically integrated AID transgene were similar (Tables 1 and 2), and a striking bias for mutations in RGYW/WRCY motifs was observed in both genes: 62% and 47% of all mutations were within RGYW/WRCY motifs in the V region and the AID transgene, respectively (Table 2). In addition, like in the V region, most mutations occurred at G⋅C base pairs. The few A⋅T mutations in the AID transgene may have arisen by a non-AID-related processes, such as during the integration of the transgene into the genome (27). These data indicate that AID mutates both itself and the Ig gene in B cell lines.
The data presented above suggest that hypermutation induced by AID does not require specific cis-acting sequences to localize mutation to a specific gene. This is because the AID transgene is not expected to share regulatory sequences with the Ig loci. However, it is formally possible that the hAID transgene and the CMV promoter-enhancer in particular contain sequences similar to the Ig loci that are required for targeting SHM. It is also possible that the sites of integration that allow high expression of AID contain regulatory sequences that share motifs with the antibody gene. Because non-B cells are not considered to have B cell-specific transacting factors, expressing the AID transgene in a non-B cell should cause any putative B cell-specific cis-acting sequence to be silent. Thus, mutation of the AID transgene in non-B cells would argue against the requirement of regulatory B cell-specific cis-acting sequences for targeting SHM. We therefore tested whether the AID transgene can mutate in non-B cells.
To confirm that human AID can activate SHM in CHO cells, CHO cells were first stably transfected with murine heavy and light chain Ig genes (Fig. 2A). The murine heavy chain construct used in this experiment (i.e., Vn/ECMV γ2a construct; Fig. 2A) has two unique features. First, the intronic μ enhancer was replaced with the CMV enhancer to ensure that the Ig heavy chain gene was expressed in CHO cells. Northern blots confirm that the Vn/ECMV γ2a transgene was expressed (data not shown). Second, a nonsense codon was introduced into an RGYW hotspot in the V region of the heavy chain construct (Fig. 2A), which allows SHM to be measured by reversion of the nonsense codon that would result in the production and secretion of IgG2a that could be detected at the single-cell level with the ELISA-spot assay.
CHO cells stably expressing the murine Ig genes were transfected with the hAID transgene, the antisense hAID transgene, and the empty vector control. Independent transfectants were grown to ≈2 × 106 cells and distributed into ELISA plates coated with anti-murine γ2a antibody. After 20 h, the ELISA plates were developed for secreted antibody. The revertant frequency for each individual clone was then plotted (Fig. 2B Left). Because some hAID-transfected CHO cells did not express hAID (i.e., CHO clones A.4, A.8, A.13, A.16, A.17, A.19, and A.21; Fig. 2C), the revertant frequencies for each individual CHO clone were plotted in the relevant AID-negative (AID−) and AID-positive (AID+) columns (Fig. 2B). As shown in Fig. 2B Left, clones that express hAID reverted the nonsense codon in the Vn/ECMV γ2a transgene ≈15-fold more frequently than clones that did not express AID (P < 0.01). To more accurately determine the mutation rates at the nonsense codon, two AID+ clones (i.e., CHO A.3 and A.9) were subcloned. Ten subclones of each were assayed by the ELISA spot assay, and mutation rates were calculated by fluctuation analyses (19). CHO clones A.3 and A.9 displayed mutation rates of 4.4 × 10−6 and 5.0 × 10−6 mutations/bp per generation, respectively (Fig. 2B Right). Although these two clones chosen for further analysis initially reverted at high frequencies (Fig. 2B Left), the corresponding subclones displayed a similar range of reversion frequencies and mutation rates to that of the larger group of independently transfected AID+ CHO clones. It is unclear why CHO clones that do not express AID have such a high background of reversion frequencies (AID−; Fig. 2B Left). Nevertheless, these data support findings (9) that AID can induce SHM in non-B cells.
To test whether the AID transgene was mutating in CHO cells, AID was sequenced from 2-month-old cultures of CHO clones A.3 and A.9. The AID transgene was found to contain many mutations in the sense (CHO clones A.3 and A.9; Fig. 1 and Table 1) but not in the antisense orientation (CHO clones αA.1 and αA.5; Table 1). The calculated rates of mutation of the AID transgene were similar between the CHO clones and the B cell lines (Table 2), and the characteristics of the mutations displayed a similar pattern typical to SHM in cultured cells, namely a bias toward mutations in G⋅C base pairs, a preference for transition mutations, and RGYW hotspot targeting (Table 2).
Discussion
The work reported here suggests that the SHM process does not depend on a specific cis-acting sequence(s) to target mutation to the Ig gene and will proceed with any cis-acting sequence that confers a high rate of transcription to the target gene. Two of our findings support this hypothesis: (i) an Ig transgene mutates in a non-B cell, and (ii) the AID transgene driven by a strong promoter mutates in B and non-B cells. Although it is possible that the AID transgene has a B cell-specific cis-acting sequence(s), if this sequence element were to exist it should be inactive in non-B cells because non-B cells lack B cell-specific transacting factors. Similar findings to those reported here were described in fibroblasts activated to mutate a substrate when AID was overexpressed (9). The lack of a requirement of specific cis-acting sequences is also supported by findings that other genes undergo SHM in B cells (10–13), and other genes essential for cell viability might also be mutated because constitutive SHM appears to decrease the viability of cultured cells (19). Although the notion that SHM can occur in any highly transcribed gene is unsettling, this model may explain why many types of lymphomas arise from B cells that are undergoing SHM (11, 28).
On the other hand, some observations support the notion that SHM is regulated by cis-acting sequences. First, other transcribed genes in germinal center B cells do not undergo SHM (11, 15, 16). The critical issue here is whether these genes are in fact mutating, but at levels that are below the PCR error rate, or whether they are not mutating at all. Because mutation rates are positively correlated with transcription rates (22) and RGYW/WRCY hotspot density (29), other genes might in fact be mutating, but at rates that simply correlate with the quantity of these other features. In this regard, the Ig gene might be mutated at a higher rate than other genes because it is transcribed at very high rates. In addition, it must be considered that accumulation of mutations downstream of promoters will occur only in regions that do not confer a selective disadvantage, such as regions that do not contain ORFs or regulatory sequences for housekeeping genes. Mutations in these regions should reduce the viability of the cell, and as a consequence, the apparent rate of mutation at these loci will seem to be low or absent.
Second, the classical observation that the V region mutates at higher rates than the constant (C) region also supports the idea that cis-acting sequences are involved in targeting SHM. For example, a B cell-specific cis-acting sequence might affect the chromatin structure over the V region, allowing the “mutator” to gain access to the DNA. However, other explanations exist that could account for this differential mutation of the V versus C regions without the requirement of B cell-specific cis-acting sequences. One possibility is that the mutator associates with the RNA polymerase II complex to produce mutations during the initiation phase, but eventually falls off this complex during the elongation phase (30, 31). Another possibility is that mutation depends on the availability of single-stranded DNA (see below) and that there is more single-stranded DNA in the V region than in the C region, caused by (i) stable RNA–DNA hybrids in the V region as a result of transcription that leaves the nontranscribed strand single-stranded, (ii) a higher RNA polymerase II density in the V region than in the C region, or (iii) transcription inducing stable secondary DNA structures in the V region with single-stranded loops of DNA (6). Although these models suggest that mutation can be focused on the V region without the requirement of cis-acting sequences, there are presently no data to support these beliefs.
Many of the findings that support the notion that SHM is not regulated by B cell-specific cis-acting sequences come from reports, including this one, where AID is overexpressed at levels that are believed to be higher than in centroblasts (8, 9, 32), although this value is not known. Because overexpression of APOBEC-1, which is homologous to AID, resulted in hyperediting of its target substrate ApoB mRNA (33), it is possible that targeting of SHM has been deregulated in cells that overexpress AID. Thus, caution must be exercised when interpreting these data. In addition, mutation rates induced by expression of AID in cell lines are ≈10-fold lower than the rates observed in the V region in vivo (14). Thus other B cell-specific factors might indeed help focus mutation over the V region.
AID has been postulated to function directly (i.e., to directly deaminate cytidines in DNA) (6, 8) and indirectly (i.e., via its putative mRNA editing activity) (6) in the SHM process. The fact that human AID can activate SHM in a hamster ovary cell line and E. coli (34) argues against AID having an indirect role in SHM because this would require that the transcript edited by AID be expressed ubiquitously and have the same recognition motif in different species. Further support to the notion that AID is a DNA-specific cytidine deaminase was provided by the finding that AID predominantly caused transition mutations at G⋅C base pairs in the P1–5 hybridoma (8), fibroblasts (9), and E. coli (34). In addition, the mutation rates induced by AID in E. coli are increased slightly in the absence of uracil DNA glycosylase (34), suggesting that uracil is an intermediate in the SHM process. Thus, AID might initiate SHM by deaminating cytidines on DNA, resulting in the recruitment of the mismatch repair system and/or uracil DNA glycosylases (14, 34, 35), which in turn could cause mutations at other base pairs during the repair phase. Because enzyme-catalyzed cytidine deamination probably requires single-stranded DNA (because the amino group on cytidine is hydrogen-bonded to the carboxyl of guanosine), AID therefore might choose its target based on the availability of single-stranded DNA (14).
The findings presented in this article also have practical implications. Because the AID transgene was found to mutate, it is likely that any transgene under the regulation of a strong promoter will mutate as long as AID is expressed in that cell. Semirandom mutagenesis can facilitate the characterization of the gene products for structure/function analysis.
Acknowledgments
We thank Drs. M. Iglesias-Ussel, M. Sadofsky, B. Diamond, and R. Laskov for critical review of the manuscript. We thank M. Lin for generating the CHO lines used in this study and M. Fan for technical help. This work was supported by National Institutes of Health Grant CA72649 to M.D.S., who is also supported by the Harry Eagle chair provided by the National Women's Division of the Albert Einstein College of Medicine. A.M. is a recipient of Cancer Research Institute and Harry Eagle Fellowships.
Abbreviations
AID, activation-induced cytidine deaminase
hAID, human AID
SHM, somatic hypermutation
CHO, Chinese hamster ovary
V region, variable region
CMV, cytomegalovirus
References
- 1.Muramatsu M., Kinoshita, K., Fagarasan, S., Yamada, S., Shinkai, Y. & Honjo, T. (2000) Cell 102, 553-563. [DOI] [PubMed] [Google Scholar]
- 2.Revy P., Muto, T., Levy, Y., Geissmann, F., Plebani, A., Sanal, O., Catalan, N., Forveille, M., Dufourcq-Labelouse, R., Gennery, A., et al. (2000) Cell 102, 565-575. [DOI] [PubMed] [Google Scholar]
- 3.Arakawa H., Hauschild, J. & Buerstedde, J. M. (2002) Science 295, 1301-1306. [DOI] [PubMed] [Google Scholar]
- 4.Harris R. S., Sale, J. E., Petersen-Mahrt, S. K. & Neuberger, M. S. (2002) Curr. Biol. 12, 435-438. [DOI] [PubMed] [Google Scholar]
- 5.Muramatsu M., Sankaranand, V. S., Anant, S., Sugai, M., Kinoshita, K., Davidson, N. O. & Honjo, T. (1999) J. Biol. Chem. 274, 18470-18476. [DOI] [PubMed] [Google Scholar]
- 6.Kinoshita K. & Honjo, T. (2001) Nat. Rev. Mol. Cell. Biol. 2, 493-503. [DOI] [PubMed] [Google Scholar]
- 7.Golding G. B., Gearhart, P. J. & Glickman, B. W. (1987) Genetics 115, 169-176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Martin A., Bardwell, P. D., Woo, C. J., Fan, M., Shulman, M. J. & Scharff, M. D. (2002) Nature (London) 415, 802-806. [DOI] [PubMed] [Google Scholar]
- 9.Yoshikawa K., Okazaki, I. M., Eto, T., Kinoshita, K., Muramatsu, M., Nagaoka, H. & Honjo, T. (2002) Science 296, 2033-2036. [DOI] [PubMed] [Google Scholar]
- 10.Pasqualucci L., Migliazza, A., Fracchiolla, N., William, C., Neri, A., Baldini, L., Chaganti, R. S. K., Klein, U., Kuppers, R., Rajewsky, K. & Dalla-Favera, R. (1998) Proc. Natl. Acad. Sci. USA 95, 11816-11821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pasqualucci L., Neumeister, P., Goossens, T., Nanjangud, G., Chaganti, R. S., Kuppers, R. & Dalla-Favera, R. (2001) Nature (London) 412, 341-346. [DOI] [PubMed] [Google Scholar]
- 12.Shen H. M., Peters, A., Baron, B., Zhu, X. & Storb, U. (1998) Science 280, 1750-1752. [DOI] [PubMed] [Google Scholar]
- 13.Muschen M., Re, D., Jungnickel, B., Diehl, V., Rajewsky, K. & Kuppers, R. (2000) J. Exp. Med. 192, 1833-1840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Martin A. & Scharff, M. D. (2002) Nat. Rev. Immunol. 2, 605-614. [DOI] [PubMed] [Google Scholar]
- 15.Storb U., Peters, A., Klotz, E., Kim, N., Shen, H. M., Hackett, J., Rogerson, B. & Martin, T. E. (1998) Immunol. Rev. 162, 153-160. [DOI] [PubMed] [Google Scholar]
- 16.Shen H. M., Michael, N., Kim, N. & Storb, U. (2000) Int. Immunol. 12, 1085-1093. [DOI] [PubMed] [Google Scholar]
- 17.Lin M. M., Green, N. S., Zhang, W. & Scharff, M. D. (1998) Int. Immunol. 10, 1121-1129. [DOI] [PubMed] [Google Scholar]
- 18.Zhu M., Rabinowitz, J. L., Green, N. S., Kobrin, B. J. & Scharff, M. D. (1995) Proc. Natl. Acad. Sci. USA 92, 2810-2814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang W., Bardwell, P. D., Woo, C. J., Poltoratsky, V., Scharff, M. D. & Martin, A. (2001) Int. Immunol. 13, 1175-1184. [DOI] [PubMed] [Google Scholar]
- 20.Bemark M. & Neuberger, M. S. (2000) Oncogene 19, 3404-3410. [DOI] [PubMed] [Google Scholar]
- 21.Zan H., Li, Z., Yamaji, K., Dramitinos, P., Cerutti, A. & Casali, P. (2000) J. Immunol. 165, 830-839. [DOI] [PubMed] [Google Scholar]
- 22.Bachl J., Carlson, C., Gray-Schopfer, V., Dessing, M. & Olsson, C. (2001) J. Immunol. 166, 5051-5057. [DOI] [PubMed] [Google Scholar]
- 23.Rogozin I. B. & Kolchanov, N. A. (1992) Biochim. Biophy. Acta 1171, 11-18. [DOI] [PubMed] [Google Scholar]
- 24.Rogozin I. B., Pavlov, Y. I., Bebenek, K., Matsuda, T. & Kunkel, T. A. (2001) Nat. Immunol. 2, 530-536. [DOI] [PubMed] [Google Scholar]
- 25.Zeng X., Winter, D. B., Kasmer, C., Kraemer, K. H., Lehmann, A. R. & Gearhart, P. J. (2001) Nat. Immunol. 2, 537-541. [DOI] [PubMed] [Google Scholar]
- 26.Rada C., Ehrenstein, M. R., Neuberger, M. S. & Milstein, C. (1998) Immunity 9, 135-141. [DOI] [PubMed] [Google Scholar]
- 27.Wilkie T. M. & Palmiter, R. D. (1987) Mol. Cell. Biol. 7, 1646-1655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kuppers R. & Dalla-Favera, R. (2001) Oncogene 20, 5580-5594. [DOI] [PubMed] [Google Scholar]
- 29.Michael N., Martin, T. E., Nicolae, D., Kim, N., Padjen, K., Zhan, P., Nguyen, H., Pinkert, C. & Storb, U. (2002) Immunity 16, 123-134. [DOI] [PubMed] [Google Scholar]
- 30.Maizels N. (1995) Cell 83, 9-12. [DOI] [PubMed] [Google Scholar]
- 31.Storb U., Peters, A., Klotz, E., Kim, N., Shen, H. M., Kage, K., Rogerson, B. & Martin, T. E. (1998) Curr. Top. Microbiol. Immunol. 229, 11-19. [DOI] [PubMed] [Google Scholar]
- 32.Okazaki I. M., Kinoshita, K., Muramatsu, M., Yoshikawa, K. & Honjo, T. (2002) Nature (London) 416, 340-345. [DOI] [PubMed] [Google Scholar]
- 33.Davidson N. O. & Shelness, G. S. (2000) Annu. Rev. Nutr. 20, 169-193. [DOI] [PubMed] [Google Scholar]
- 34.Petersen-Mahrt S. K., Harris, R. S. & Neuberger, M. S. (2002) Nature (London) 418, 99-104. [DOI] [PubMed] [Google Scholar]
- 35.Poltoratsky V., Goodman, M. F. & Scharff, M. D. (2000) J. Exp. Med. 192, F27-F30. [DOI] [PMC free article] [PubMed] [Google Scholar]