Abstract
Divalent metal transporter 1 (DMT1) mediates apical iron uptake into duodenal enterocytes and also transfers iron from the endosome into the cytosol after cellular uptake via the transferrin receptor. Hence, mutations in DMT1 cause systemic iron deficiency and anemia. DMT1 mRNA levels are increased in the duodenum of iron-deficient animals. This regulation has been observed for DMT1 mRNA harboring an iron–responsive element (IRE) in its 3′ UTR, but not for a processing variant lacking a 3′UTR IRE, suggesting that the IRE regulates the expression of DMT1 mRNA in response to iron levels. Here, we show that iron regulation of DMT1 involves the expression of a previously unrecognized upstream 5′ exon (exon 1A) of the human and murine DMT1 gene. The expression of this previously uncharacterized 5′ exon is tissue-specific and particularly prevalent in the duodenum and kidney. It adds an in-frame AUG translation initiation codon extending the DMT1 ORF by a conserved sequence of 29–31 amino acids. In combination with the IRE- and non-IRE variants in the 3′UTR, our results reveal the existence of four DMT1 mRNA isoforms predicting the synthesis of four different DMT1 proteins. We show that two regulatory regions, the 5′ promoter/exon 1A region and the IRE-containing terminal exon participate in iron regulation of DMT1 expression, which operate in a tissue-specific way. These results uncover an unexpected complexity of DMT1 expression and regulation, with implications for understanding the physiology, cell biology, and pathophysiology of mammalian iron metabolism.
Iron is an essential nutrient for nearly all living cells. It is required by a large number of heme and non-heme enzymes, and it plays an essential function in oxidative phosphorylation and oxygen transport. On the other hand, iron excess can cause irreversible cell and organ damage, because it can catalyze free radical formation which injures cellular membranes, nucleic acids, and proteins. Therefore, iron homeostasis must be tightly regulated to avoid iron deficiency and overload both at the systemic and at the cellular level.
The transferrin receptor (TfR) plays a key role for cellular iron uptake. It captures iron-loaded diferric transferrin at the plasma membrane and internalizes it by receptor-mediated endocytosis into endosomes (1). In iron-deficient cells, TfR expression is increased transcriptionally and posttranscriptionally (2). The latter response is mediated by iron-responsive elements (IREs) located in the 3′ UTR of TfR mRNA. Iron deficiency induces the binding of iron regulatory protein 1 or 2 to these IREs, which, in turn, protects the mRNA against nucleolytic degradation (3). Fusion of the 3′ UTR of TfR mRNA to heterologous transcripts suffices to confer iron regulation to reporter mRNAs (4, 5).
Iron taken up by means of the TfR must be transported across the endosomal membrane to be released into the cytosol. This transport is mediated by the divalent metal transporter 1 (DMT1, also known as Nramp2, SLC11A2, or DCT1; ref. 6), a conserved membrane protein. Remarkably, DMT1 not only participates in cellular iron absorption, but it also transports iron from the lumen of the gut across the apical membrane into the duodenal enterocytes, and hence also plays a critical role for systemic iron absorption (7). Rodents carrying a missense mutation in DMT1 (Gly-185-Arg), such as the “microcytic anemia (mk) mouse” and the “Belgrade rat” thus suffer from a recessively inherited systemic iron deficiency and from microcytic anemia (6, 8).
When systemic iron requirements are augmented, such as in iron deficiency, hypoxia, or stimulated erythropoiesis, the expression of DMT1 mRNA and protein in the duodenum is increased (9, 10), and more iron is absorbed. Cloning of DMT1 cDNAs has shown that alternative splicing and 3′ end processing yields two DMT1 mRNA variants that differ with regard to their 3′ terminal sequences of the ORF and their 3′ UTRs (6, 11, 12). One of these variants carries a conserved IRE (“IRE form”), whereas the other one lacks it (“non-IRE form”; ref. 12). Interestingly, only the IRE form has been found to be up-regulated in the duodenum of iron-deficient animals, suggesting that the IRE plays a similar role as in TfR mRNA and mediates iron regulation of DMT1 mRNA stability (7, 13). However, the IRE-containing DMT1 mRNA has displayed only a minor degree of iron regulation in Hep3B cells (14), and no change of endogenous DMT1 mRNA expression was observed in LMTK fibroblasts after changes in iron concentration or exposure to NO, although these stimuli effectively regulated TfR expression in these cells (15).
To shed light on the regulation of DMT1 expression by iron, we analyzed a cell culture model system for duodenal iron uptake, human Caco-2 cells, and samples taken from the duodenum of iron-deficient and control mice. Because DMT1 expression is thought to be critical to maintain both systemic and cellular iron homeostasis and to be altered in diseases of iron metabolism (16, 17), we investigated how changes in cellular and systemic iron levels alter DMT1 expression. We also wanted to understand better the role of the DMT1 IRE and to identify regulatory elements involved in controlling DMT1 expression. We show that a previously uncharacterized 5′ end mRNA processing variant of the human, mouse, and rat DMT1 gene affects both iron regulation and the deduced amino acid composition of the DMT1 proteins.
Materials and Methods
Animals.
Experiments were performed with the C57BL6 inbred mouse strain. For this study, weaned animals (about 4 weeks old) were fed for 4 weeks with a control diet (C1000) or an iron-deficient diet (C1038; Altromin, Lage, Germany). Efficiency of iron deprivation was ascertained by comparison of serum iron levels (29.3 − 6.8 μmol/liter receiving the control and 5.0 − 1.8 μmol/liter receiving the iron deficient diet) and of hemoglobin levels (15.0 ± 1.0 g/dl and 11.9 ± 0.7 g/dl, respectively).
Cell Culture and Iron Treatment.
Caco-2 cells were cultured in DMEM supplemented with 10% (vol/vol) FBS/100 μM nonessential amino acids/1 mM sodium pyruvate/100 units/liter penicillin/100 unit/liter streptomycin. 293 cells were cultured in DMEM supplemented with 10% (vol/vol) FBS/2 mM l-glutamine/100 units/liter penicillin/100 units/liter streptomycin. For iron regulation experiments, Caco-2 cells were seeded at a density of 6 × 105 cells in 10-cm plates precoated with 0.1% gelatin. After 6 days of culture, the medium was supplemented with 100 μM hemin or 100 μM desferrioxamine for 10–12 h.
5′ Rapid Amplification of cDNA Ends.
Total RNA was isolated from Caco-2 cells treated with desferrioxamine by using the RNA clean reagent (Hybaid) according to the manufacturer's instructions. Five μg of total RNA from Caco-2 cells treated with desferrioxamine was used to amplify the 5′ end of the human DMT1 mRNA with the Gene Racer kit (Invitrogen). The full-length mRNA was ligated to the Gene Racer oligo and reverse-transcribed by using avian myeloblastosis virus-reverse transcriptase (AMV-RT) and a gene-specific primer 5′ CACGGGTGGCTTCTTCTGTCAGCAG 3′. The amplification of 5′ ends of the cDNA was performed by using the Advantage 2 polymerase Mix (CLONTECH) with the Gene Racer 5′ Primer and the DMT1 specific primer described above. The cycling parameters were as follows: 1 min at 95°C, 25 cycles of 30 sec at 95°C, 90 sec at 68°C, and 2 min at 68°C. A nested PCR was performed by using the Gene Racer 5′ Nested Primer and a second DMT1-specific primer 5′ TTCTTCTGTCAGCAGGCCTTTAGAGATGC 3′. The amplification product was gel purified and cloned into pCR4-TOPO (Invitrogen). The relevant clones were sequenced by using the USB DNA sequencing kit. The sequence data for the exon 1A variants have been deposited in the GenBank database (accession nos. AJ493662 for the human, AJ493663 for the mouse).
For amplification of the 5′ end of the murine DMT1 mRNA, total RNA was prepared from mouse duodenum by using the Trizol reagent (GIBCO/BRL), according to the manufacturer's instructions. The cDNA was synthesized from 5 μg of total RNA by using Random Primers according to the Gene Racer kit‘s instructions. The first PCR amplification was performed by using the Gene Racer 5′ Primer and the mouse DMT1-specific primer 5′ CTAGGTAGGCAATGCTCATAAGAAAGCCAGG 3′ for 35 cycles with the settings described above. The second amplification was performed for 30 cycles with the Gene Racer 5′ Nested Primer and a second mouse DMT1-specific primer 5′ CATAAGAAAGCCAGGCCCCGTGAACGCC 3′. The PCR products were processed as described for the 5′ rapid amplification of cDNA ends of human DMT1.
Semiquantitative RT-PCR.
Total RNA was isolated by using the RNA clean reagent (Hybaid, Middlesex, U.K.) for cultured cell or Trizol reagent (GIBCO/BRL) for mouse tissue samples. The first strand of cDNA was obtained by reverse transcription from 5 μg of total RNA by using 1 μg of oligodT(18) primers and the Superscript II RT kit (GIBCO/BRL) according to the manufacturer’s instructions. After RNase H treatment, 5% of the reaction was used for PCR amplification with the Amplitaq DNA Polymerase (Perkin–Elmer). The amplification of the cDNA was performed at 94°C for 30 sec, 55°C for 40 sec, and 72°C for 40 sec for 16–35 cycles depending on the concentration of cDNA and the sets of primers used (listed in Table 1). The PCR products were resolved on ethidium bromide-stained gels, scanned, and quantified by using the Fuji FLA2000 FluorImager. The reported quantifications for mRNA levels are normalized with respect to β-actin mRNA. For each set of primers, the number of PCR cycles was optimized by using as control for linearity a titration of representative samples.
Table 1.
Sets of primers used in this study
| cDNA | Specificity | Forward primers, 5′–3′ | Reverse primers, 5′–3′ |
|---|---|---|---|
| hβactin | CTTCTACAATGAGCTGCGTG | GAGGATCTTCATGAGGTAGTC | |
| hTfR | CGATAACAGTCATGTGGA | AGTAACTGTTGCAGCCTTAC | |
| hDMT1 | 1A | GGAGCTGGCATTGGGAAAGTC | GGAGATCTTCTCATTAAAGTAAG |
| 1B | GTTGCGGAGCTGGTAAGAATC | GGAGATCTTCTCATTAAAGTAAG | |
| IRE | GAGCCAGTGTGTTTCTATGG | CCTAAGCCTGATAGAGCTAG | |
| Non-IRE | GGGAAGGGTGTTCAAAACTG | CAATGCAGCACGGAAAACTG | |
| 1A-IRE | GGAGCTGGCATTGGGAAAGTC | GTGGCTTCTTCTGTCAGCAG | |
| 1A-nonIRE | GGAGCTGGCATTGGGAAAGTC | CTGAGCTGTCAATCCCAGATG | |
| 1B-IRE | GTTGCGGAGCTGGTAAGAATC | GTGGCTTCTTCTGTCAGCAG | |
| 1B-nonIRE | GTTGCGGAGCTGGTAAGAATC | CTGAGCTGTCAATCCCAGATG | |
| mβactin | GACGACATGGAGAAGATCTG | TGAAGCTGTAGCCACGCTC | |
| mDMT1 | 1A | GTACTCCTCTGCATATATAGAGG | CTAGGTAGGCAATGCTCATAAGAAAGCCAGG |
| 1B | CAATCACGGGAGGGCAGGAG | CTAGGTAGGCAATGCTCATAAGAAAGCCAGG | |
| IRE | CTGCTGAGCGAAGATACCAG | CTCAGGAGCTTAGGTCAGAAG | |
| Non-IRE | CGCCCAGATTTTACACAGTG | AAGCTTCACTACCTGCACAC | |
| 1A-IRE | GTACTCCTCTGCATATATAGAGG | CTTCGCTCAGCAGGACTTTCG | |
| 1A-nonIRE | GTACTCCTCTGCATATATAGAGG | GACCCATACGCAACTGTCAG | |
| 1B-IRE | AGGTGGCGGAGCCGAATC | CTTCGCTCAGCAGGACTTTCG | |
| 1B-nonIRE | AGGTGGCGGAGCCGAATC | GACCCATACGCAACTGTCAG |
Results and Discussion
Cell-Specific Iron Regulation of DMT1.
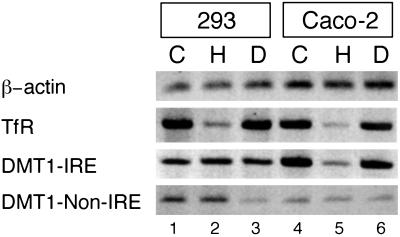
To gain insight into the molecular mechanism of iron regulation of the DMT1 gene, we first investigated human Caco-2 cells, a cellular model system that recapitulates the DMT1 regulation observed in dietary iron-deficient animals (14). Cells were treated with the iron donor, hemin (H), with the iron chelator desferrioxamine (D), or left untreated (C). The effect of the iron treatment on DMT1 mRNA expression was assessed by RT-PCR, using specific primers to distinguish between the IRE and the non-IRE isoforms. The TfR and β-actin mRNAs were assessed as positive and negative controls, respectively. As shown in Fig. 1 (lanes 4–6), the IRE form of DMT1 is iron regulated, whereas the non-IRE form is not. This result is consistent with earlier findings (13, 14). Interestingly, human 293 cells (lanes 1–3) and HeLa cells (data not shown) display no regulation of either the IRE or the weakly expressed non-IRE form (lanes 1–3), although TfR mRNA as a positive control is clearly iron regulated. Thus, the IRE form of DMT1 mRNA is regulated by iron in Caco-2 but not in 293 or HeLa cells. These results reflect the existence of cell-specific differences and demonstrate that the 3′ UTR containing the IRE is not sufficient to mediate iron regulation.
Fig 1.
Iron regulation of 3′ end variants of DMT1 mRNA in 293 and Caco-2 cells. Semiquantitative RT-PCR was performed by using mRNA from 293 cells (lanes 1–3) or Caco-2 cells (lanes 4–6) that were treated with hemin (H) or desferrioxamine (D), or the remained untreated (C) during 12 h, as described in Materials and Methods. PCR amplification (16 cycles) was performed with hβ-actin primers, and 23 cycles with hTfR, hDMT1-IRE primers, and hDMT1-non-IRE primers. All PCR products have the respective predicted size.
A Conserved 5′ Exon Extends the ORF of DMT1.
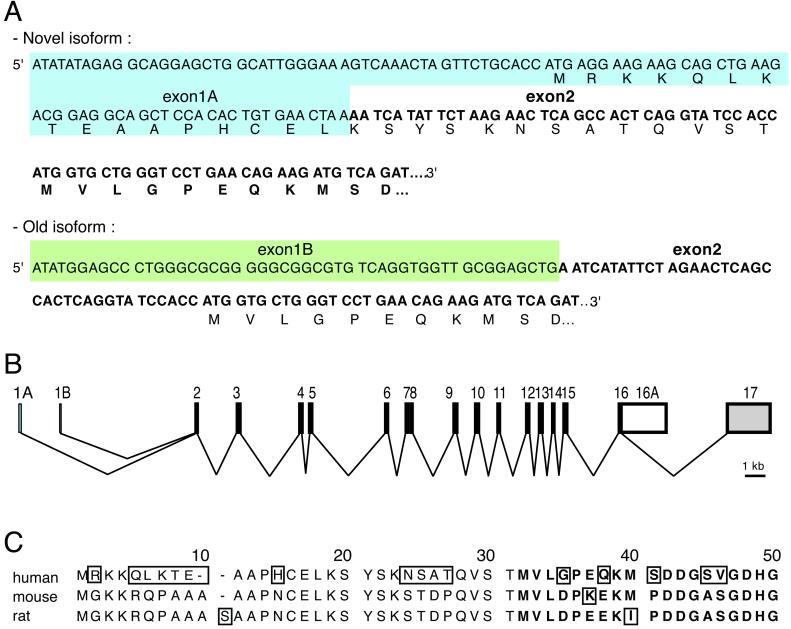
In principle, the observed cell-specific differences could arise from the presence or absence of transacting regulatory factors in Caco-2 and 293/HeLa cells, respectively, or from heterogeneity between the IRE-containing DMT1 mRNAs themselves. To explore the latter possibility and to determine whether regulatory elements could be present in the 5′ end of the DMT1 mRNA, a rapid amplification of cDNA ends was performed by using mRNA from desferrioxamine-treated Caco-2 cells. Sequencing of the amplification products revealed the presence of two species of DMT1 cDNA (Fig. 2A). One corresponds perfectly to the published DMT1 cDNA sequence (ref. 18; GenBank accession no. AB004857); the second aligns precisely with the published DMT1 cDNA sequence downstream from the beginning of exon 2 but totally differs from the published exon 1 sequence (AJ493662). An alignment of this previously uncharacterized 5′ end sequence with the human DMT1 gene (NT-009782) shows that these 99 nucleotides correspond exactly to a sequence located 1.9 kb upstream from the previously identified exon 1. This genomic sequence is immediately followed by a consensus splice donor sequence (5′ GTaag 3′), suggesting that two variant DMT1 mRNAs are generated from alternative promoters with subsequent, mutually exclusive splicing of the respective first exons to exon 2. These findings reveal a genomic organization of the human DMT1 gene (Fig. 2B) with two alternative transcription start sites, one for the previously uncharacterized exon 1, called “exon 1A,” and the second for the previously described first exon, renamed “exon 1B.”
Fig 2.
Identification of a previously uncharacterized isoform of the DMT1 mRNAs. (A) Sequences of the alternative 5′ regions of the human DMT1 mRNA. The sequences of the two DMT1 variants are identical after reaching the sequence corresponding to exon 2. The specific sequences of each isoform are boxed in blue for the newly identified exon 1 (exon 1A) and in green for the previously described exon 1 (exon1B). (B) Revised genomic organization of the human DMT1 gene. The sequence of the exon 1A has been mapped 1.9 kb upstream of exon 1B in the human genomic sequence (NT-009782). The two 5′ DMT1 variants are generated by alternative promoter usage followed by splicing to exon 2. (C) Alignment of the predicted N-terminal extensions encoded by the exon 1A isoforms of human, mouse, and rat (AF008439) DMT1. Amino acids that are boxed are not conserved in all three species.
Interestingly, exon 1A possesses an AUG codon that is in frame with the previously assigned translation initiation codon in exon 2 (Fig. 2A). This 1A isoform of DMT1 mRNA thus may encode a version of the DMT1 protein with an additional N-terminal domain.
To determine whether the alternative exon 1A is conserved in other species, we next performed a 5′ rapid amplification of cDNA ends-PCR with RNA from mouse duodenum. As in Caco-2 cells, we discovered two 5′ variants of the DMT1 mRNA (data not shown). One of these 5′ variants corresponds to the formerly reported sequence of the mouse DMT1 cDNAs (AF029758, L33415) and extends farther upstream than the sequence present in the database. The exon 1 sequence of the second 5′ variant of the mouse DMT1 mRNA does not align with any published murine cDNA or genomic sequence (AJ493663). Interestingly, this 5′ end variant also harbors an AUG triplet that may specify an N-terminal extension of 30 amino acids of the mouse DMT1 protein, similar to what we observed for the human exon 1A isoform (Fig. 2C). Although the DNA sequences of the human and the mouse DMT1 exons 1A only display 30% identity (data not shown), the amino acid sequences of the predicted N-terminal extensions are 66% identical (Fig. 2C). This conservation strongly supports the notion that the exon 1A of DMT1 mRNA specifies an isoform of the DMT1 protein.
In further support of this conclusion, database searches revealed the presence of rat DMT1 cDNA sequences with two different 5′ end regions (AF008439, AF029758). One of these (AF008439) predicts a 5′ extension of the ORF before the annotated coding region, which is in-frame with the rest of the polypeptide sequence of DMT1. This additional amino acid sequence shares more than 80% similarity with the mouse N-terminal extension and more than 60% with the human sequence (Fig. 2C). In all three cases, the 5′ AUG codon is framed by a favorable context to initiate translation (gccA/GccAUGA/G; ref. 19).
The additional N-terminal peptide sequences do not show significant similarity with any known protein domains during BLAST or SMART searches. Further analysis by different search algorithms (SIGNAL P or PSPORTII) failed to predict a signal peptide sequence or cleavage site. The predicted subcellular localization for the extended form of DMT1 is the plasma membrane (73.9% by PSPORTII), which corresponds to the localization of the DMT1 protein in the duodenum (13, 20, 21).
The Expression of the Exon 1A DMT1 mRNA Is Tissue-Specific.
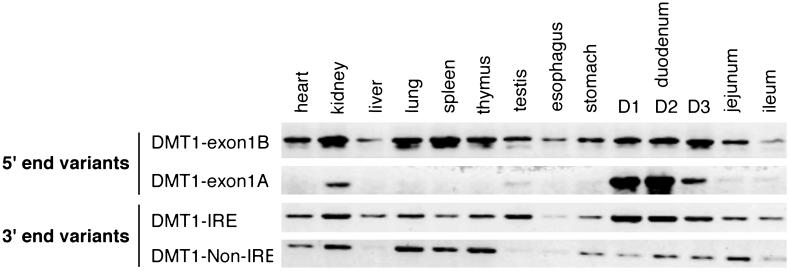
Next, we examined different mouse tissues for the expression of the 5′ (1A or 1B) and 3′ (IRE or non-IRE) variants by RT-PCR. For the former, we used forward primers specific for exons 1A or 1B, respectively, and a common reverse primer which hybridizes to exon 3; the latter was determined with specific primers for the IRE and non-IRE containing last exons. As shown in Fig. 3, the exon 1B isoform appears to be ubiquitously expressed. By contrast, the expression of the exon 1A isoform is tissue-specific, with the highest expression in the duodenum and the kidney, and lower expression in the testis and other parts of the gastrointestinal tract.
Fig 3.
Expression pattern of the DMT1 5′ and 3′ end variants in mouse tissues. Total RNA from organs and tissue obtained from a C57BL6 mouse were reverse-transcribed by using random primers. PCR amplifications were performed by using specific primers of the alternative 5′ exon (DMT1-1A vs. DMT1-1B) or the alternative 3′ terminal exon (DMT1-IRE vs. DMT1-non-IRE). D1 refers to the 1st cm of the duodenum, D2 and D3 refer to the 2nd and the 3rd cm respectively. All depicted PCR products have the respective predicted sizes. Analysis of total RNA from mouse brain homogenate revealed the expression of the 1B isoform as well as of the IRE and non-IRE 3′ variants, but no evidence for the expression of the 1A isoform in brain could be obtained (data not shown).
Interestingly, both the IRE and the non-IRE isoforms are expressed in most of the tested organs and tissues. Thus, the presence or absence of the IRE in the 3′ UTR is unlikely to explain the observed cell type-dependent differences in iron regulation. Earlier studies (7, 13) found the strongest regulation of DMT1 mRNA in the duodenum of iron-deficient animals. Thus, we wondered whether the strong duodenal expression of the exon 1A isoform may play a role in the iron regulation of DMT1 mRNAs.
Iron Regulation of the Exon 1A Isoform in Caco-2 Cells.
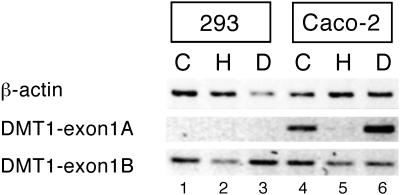
To obtain further information regarding the expression pattern of the exon 1A/1B isoforms and to complement the analysis of iron regulation of the DMT1 3′ variants in Fig. 1, iron regulation of the exon 1A vs. 1B isoforms was assessed in 293, HeLa, and Caco-2 cells. Semiquantitative RT-PCR was performed by using a specific set of forward primers for each 5′ variant of human DMT1 mRNA (Fig. 4). Although the exon 1B isoform is well expressed in these cell lines, it is poorly regulated by iron: the D/H ratio that reflects iron regulation is less than 1.5. Interestingly, the exon 1A isoform is not detectably expressed in 293 cells (or HeLa cells; data not shown), but it is well expressed in the Caco-2 cells. This finding is consistent with the restricted expression pattern of the 1A isoform in mouse tissues (Fig. 3). Notably, the exon 1A isoform in Caco-2 cells is strongly regulated by iron. In view of the regulatory patterns of the DMT1 3′ variants (Fig. 1), these observations suggest that the iron regulation of DMT1 mRNA in Caco-2 cells may involve the expression of exon 1A in addition or alternatively to the expression of the IRE 3′ variant.
Fig 4.
Iron regulation of 5′ end variants of DMT1 mRNA in human cell lines. Semiquantitative RT-PCR was performed on total RNA from cells treated as in Fig. 1 by using 16 PCR cycles for hβ-actin and 25 cycles for DMT1 1A and 1B. The forward primers used to amplify the two 5′ variants of DMT1 hybridized to the respective exon 1 (A or B) and the reverse primer to exon 3. The PCR products have a size of 292 and 223 bp, respectively. Shown is one representative experiment of three.
Iron Regulation of the Four Isoforms of DMT1 mRNA in Cell Lines and Mouse Tissues.
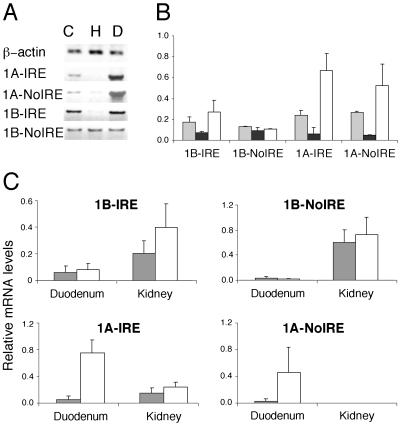
The combination of alternative 5′ and 3′ exons (1A or 1B and IRE or non-IRE, respectively) could specify up to four distinct DMT1 mRNAs encoding four DMT1 polypeptides in different cells and tissues. Although the different 5′ and 3′ exons are separated by 1.7 kb, we decided to optimize the PCR conditions to be able to reliably and specifically distinguish between these variants. To this end, exon 1A- or 1B-specific forward primers were combined with IRE- or non-IRE-specific reverse primers, as described in Materials and Methods; β-actin was used as a calibration standard. As shown in Fig. 5A, the four conceptual isoforms of DMT1 mRNA are indeed all expressed in Caco-2 cells, and they respond differently to iron treatment. As expected from Figs. 1 and 4, the expression of the 1B-non-IRE isoform displays no iron regulation (A and B). The 1A-IRE and the 1A-non-IRE isoforms are both increased in desferrioxamine-treated cells. The regulation of the 1A-non-IRE isoform clearly shows that the 3′ UTR IRE is not mandatory for iron regulation. Rather, the presence of exon 1A is itself associated with iron regulation.
Fig 5.
Iron regulation of the four alternative isoforms of DMT1 mRNA in Caco-2 cells (A and B) as well as mouse kidney and duodenum (C). (A) Caco-2 cells were treated as described in Fig. 1. Total RNA was analyzed by semiquantitative RT-PCR using isoform-specific forward and reverse primers, as described in the text. PCR cycles (16) were used to amplify hβ-actin cDNA, 27 cycles for 1A-IRE, 1B-IRE, and 1B-non-IRE cDNAs, and 31 cycles for 1A-non-IRE cDNA. (B) Quantification of three experiments like the one shown in A by RT-PCR. Gray bars correspond to control, black to hemin, and white to desferrioxamine to treated samples. (C) Quantification of the expression of the four mDMT1 mRNA isoforms in duodenum and kidney from mice fed with an iron deficient diet (white bars) or with a control diet (gray bars). Bars represent average values obtained from four mice each ± SD. No expression of the 1A-non-IRE isoform was detected in kidneys.
The 1B-IRE isoform of DMT1 is also regulated by iron (Fig. 5 A and B), showing that transcription from the 1A promoter is not obligatory for iron regulation. This finding and the lack of regulation of the 1B-non-IRE variant rather indicate that the 3′ UTR containing the IRE contributes to regulation. It is important to note that the number of PCR cycles needed to obtain a detectable signal for the 1A-non-IRE isoform (31 cycles) is significantly higher than for 1B-non-IRE (27 cycles). This fact suggests that the 1B-non-IRE mRNA is more abundant and, hence, dominates the overall regulation pattern observed for the non-IRE forms in Caco-2 cells (Fig. 1). Collectively, these data argue that the iron regulation of DMT1 mRNAs in Caco-2 cells involves at least two regulatory regions, one associated with the expression of the 1A isoform (the 1A promoter or the exon itself) and the other mapping to the IRE-containing 3′ UTR.
Finally, we assessed the expression and regulation of the four mRNAs in the two tissues in which the exon 1A form is most actively expressed in the mouse, the proximal duodenum, and the kidney (Fig. 3). To this end, RNA was prepared from mice fed an iron-deficient vs. a control diet, and a PCR strategy analogous to the analysis of Caco-2 cells was followed. In essence, the findings with mRNA from the proximal duodenum of mice closely reflect the results from human Caco-2 cells (Fig. 5C). Therefore, the conclusions drawn above apply to both the physiological site and a cell culture model system for intestinal iron absorption. Collectively, they suggest that the reported strong iron regulation of DMT1 mRNA (7, 13) can mainly be attributed to the expression of the 1A-IRE isoform.
Analysis of the kidney, where DMT1 may play a role in iron reabsorption (22), yields quite different results (Fig. 5C). First, the 1A-non-IRE isoform is not detectably expressed. Second, the overall range of iron regulation of 1A-IRE and 1B-IRE is less than in the duodenum. This difference is particularly striking for the 1A-IRE mRNA, which is only 1.5-fold more abundant in the kidney of iron-deficient than control animals. Thus, both IRE-containing DMT1 isoforms are iron regulated, irrespective of the nature of exon 1. Taken together, these observations suggest that DMT1 regulation in the kidney is associated with the presence of IRE in the 3′ UTR, whereas in the duodenum, iron regulation is most strongly associated with the presence of exon 1A. Thus, not only do at least two regulatory regions seem to be involved in iron regulation, but their importance seems to differ in a tissue-specific way.
Implications for DMT1 Function and Regulation.
The discovery of exon 1A-bearing isoforms of DMT1 sheds light on the complexity of DMT1 expression, function, and regulation. Our findings show that alternative promoters and alternative RNA processing creates a combination of four DMT1 mRNA variants with different first and last exons. All of these mRNA variants are expressed in the proximal duodenum, where systemic iron absorption is regulated. These variants specify four different ORFs that encode four distinct polypeptides. It will be important to consider this heterogeneity in the context of DMT1 function(s) in different tissues. Because DMT1 can transport multiple divalent metals (7, 23, 24), it is conceivable that the isoproteins could differ in their metal selectivity and/or transport capacity for divalent metals.
In the light of the different subcellular localizations of DMT1 (13, 22, 25, 26), it is also important to consider possible roles of the N and/or C terminus on the intracellular localization of the DMT1 isoforms. Strikingly, the 1A isoform of DMT1 mRNA is most actively expressed in duodenal and kidney cells, which are polarized. In these polarized cells, DMT1 localizes to the apical membrane (13, 22). By contrast, the broad distribution of the 1B isoforms correlates with the role of DMT1 in the release of iron from endosome, following iron uptake by the transferrin cycle (26). We suggest that the exclusive N-terminal peptide sequence encoded by exon 1A may play a role in targeting DMT1 to the apical membrane of polarized cells. Furthermore, different DMT1 polypeptides may interact with distinct sets of partner or regulatory proteins, adding further complexity to the biology of the transport of divalent metals into and within the body.
Analysis of cell lines and murine tissues also shows that the variant forms of DMT1 mRNA differ in terms of iron regulation. At least two regulatory regions seem to be involved, one being the IRE-containing 3′ UTR exon. The other region is defined by the presence of exon 1A, and may include the promoter and/or the exon itself. We have analyzed the human DMT1 gene sequences extending 2.8 kb upstream of the exon 1A transcription start site. This analysis yielded no informative consensus sequences. In particular, no close matches were found for the consensus binding sequence of Hif-1 (G/Y A C G T G C G/T; ref. 27) or for the metal response elements (MRE; T G C R C N C followed by a G/C-rich region; ref. 28). Similarly, and in contrast to an earlier report (12), we did not identify MREs in the genomic sequence upstream of exon 1B. We conclude that DMT1 expression may be controlled both at the transcriptional and at the posttranscriptional level by iron. These findings may help to illuminate earlier and seemingly contradictory results: the IRE-containing 3′ UTR was found to play a role in iron regulation (7, 13), but not to mediate profound iron regulation of reporter mRNAs (29) nor to suffice for iron regulation of endogenous DMT1 mRNA in Hep 3B hepatoma cells or LMTK-fibroblasts (14, 15). The two regulatory regions also seem to be used in different ways in distinct tissues, as indicated by the predominant regulatory role of the exon 1A related region in the duodenum and the IRE-containing final exon in the kidney. Having identified these regulatory regions and their use, it is now possible to address their mechanism(s) of function and biological role(s).
Acknowledgments
We thank Dr. Martina Muckenthaler for providing mouse tissue samples and Karen Brennan for help with the mouse work. We thank Dr. Bruno Galy for fruitful discussions. Funds from the Gottfried Wilhem Leibniz Prize were awarded by the Deutsche Forschungsgemeinschaft (to M.W.H.). N.H. was supported by the Association de Recherche contre le Cancer and by a European Molecular Biology Laboratory long-term fellowship.
Abbreviations
TfR, transferrin receptor
IRE, iron-responsive element
DMT1, divalent metal transporter 1
References
- 1.Andrews N. C. (2000) Nat. Rev. Genet. 1, 208-217. [DOI] [PubMed] [Google Scholar]
- 2.Casey J. L., Di Jeso, B., Rao, K., Klausner, R. D. & Harford, J. B. (1988) Proc. Natl. Acad. Sci. USA 85, 1787-1791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hentze M. W. & Kühn, L. C. (1996) Proc. Natl. Acad. Sci. USA 93, 8175-8182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Casey J. L., Hentze, M. W., Koeller, D. M., Caughman, S. W., Rouault, T. A., Klausner, R. D. & Harford, J. B. (1988) Science 240, 924-928. [DOI] [PubMed] [Google Scholar]
- 5.Müllner E. W. & Kühn, L. C. (1988) Cell 53, 815-825. [DOI] [PubMed] [Google Scholar]
- 6.Fleming M. D., Romano, M. A., Su, M. A., Garrick, L. M., Garrick, M. D. & Andrews, N. C. (1998) Proc. Natl. Acad. Sci. USA 95, 1148-1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gunshin H., Mackenzie, B., Berger, U. V., Gunshin, Y., Romero, M. F., Boron, W. F., Nussberger, S., Gollan, J. L. & Hediger, M. A. (1997) Nature (London) 388, 482-488. [DOI] [PubMed] [Google Scholar]
- 8.Fleming M. D., Trenor, C. C., 3rd, Su, M. A., Foernzler, D., Beier, D. R., Dietrich, W. F. & Andrews, N. C. (1997) Nat. Genet. 16, 383-386. [DOI] [PubMed] [Google Scholar]
- 9.Canonne-Hergaux F., Fleming, M. D., Levy, J. E., Gauthier, S., Ralph, T., Picard, V., Andrews, N. C. & Gros, P. (2000) Blood 96, 3964-3970. [PubMed] [Google Scholar]
- 10.Canonne-Hergaux F., Levy, J. E., Fleming, M. D., Montross, L. K., Andrews, N. C. & Gros, P. (2001) Blood 97, 1138-1140. [DOI] [PubMed] [Google Scholar]
- 11.Gruenheid S., Cellier, M., Vidal, S. & Gros, P. (1995) Genomics 25, 514-525. [DOI] [PubMed] [Google Scholar]
- 12.Lee P. L., Gelbart, T., West, C., Halloran, C. & Beutler, E. (1998) Blood Cells Mol. Dis. 24, 199-215. [DOI] [PubMed] [Google Scholar]
- 13.Canonne-Hergaux F., Gruenheid, S., Ponka, P. & Gros, P. (1999) Blood 93, 4406-4417. [PubMed] [Google Scholar]
- 14.Gunshin H., Allerson, C. R., Polycarpou-Schwarz, M., Rolfs, A., Rogers, J. T., Kishi, F., Hentze, M. W., Rouault, T. A., Andrews, N. C. & Hediger, M. A. (2001) FEBS Lett. 509, 309-316. [DOI] [PubMed] [Google Scholar]
- 15.Wardrop S. L. & Richardson, D. R. (1999) Eur. J. Biochem. 263, 41-49. [DOI] [PubMed] [Google Scholar]
- 16.Zoller H., Pietrangelo, A., Vogel, W. & Weiss, G. (1999) Lancet 353, 2120-2123. [DOI] [PubMed] [Google Scholar]
- 17.Zoller H., Koch, R. O., Theurl, I., Obrist, P., Pietrangelo, A., Montosi, G., Haile, D. J., Vogel, W. & Weiss, G. (2001) Gastroenterology 120, 1412-1419. [DOI] [PubMed] [Google Scholar]
- 18.Kishi F. & Tabuchi, M. (1997) Mol. Immunol. 34, 839-842. [DOI] [PubMed] [Google Scholar]
- 19.Kozak M. (1999) Gene 234, 187-208. [DOI] [PubMed] [Google Scholar]
- 20.Griffiths W. J., Kelly, A. L., Smith, S. J. & Cox, T. M. (2000) Q. J. Med. 93, 575-587. [DOI] [PubMed] [Google Scholar]
- 21.Trinder D., Oates, P. S., Thomas, C., Sadleir, J. & Morgan, E. H. (2000) Gut 46, 270-276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ferguson C. J., Wareing, M., Ward, D. T., Green, R., Smith, C. P. & Riccardi, D. (2001) Am. J. Physiol. Renal. Physiol. 280, F803-F814. [DOI] [PubMed] [Google Scholar]
- 23.Tallkvist J., Bowlus, C. L. & Lonnerdal, B. (2001) Toxicol. Lett. 122, 171-177. [DOI] [PubMed] [Google Scholar]
- 24.Yamaji S., Tennant, J., Tandy, S., Williams, M., Singh Srai, S. K. & Sharp, P. (2001) FEBS Lett. 507, 137-141. [DOI] [PubMed] [Google Scholar]
- 25.Roth J. A., Horbinski, C., Feng, L., Dolan, K. G., Higgins, D. & Garrick, M. D. (2000) J. Neurosci. 20, 7595-7601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tabuchi M., Yoshimori, T., Yamaguchi, K., Yoshida, T. & Kishi, F. (2000) J. Biol. Chem. 275, 22220-22228. [DOI] [PubMed] [Google Scholar]
- 27.Semenza G. L., Roth, P. H., Fang, H. M. & Wang, G. L. (1994) J. Biol. Chem. 269, 23757-23763. [PubMed] [Google Scholar]
- 28.Culotta V. C. & Hamer, D. H. (1989) Mol. Cell. Biol. 9, 1376-1380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tchernitchko D., Bourgeois, M., Martin, M. E. & Beaumont, C. (2002) Biochem. J. 363, 449-455. [DOI] [PMC free article] [PubMed] [Google Scholar]