Abstract
A sensor protein ChvG is part of a chromosomally encoded two-component regulatory system ChvG/ChvI that is important for the virulence of Agrobacterium tumefaciens. However, it is not clear what genes ChvG regulates or what signal(s) it senses. In this communication, we demonstrate that ChvG is involved in the regulation of acid-inducible genes, including aopB and katA, residing on the circular and linear chromosomes, respectively, and the tumor-inducing (Ti)-plasmid-harbored vir genes, virB and virE. ChvG was absolutely required for the expression of aopB and very important for the expression of virB and virE. However, it was responsible only for the responsiveness of katA and, to a limited extent, the vir genes to acidic pH. ChvG appears to play a role in katA expression by repressing katA at neutral pH. ChvG had no effect on the expression of two genes that were not acid-inducible. Because ChvG regulates unlinked acid-inducible genes encoding different functions in different ways, we hypothesize that ChvG is a global sensor protein that can directly or indirectly sense extracellular acidity. We also analyzed the re-sequenced chvG and found that ChvG is more homologous to its Sinorhizobium meliloti counterpart ExoS than was previously thought. Full-length ChvG is conserved in members of the α-proteobacteria, whereas only the C-terminal kinase domain is conserved in other bacteria. Sensing acidity appears to enable Agrobacterium to coordinate its coping with the environment of wounded plants to cause tumors.
Bacteria are subjected continually to various rapid and unexpected environmental changes, such as fluctuating nutrient and toxin levels, acidity, temperature, cell density, and water availability. To survive in their ever changing environments, the cells must constantly monitor external conditions and adjust their structure, physiology, and behavior accordingly. To achieve this, they must be able to alter their gene expression rapidly and efficiently in response to environmental signals. Bacterial two-component sensory transduction systems are common components of complex regulatory networks and cascades. They consist of two protein components: a sensor that monitors some environmental parameter and a response regulator that mediates a change in gene expression in response to sensor signals. The two-component regulatory systems are of prime importance in transmitting environmental signals and regulating adaptive responses (for reviews, see refs. 1 and 2). The ability to respond to environmental stimuli is especially important for plant-associated bacteria that compete with other organisms both in the soil and in the plant. Many two-component sensory transduction systems operate in plant-associated bacteria (3), and these often control complex regulatory networks and cascades. For example, Agrobacterium tumefaciens genome contains at least 25 two-component pathways (4, 5). The best studied is VirA/VirG (3, 6), which controls the expression of the virulence (vir) genes that are required for causing crown gall tumors on plants.
A. tumefaciens is a Gram-negative plant pathogen that infects a wide range of plants. The infection occurs at plant wound sites and involves the transfer of oncogenic DNA (T-DNA) from the bacterium into the plant cell nucleus. The T-DNA and the vir genes required for T-DNA transfer are located on a large plasmid called the tumor-inducing (Ti) plasmid. In addition, some chromosomal genes are involved in the infection process (for reviews, see refs. 7–9).
In an attempt to identify additional chromosomal virulence genes, Cangelosi et al. (10) used TnphoA to identify insertion mutations in genes that encode proteins with extracytoplasmic domains, the rationale being that certain virulence determinants are likely to be associated with the cell envelope. A number of avirulent mutants were identified and characterized. Charles and Nester (11) found that two of the mutants had lesions in a gene encoding a putative sensor protein, which was designated chvG; an adjacent cognate response regulator (chvI) was identified by additional sequencing of the region. The same two-component system chvG/chvI was identified independently by Mantis and Winans (12) by complementing an Escherichia coli phoB mutant with members of an Agrobacterium clone bank. Interestingly, mutations at either chvG or chvI abolished tumor-forming ability, suggesting that ChvG and ChvI, the first chromosomal two-component system identified in A. tumefaciens, are directly or indirectly required for virulence. Elucidating the environmental signal(s) that activates ChvG and identifying ChvI target genes should shed light on the role these proteins play in regulating A. tumefaciens virulence.
The Sinorhizobium meliloti exoS and chvI genes (13, 14) are highly homologous to the A. tumefaciens chvG and chvI genes, respectively. In S. meliloti, ExoS and ChvI appear to regulate the production of succinoglycan, which plays a crucial role in establishing the symbiosis between S. meliloti and its host plant alfalfa. Brucella abortus BvrS and BvrR, which control the cell invasion and virulence of B. abortus on animal cells (15), are also homologous to ChvG and ChvI, respectively.
The responses of bacteria to pH are of interest for many reasons (16, 17). Many bacteria, including A. tumefaciens, E. coli, and Salmonella typhimurium, grow best at neutral pH, but they can also grow in moderate acid or base, with a positive or negative transmembrane pH difference. Pathogenic bacteria often encounter extremes of pH both within and outside their hosts. During the infection process, the cells are exposed to low pH in the host. Consequently, low pH is often one of the signals that induce virulence factors that contribute to pathogenesis. However, how pH regulates bacterial gene expression is not well understood.
Acidic conditions play an important role in Agrobacterium–plant interactions. Expression of the vir genes is controlled primarily through the VirA/VirG two-component system and depends on external acidification following release of acids at the wound site of the plant (6). Two distinct pH-mediated responses of vir gene induction have been identified: the acid-dependent transcription of virG and the VirA/VirG-dependent transcription of the vir operons, which include virB and virE. Both responses are maximally expressed at acidic pH (18, 19).
Recently, we identified two chromosomal genes, katA and aopB, both of which are inducible by an acidic pH and are involved in A. tumefaciens tumorigenesis (20, 21). katA encodes a catalase that is involved in detoxification of H2O2 released during Agrobacterium–plant interaction. aopB encodes an outer membrane protein exposed on the bacterial cell surface.
Because ChvG is a sensor protein and chvG null mutants are highly sensitive to acidic pH (11), we determined whether ChvG/ChvI might be involved in the regulation of acidic pH-inducible genes. The experiments reported in this paper show that ChvG regulates acidic inducibility of chromosomal genes, aopB and katA, and the Ti-plasmid-harbored vir genes, virB and virE. We hypothesize that ChvG is a global sensor protein that directly or indirectly senses the acidity of the environment.
Materials and Methods
Bacterial Strains, Plasmids, and Media.
The bacterial strains, plasmids, and primers used in this work are listed in Table 4, which is published as supporting information on the PNAS web site, www.pnas.org. The construction of new strains and plasmids is described in the text. LB medium was used for E. coli; MG/L, AB, and IB media (10) were used for A. tumefaciens. The original IB medium is pH 5.5 (10) and is designated IB pH 5.5 in this study. When the pH of the IB medium was adjusted to pH 7.0, the medium was designated IB pH 7.0.
Cloning and Sequencing.
The genomic DNA of A. tumefaciens strain C58 was used as the template for PCR to amplify the chvG gene. The two primers ChvG-UP and ChvG-DN (see Table 4) were complementary to the sequences upstream and downstream of the chvG gene based on the published sequence (11). The PCR product (2.1 kb) was digested with BamHI and HindIII, and then cloned into BamHI–HindIII-digested pRSET-A (Invitrogen) to generate pLP30. Both strands of the DNA fragments containing chvG were sequenced from pLP30, pTC147, and pTC190 with Applied Biosystems PRISM Ready Reaction Dye Primer Cycle Sequencing Kit and ABI PRISM 377 DNA Sequencer (Perkin–Elmer) according to the manufacturer's instructions.
Complementation.
To amplify the wild-type chvG by PCR, we used the C58 genomic DNA as the template and two new primers, ChvG-H and ChvG-B (see Table 4) based on the corrected chvG sequence. The resulting 2.5-kb PCR product was digested with BamHI and HindIII, and cloned into BamHI–HindIII-digested pSW172. The resulting plasmid pLP36 was introduced into A6340 by triparental mating, producing strain A6340(pLP36).
Generation of the His-ChvG Fusion Protein and ChvG Antibody.
To generate ChvG fusion proteins that can be subsequently purified by affinity chromatography, we fused the chvG coding sequence in frame with the DNA sequence encoding the six histidine residues in the fusion vector pRSET-A. C58 genomic DNA was used as the template; two primers ChvG-N and ChvG-C (see Table 4) complementary to the corrected sequence of chvG gene were used; and a partial chvG gene fragment (containing amino acids 196–596 of ChvG) was amplified by PCR. The pRSET-A and the 1.2-kb PCR product were double-digested by BamHI and HindIII, purified, and further ligated together to generate the plasmid pLP33. The proper construction was confirmed by restriction digestion, and the proper in-frame fusion of ChvG with the His tag in the vector was confirmed by sequencing. The plasmid DNA was transformed into competent cells of BL21(DE3).
The resulting strain overproduced the fusion protein in the form of inclusion body, which was collected and solubilized in 8 M urea; the fusion protein was purified in a metal-affinity TALON (CLONTECH) resin column. The column-purified protein was dialyzed in sterile dH2O for 24 hr with stirring and three changes of water. The protein was gel purified as described by Hager and Burgess (22), and was precipitated further by TCA as described by Bollag et al. (23). The resulting His-ChvG fusion protein was used to raise the polyclonal antibody in a rabbit by intramuscular injection.
Detection of ChvG Protein in A. tumefaciens.
To detect the ChvG protein in A. tumefaciens, the bacterial membrane fraction was prepared according to the protocols described by Chervitz et al. (24) and Cheng and Walker (14). Briefly, A. tumefaciens cells were grown in liquid MG/L and then grown in liquid IB (pH 5.5) medium at 28°C with vigorous shaking for 16–18 hr. The cells were collected by centrifugation and fractionated after sonication; the fractions were then subjected to immunoblot analysis.
Gene Expression Measurements.
The aopB, katA, and rrn gene expression were measured by using the aopB:gfp, katA:gfp, and rrn:gfp fusions (see Table 4), respectively. The virB and virE gene expression were measured by using the virB:lacZ and virE:lacZ fusions (see Table 4), respectively. A. tumefaciens cells containing the appropriate fusions were cultured on AB agar plates at 28°C for 2 days and then transferred to agar plates of IB buffered at pH 5.5 or pH 7.0. For the bacterial cells containing the virB:lacZ and virE:lacZ fusions, 100 μM acetosyringone (AS) was added into the IB media to induce the vir genes. After incubation on the IB agar plates for 2 days, the bacterial cells were scraped and resuspended in water. After washing once with water by centrifugation at 10,000 × g for 2 min, the cells were resuspended in water.
To measure the aopB:gfp, katA:gfp, and rrn:gfp expression, the cell concentration was adjusted to 5 × 108 cells per ml based on the OD600. Then 200 μl of cell suspension was used to measure the relative fluorescence intensities (Ir) as described (25). A6007 and A6340 were used as the negative controls. In addition, the bacterial cells containing the aopB:gfp, katA:gfp, and rrn:gfp fusions were subjected to immunoblot analysis by using the GFP antibody, as described (26). To measure the virB:lacZ and virE:lacZ expression, the cell concentration was adjusted to 8 × 107 cells per ml based on the OD600. β-Galactosidase activity was measured as described (27).
Results
chvG Is Translated at an Alternative Initiation Codon.
Based on a corrected chvG sequence that we determined (see Fig. 4, which is published as supporting information on the PNAS web site) and which was published recently (4, 5), we predicted an ORF of 401 aa for chvG when ATG was used as the start codon. The predicted molecular weight of the protein is 44 kDa. However, this hypothetical ORF would not encompass the transposon insertion site of the chvG mutant A6340. Therefore, we considered the possibility that the alternative initiation codon GTG or TTG was the actual start codon. If true, a putative ORF of 596 or 595 aa would be generated that would cover the transposon insertion in chvG (see Fig. 4). We generated a His-ChvG fusion protein, which was then used to generate ChvG-specific antibody. Western analysis with this antibody detected a band consistent with the predicted molecular weight of the protein, 66 kDa.
Both GTG and TTG have been shown to be the start codon of virG in A. tumefaciens, although most of the strains use TTG (28). Interestingly, the S. meliloti exoS also has a TTG at the corresponding position; both ChvG and ExoS would have the same number of amino acids (595), if TTG is used as the initiation codon. We identified a putative Shine-Dalgarno sequence that was located at 9 and 12 bp upstream of the GTG and TTG codon, respectively (see Fig. 4). A Shine-Dalgarno sequence normally precedes the start codon by approximately <10 bp; in addition, GTG is more frequently used as alternative start codon than TTG in bacteria (29). It appears likely that chvG ORF uses GTG as the start codon; our subsequent analyses are based on this interpretation of the data.
The predicted protein sequence has a higher homology (81% identity and 89% similarity) with the S. meliloti counterpart ExoS than was previously thought (11). ChvG and ExoS should be the corresponding orthologs, because other homologous counterparts could be found on both sides of this locus in A. tumefaciens and S. meliloti (data not shown).
The transposon insertion of the mutant A6340 mapped 201 bp downstream of the start codon of the putative ORF (see Fig. 4). We amplified a DNA fragment containing the entire predicted ChvG ORF together with 369 bp of the upstream sequence. We cloned this fragment into a plasmid to generate pLP36, which was subsequently introduced into the chvG mutant A6340 as mutations at chvG confer an acid sensitive phenotype (11). pLP36 restored the ability of the chvG mutant A6340 to grow on an acidic minimal medium (IB, pH 5.5; data not shown). All of these observations are consistent with the notion that the chvG ORF uses an alternative start codon (see Fig. 4).
Analysis of ChvG Protein.
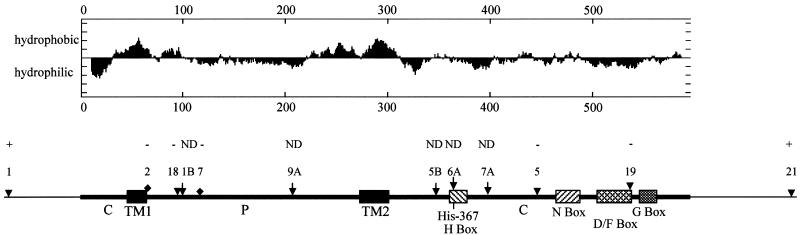
We next analyzed the ChvG protein sequence. We identified the most conserved signature sequence patterns, the H, N, D/F, and G boxes that are present in sensor histidine kinases (ref. 30; Fig. 1). Analysis of the ChvG hydrophobicity suggests that ChvG has two transmembrane domains (Fig. 1). The two TnphoA fusions identified previously (11) mapped to the periplasmic domain between the first and second transmembrane domains. The highly conserved histidine kinase domain lies in the C-terminal region that is predicted to be cytoplasmic. N-terminal periplasmic domains and C-terminal cytoplasmic domains have been found in many membrane histidine kinase sensor proteins (31).
Fig 1.
Hydropathy plot of ChvG and its predicted domains. TM1, transmembrane domain 1; TM2, transmembrane domain 2; P, the periplasmic domain; C, the cytoplasmic domain. H Box, N Box, D/F Box, and G Box are indicated. The locations of transposon insertions described previously (11) are also indicated. ▾, the site of Tn5 insertion; ⧫, the site of TnphoA insertion; ↓, the site of Tn3HoHo1 insertion. The vertical line indicates the putative phosphorylation site His-367 of ChvG. Plasmids carrying insertions marked with + are able to restore detergent resistance to the TnphoA mutants A6340 and A7678, whereas plasmids carrying insertions marked − are unable to do so; plasmids carrying insertions marked with ND indicate that the ability of the plasmids to restore the detergent resistance has not been determined (11).
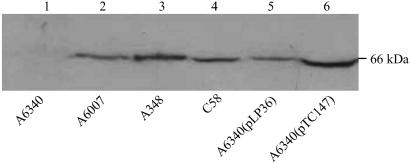
To determine whether ChvG is indeed a membrane protein, we fractionated the bacterial cells into cytoplasmic and membrane fractions; the proteins were concentrated after the fractionation procedure. Equivalent amounts of total protein from both fractions were resolved on a polyacrylamide gel, and then assayed with the ChvG-specific antibody. One strong band could be seen in the membrane fraction but not in the cytoplasmic fraction of the parent strain A6007 containing the wild-type chvG. This protein band was not detected in either the membrane or cytoplasmic fraction of the chvG mutant strain A6340. In addition, we detected the same protein band in the membrane fraction in the strains, C58, A348, and the complemented strains A6340(pLP36) and A6340(pTC147) (Fig. 2), all of which have the wild-type chvG. These data indicate that ChvG is indeed a membrane protein.
Fig 2.
Detection of the ChvG protein in the membrane fraction. The membrane fractions of A. tumefaciens strains A6340 (lane 1), A6007 (lane 2), A348 (lane 3), C58 (lane 4), A6340(pLP36) (lane 5), and A6340(pTC147) (lane 6) were prepared; proteins from the membrane fractions were electrophoresed on SDS/12% polyacrylamide gel. The ChvG protein was visualized by immunoblotting using the ChvG antibody.
We also analyzed the ChvG homologs in different bacteria. As shown in Table 1, standard blast searches for ChvG homologs in the databases revealed that ChvG is most homologous to S. meliloti ExoS (14). Strong homology was found between the two proteins throughout their entire length (data not shown). ChvG is also homologous to M. loti ExoS, B. melitensis ChvG, B. abortus BvrS (15), and a putative B. bacilliformis and C. crescentus kinase to a certain degree in all of the domains. Interestingly, all of these bacteria belong to the α-subdivision of proteobacteria. However, it was not present in two Rickettsia species, conorii and prowazekii, which are also α-proteobacteria. ChvG is weakly homologous to sensor proteins present in many other bacteria, Gram-negative or positive, with homology limited to the C-terminal half that presumably contains the kinase domain of a sensor protein.
Table 1.
Comparison of A. tumefaciens ChvG with other bacterial sensor proteins
|
|
Homology, % | |
|---|---|---|
| Identities | Positives | |
| Standard blast results with A. tumefaciens ChvG in the databases | ||
| α-Proteobacteria | ||
| S. meliloti ExoS | 78 | 86 |
| Mesorhizobium loti ExoS | 66 | 77 |
| Brucella melitensis ChvG | 58 | 70 |
| B. abortus BvrS | 58 | 71 |
| A putative Bartonella bacilliformis kinase | 53 | 69 |
| A putative Caulobacter crescentus kinase | 40 | 55 |
| Various other bacteria, Gram-negative or -positive | ||
| Standard blast hits | 22–33 | 46–52 |
| blast 2 sequences results with A. tumefaciens ChvG | ||
| S. meliloti ActS | 24 | 41 |
| S. typhimurium PhoQ | 20 | 42 |
The A. tumefaciens ChvG amino acid sequence was used to conduct the standard blast search in the databases; the results are listed in the upper part of the table. The S. meliloti ActS and S. typhimurium PhoQ were not picked up in the search; each of them was then compared with A. tumefaciens ChvG with the blast 2 sequences program, and the results are listed in the lower part.
Effects of chvG on the Regulation of Acid-Inducible Genes Located on the Two Chromosomes.
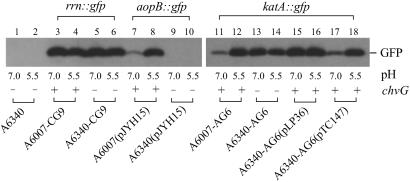
We have identified two chromosomal genes (aopB and katA) that are involved in Agrobacterium tumorigenesis and are inducible by acidic pH (20, 21); aopB is located on the circular chromosome and katA on the linear chromosome (4, 5). To determine whether ChvG is involved in the regulation of these two genes, we measured the expression of aopB:gfp and katA:gfp fusions in the presence and absence of ChvG. We constructed a plasmid containing the aopB:gfp fusion by cloning a 9.0-kb SphI DNA fragment from the genomic DNA of CGI1 (21) into pSW172 to generate pJYH15. This 9.0-kb SphI DNA fragment contained the mini-Tn5 insertion at aopB and the sequences flanking the mini-Tn5 insertion (21). We introduced pJYH15 into the chvG− mutant strain A6340 and the corresponding chvG+ strain A6007 and then measured the aopB:gfp expression by green fluorescence (Table 2) and immunoblotting (Fig. 3). We found that the aopB:gfp expression in the chvG+ strain A6007 was induced about 8-fold by the change from pH 7.0 to pH 5.5. This finding is consistent with the previous observation that aopB was induced by acidic pH (21). However, in the chvG− mutant A6340, the aopB:gfp expression was undetectable on IB buffered at pH 7.0 or pH 5.5. This finding suggests that ChvG is absolutely required for the expression of aopB.
Table 2.
The effects of chvG on the expression of acid-inducible genes located on the two chromosomes
| Strains
|
gfp reporter fusion
|
Presence of chvG
|
Ir | Fold of acidic induction
|
|
|---|---|---|---|---|---|
| IB, pH 7.0 | IB, pH 5.5 | ||||
| A6007-CG9 | rrn:gfp | + | 1,210 | 1,160 | 1.0 |
| A6340-CG9 | rrn:gfp | − | 1,172 | 1,075 | 0.9 |
| A6007(pJYH15) | aopB:gfp | + | 60 | 502 | 8.4 |
| A6340(pJYH15) | aopB:gfp | − | 0 | 0 | — |
| A6007-AG6 | katA:gfp | + | 154 | 1,406 | 9.1 |
| A6340-AG6 | katA:gfp | − | 1,387 | 1,328 | 1.0 |
| A6340-AG6(pLP36) | katA:gfp | + | 1,395 | 1,505 | 1.1 |
| A6340-AG6(pTC147) | katA:gfp | + (+chvI) | 326 | 1,383 | 4.2 |
The results presented in the table represent three independent experiments.
The relative fluorescence intensities (Ir) of A. tumefaciens strains containing the gfp reporter fusions grown on IB pH 7.0 or IB pH 5.5 were measured as described in Materials and Methods.
The fold of acidic induction for each gene was determined by dividing the Ir of the bacteria grown on IB pH 5.5 by that on IB pH 7.0.
Fig 3.
The effects of ChvG on the expression of acid-inducible genes aopB and katA as determined by Western analysis. A. tumefaciens cells containing rrn:gfp, aopB:gfp, or katA:gfp fusion were grown on IB agar plates buffered at pH 7.0 or pH 5.5. The gene expression levels in the bacterial cells were analyzed in the absence (−) [A6340, A6340-CG9, A6340(pJYH15), and A6340-AG6] or presence (+) [A6007-CG9, A6007(pJYH15), A6007-AG6, A6340-AG6(pLP36), and A6340-AG6(pTC147)] of a functional chvG. The same amount of bacterial cells was collected for each strain; total protein was electrophoresed on SDS/12% polyacrylamide gel. The GFP protein was visualized by immunoblotting using the GFP antibody.
To study the effect of chvG on katA expression, we examined katA:gfp expression in the absence of any functional katA gene, because the KatA protein represses katA:gfp expression (26). We created different genetic backgrounds for the katA:gfp fusion construct by introducing the total DNA of AG6, which is a katA− mutant containing the katA:gfp fusion (26), into A6007 and A6340 and selecting for homologous recombinants (32). This resulted in two recombinants A6007-AG6 and A6340-AG6, which lack a functional katA gene but contain the katA:gfp fusion in the A6007 and A6340 background, respectively. This was verified by Southern analysis (data not shown). We measured katA:gfp expression in these strains (Table 2, Fig. 3) and found that the katA:gfp expression in the chvG+ background was induced about 9-fold by the change from neutral to acidic pH, consistent with the previous observation that katA was inducible by acidic pH (26). In the chvG− background, the katA:gfp was still expressed, but at the same level at pH 7.0 and pH 5.5. These data suggest that chvG is required for the responsiveness of katA gene expression to low pH, but not for expression of the katA gene. In addition, the katA:gfp expression level in the chvG− cells grown at pH 7.0 or pH 5.5 was comparable to that in the chvG+ cells grown at pH 5.5. These data suggest that katA expression is repressed at neutral pH and chvG derepresses katA expression at a low pH. Thus, chvG appears to regulate aopB and katA genes differently.
We tried to complement the chvG− mutation with the plasmid pLP36 containing only the chvG gene. Although the ChvG protein was detected in strain A6340(pLP36) by western analysis (Fig. 2), the responsiveness of katA expression to a pH change was not restored (Table 2, Fig. 3). When the plasmid pTC147 containing both chvG and chvI (11) was introduced into A6340-AG6, the ability to respond to the pH change was partially restored (Table 2, Fig. 3). It is not apparent why the chromosomal chvG mutation could not be fully complemented by a plasmid-harbored chvG gene, although both pLP36 and pTC147 restored the ability of bacteria to grow on acidic media. Notably, the expression of both B. abortus bvrS and bvrR (homologous to chvG and chvI, respectively) is required to complement a bvrR mutation (15).
The Effect of chvG on the Expression of vir Genes.
We next explored the possibility that ChvG might affect the expression of vir genes encoded on the Ti plasmid, as the maximal expression of these genes requires an acidic pH environment in addition to plant phenolic compounds, such as acetosyringone (AS) (6). The requirement for the low pH is a function of VirG, which requires both an acidic pH and plant signal molecules for maximal activity (6). We introduced the plasmid pSM243cd (containing a virB:lacZ fusion) and pSM358cd (containing a virE:lacZ fusion) (33) into A6007 and A6340. A6340 is hypersensitive to carbenicillin (Cb), even when the plasmid containing a Cb-resistance gene was introduced into A6340. Thus, we used a low concentration of Cb (5 μg/ml) to select for A6340(pSM243cd) and A6340(pSM358cd), which were subsequently verified by triparental mating to be Cb resistant. We measured the expression of virB:lacZ or virE:lacZ in the bacteria grown on IB buffered at pH 7.0 or pH 5.5; 100 μM of AS was included in the media buffered at both pH 7.0 and pH 5.5 to induce vir gene expression. Thus, any difference in the vir gene expression between pH 7.0 and pH 5.5 should be due to the change in pH. As shown in Table 3, the virB:lacZ and virE:lacZ gene expression in the chvG+ strain A6007 were induced about 15- and 10-fold, respectively, by acid (pH 5.5). However, the virB:lacZ and virE:lacZ expression in the chvG− mutant A6340, although measurable, was much lower than the activity in the chvG+ strain A6007. This finding is consistent with the previous observation that ChvI was required for the expression of virB and virG (12). In addition, the induction of both virB:lacZ and virE:lacZ expression by acidic pH was marginal in the chvG− mutant. Thus, ChvG plays an important role in the expression of virB and virE, although it is not essential for the expression of virB and virE, whereas it is for aopB. ChvG also regulates the acidic responsiveness of the vir genes.
Table 3.
The effect of chvG on the expression of vir genes located on the Ti plasmid
| Strains
|
lacZ reporter fusions
|
Presence of the chromosomal chvG
|
β-Galactosidase activity | Induction fold
|
|
|---|---|---|---|---|---|
| IB + AS; pH 7.0 | IB + AS; pH 5.5 | ||||
| A6007 | NA | + | 1 | 3 | NA |
| A6340 | NA | − | 2 | 2 | NA |
| A6007(pSM243cd) | virB:lacZ | + | 200 | 2,959 | 14.8 |
| A6340(pSM243cd) | virB:lacZ | − | 15 | 27 | 1.8 |
| A6007(pSM358cd) | virE:lacZ | + | 713 | 6,777 | 9.5 |
| A6340(pSM358cd) | virE:lacZ | − | 83 | 192 | 2.3 |
| A6007(pSHM4) | chvH:lacZ | + | 86 | 80 | 0.9 |
| A6340(pSHM4) | chvH:lacZ | − | 93 | 82 | 0.9 |
The results presented in the table represent three independent experiments. NA, not applicable.
The β-galactosidase activity of A. tumefaciens strains containing the chvH:lacZ, virB:lacZ, or virE:lacZ fusion on IB pH 7.0 or IB pH 5.5 in the presence of 100 μM acetosyringone (AS) were measured as described in Materials and Methods.
The induction fold was calculated by dividing the gene expression level of the bacteria grown on IB pH 5.5 by that on IB 7.0.
ChvG Did Not Affect Genes That Were Not Acid-Inducible.
To determine whether the effects of chvG on katA and aopB gene expression were specific under the test conditions, we studied the expression of a 16S ribosomal RNA gene (designated as rrn) in CG9. This strain contains an rrn:gfp fusion with the mini-Tn5 transposon inserted onto an rrn gene encoding 16S ribosomal RNA. This fusion was identified in the same screening program as CGI1 and AG6 (21, 26, 34). We created different genetic backgrounds for the rrn:gfp fusion by introducing the total DNA of CG9 into A6007 and A6340 and selecting for homologous recombinants in the same way when A6007-AG6 and A6340-AG6 were created. The resulting strains A6007-CG9 and A6340-CG9 contained the rrn:gfp fusion in the A6007 and A6340 background, respectively. We examined the GFP expression in these strains and found that the 16S ribosomal RNA expression was constant in chvG+ and chvG− backgrounds (Table 2, Fig. 3), suggesting that the 16S ribosomal RNA gene is not acid-inducible and not affected by ChvG. This finding suggests that the effects of ChvG on aopB and katA are specific to acid-inducible genes, which is consistent with the previous observation that a lac:lacZ fusion was not significantly affected by a chvI mutation (12).
To determine whether the effects of chvG on the virB and virE gene expression were specific under the test conditions, we also examined the expression of a chromosomal gene chvH that is also involved in virulence (35), by using the reporter construct chvH:lacZ. This construct was harbored on a plasmid pSMH4, which carries the 507 bp chvH sequence upstream of the start codon and 12 bp of chvH coding sequence fused in frame with the lacZ coding sequence. The lacZ was amplified by PCR using pTn3HoHo1 (27) as the template with the primers LacZ-1 and LacZ-2 (see Table 4). Our results show that the chvH:lacZ expression was not affected either by ChvG or acidic pH, supporting our observation that ChvG is involved in the induction of Ti-plasmid harbored virB and virE by an acidic pH. It seems likely that ChvG regulates all vir genes harbored on the Ti-plasmid, because they are regulated as one regulon through the virA/virG two-component system (6).
Discussion
To efficiently infect host plants, A. tumefaciens must coordinate the expression of a variety of genes encoding different functions, which are concerned with the infection process. Most of these genes appear to function best in the acidic environment of a wounded plant. The data in this paper suggest that the two-component regulatory system chvG/chvI plays a role in coordinating the expression of acid-inducible genes in A. tumefaciens. The Ti-plasmid-harbored vir genes are directly involved in causing tumors on plants, whereas katA is involved in defending against a plant defense response (20). The role of aopB in virulence is not known (21), although different surface-associated proteins have been identified as virulence factors in pathogenic bacteria (36).
The expression of several chromosomal genes that are not acid-inducible was not affected by ChvG. This included a 16S ribosomal RNA gene (Table 2) and the virulence-associated gene, chvH (Table 3). In addition, ChvG was not required for the accumulation of the virulence factor ChvE as detected by immunoblot analysis. In fact, ChvE accumulated to a higher level in the absence of ChvG (data not shown). ChvE is required for maximal induction of the Ti-plasmid-encoded vir genes. The results suggest that ChvG is specifically required for the expression and induction of acid-inducible genes.
Previously we observed that the chvG mutants A6340 and A7678 are much more sensitive to detergents and antibiotics than the wild-type strain (11). This suggests that the permeability of the cell envelope is affected by chvG/chvI. One possible explanation is that the expression of some envelope proteins is affected by ChvG. Indeed, ChvG is required for the expression of aopB, which encodes an outer membrane protein exposed on the bacterial cell surface (21). Consistent with this observation is the finding that bvrS/bvrR of B. abortus (homologous to A. tumefaciens chvG/chvI) is required for expression of several outer membrane proteins (39), although it is unknown whether the genes encoding these proteins are acid-inducible.
ChvG appears to regulate acid-inducible genes in different ways, depending on the target genes. The simplest mechanism that one can envision is the following. ChvG can activate a transcriptional regulator that activates target genes. This is clearly illustrated by the data on aopB, virB, and virE. No gene expression can be observed of aopB in a chvG mutant. In the chvG mutant, the virB gene expression drops 100-fold, whereas virE expression drops 30-fold. We hypothesize that chvG is required for the activation of virG. In a chvG mutant, the virG expression is very low, which then accounts for the very reduced levels of virB and virE expression.
The explanation for the effect of ChvG on katA seems indirect. ChvG may activate a repressor which represses katA at a neutral pH. If the repressor is only active at pH 7.0, this would account for the low level of katA expression at pH 7.0 and its increased level at pH 5.5. In the absence of chvG, the level of katA expression increases to the level seen at pH 5.5 in the presence of chvG.
A comparison of the Agrobacterium ChvG sequence with the genomes of other α-proteobacteria demonstrates that the entire ChvG length is homologous to the corresponding protein sensors in Sinorhizobium, Mesorhizobium, Brucella, Bartonella, and Caulobacter (data not shown). Interestingly, some α-proteobacteria like R. conorii and R. prowazekii lack a ChvG homolog, based on blast searches. Because R. conorii and R. prowazekii are obligate intracellular bacteria, it remains unclear whether ChvG homologs will only be found in free-living α-proteobacteria. Additional genomes of α-proteobacteria must be sequenced to answer this question. As a ChvG homolog, S. meliloti ExoS plays a role in regulating the production of succinoglycan, which is important for establishing symbiosis. It remains to be determined whether pH plays a role in this process and whether ExoS is involved in regulating the expression of any acid-inducible genes in S. meliloti. A S. meliloti kinase, ActS, has been hypothesized to be a pH sensor, but it does not affect the expression of a low pH-inducible gene phrR (37). The homology between ActS and ChvG is very low (Table 1). Brucella and Bartonella species are human and animal pathogens that cause brucellosis and bartonellosis, respectively. The BvrS protein, a ChvG homolog in B. abortus, is required for the inhibition of lysosome fusion and replication inside animal cells (15). Once bacteria like Brucella and Bartonella are internalized into animal cells, it is possible that they will encounter an acidic pH environment within the vesicles containing them. However, it is not known whether ChvG homologs of these bacteria sense intracellular acidity. C. crescentus lives in a dilute aquatic environment. It would be of interest to determine whether this ChvG homolog senses aquatic pH.
A sensor kinase protein PhoQ in S. typhimurium has been previously implicated in sensing both Mg2+ and pH. PhoQ appears responsible for the low pH induction of only a few of many acid shock proteins (38). It is not clear whether PhoQ senses pH independently of Mg2+ or whether pH affects the interaction between Mg2+ and the Mg2+-sensing site on PhoQ (38). The homology between PhoQ and ChvG is very low (Table 1) and limited to the kinase domain, suggesting that ChvG senses pH by a mechanism different from PhoQ. Nevertheless, it is important to determine how ChvG senses pH and whether it senses another signal(s).
Supplementary Material
Acknowledgments
We thank H. M. Soo and L. W. Tan for technical support, and X. Q. Xu, X. Tang, L. M. Chang, and B. F. Lu for discussion. This work was supported by the National University of Singapore Academic Research Grants R-154-000-082-112 and R-154-000-107-112 (to S.Q.P.) and National Institutes of Health Public Health Service Grant GM32618 (to E.W.N.).
Abbreviations
Ti, tumor-inducing
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AY144331).
References
- 1.Parkinson J. S. & Kofoid, E. C. (1992) Annu. Rev. Genet. 26, 71-112. [DOI] [PubMed] [Google Scholar]
- 2.Parkinson J. S. (1993) Cell 73, 857-871. [DOI] [PubMed] [Google Scholar]
- 3.Charles T., Jin, S. & Nester, E. W. (1992) Annu. Rev. Phytopathol. 30, 463-484. [DOI] [PubMed] [Google Scholar]
- 4.Wood D. W., Setubal, J. C., Kaul, R., Monks, D. E., Kitajima, J. P., Okura, V. K., Zhou, Y., Chen, L., Wood, G. E., Almeida, N. F., Jr., et al. (2001) Science 294, 2317-2323. [DOI] [PubMed] [Google Scholar]
- 5.Goodner B., Hinkle, G., Gattung, S., Miller, N., Blanchard, M., Qurollo, B., Goldman, B. S., Cao, Y., Askenazi, M., Halling, C., et al. (2001) Science 294, 2323-2328. [DOI] [PubMed] [Google Scholar]
- 6.Winans S. C. (1992) Microbiol. Rev. 56, 12-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gelvin S. B. (2000) Annu. Rev. Plant Physiol. Plant Mol. Biol. 51, 223-256. [DOI] [PubMed] [Google Scholar]
- 8.Zhu J., Oger, P. M., Schrammeijer, B., Hooykaas, P. J., Farrand, S. K. & Winans, S. C. (2000) J. Bacteriol. 182, 3885-3895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zupan J., Muth, T. R., Draper, O. & Zambryski, P. (2000) Plant J. 23, 11-28. [DOI] [PubMed] [Google Scholar]
- 10.Cangelosi G. A, Best, E. A., Martinetti, G. & Nester, E. W. (1991) Methods Enzymol. 204, 384-397. [DOI] [PubMed] [Google Scholar]
- 11.Charles T. C. & Nester, E. W. (1993) J. Bacteriol. 175, 6614-6625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mantis N. J. & Winans, S. C. (1993) J. Bacteriol. 175, 6626-6636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Østeras M., Stanley, J. & Finan, T. M. (1995) J. Bacteriol. 177, 5485-5494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cheng H. P. & Walker, G. C. (1998) J. Bacteriol. 180, 20-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sola-Landa A., Pizarro-Cerda, J., Grillo, M. J., Moreno, E., Moriyon, I., Blasco, J. M., Gorvel, J. P. & Lopez-Goni, I. (1998) Mol. Microbiol. 29, 125-138. [DOI] [PubMed] [Google Scholar]
- 16.Hall H. K., Karem, K. & Foster, J. W. (1995) Adv. Microb. Physiol. 37, 229-272. [DOI] [PubMed] [Google Scholar]
- 17.Foster J. W. (1999) Curr. Opin. Microbiol. 2, 170-174. [DOI] [PubMed] [Google Scholar]
- 18.Winans S. C. (1990) J. Bacteriol. 172, 2433-2438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen C. Y. & Winans, S. C. (1991) J. Bacteriol. 173, 1139-1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xu X. Q. & Pan, S. Q. (2000) Mol. Microbiol. 35, 407-414. [DOI] [PubMed] [Google Scholar]
- 21.Jia Y. H., Li, L., Hou, Q. M. & Pan, S. Q. (2002) Gene 284, 113-124. [DOI] [PubMed] [Google Scholar]
- 22.Hager D. & Burgess, R. (1980) Anal. Biochem. 109, 76-86. [DOI] [PubMed] [Google Scholar]
- 23.Bollag D. M., Rozycki, M. D. & Edelstein, S. J. (1996) in Protein Methods, eds. Bollag, D. M., Rozycki, M. D. & Edelstein, S. J. (Wiley-Liss, New York), pp. 83–106.
- 24.Chervitz S. A., Lin, C. M. & Falke, J. J. (1995) Biochemistry 34, 9722-9733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tang X., Lu, B. F. & Pan, S. Q. (1999) FEMS Microbiol. Lett. 179, 37-42. [DOI] [PubMed] [Google Scholar]
- 26.Xu X. Q., Li, L. P & Pan, S. Q. (2001) Mol. Microbiol. 42, 645-657. [DOI] [PubMed] [Google Scholar]
- 27.Stachel S. E., An, G., Flores, C. & Nester, E. W. (1985) EMBO J. 4, 891-898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee S. H. & Sim, W. S. (1998) Mol. Cells 8, 393-400. [PubMed] [Google Scholar]
- 29.Lewin B., (1994) Genes V (Oxford Univ. Press, Oxford), pp. 179–268.
- 30.Ninfa A. J. (1996) in Escherichia coli and Salmonella typhimurium: Cellular and Molecular Biology, eds. Neidhardt, F. C., Curtiss, R., III, Ingraham, J. L., Lin, E. C. C., Low, K. B., Magasanik, B., Reznikoff, W. S., Riley, M., Schaechter, M. & Umbarger, H. E. (Am. Soc. Microbiol., Washington, DC), pp. 1246–1262.
- 31.Dutta R., Qin, L. & Inouye, M. (1999) Mol. Microbiol. 34, 633-640. [DOI] [PubMed] [Google Scholar]
- 32.Charles T. C., Doty, S. L. & Nester, E. W. (1994) Appl. Environ. Microbiol. 60, 4192-4194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stachel S. E. & Zambryski, P. C. (1986) Cell 46, 325-333. [DOI] [PubMed] [Google Scholar]
- 34.Li L., Li, Y., Lim, T. M. & Pan, S. Q. (1999) FEMS Microbiol. Lett. 179, 141-146. [DOI] [PubMed] [Google Scholar]
- 35.Peng W. T., Banta, L. M., Charles, T. C. & Nester, E. W. (2001) J. Bacteriol. 183, 36-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Klemm P. & Schembri, M. A. (2000) Int. J. Med. Microbiol. 290, 27-35. [DOI] [PubMed] [Google Scholar]
- 37.Reeve W. G., Tiwari, R. P., Wong, C. M., Dilworth, M. J. & Glenn, A. R. (1998) Microbiology 144, 3335-3342. [DOI] [PubMed] [Google Scholar]
- 38.Bearson B. L., Wilson, L. & Foster, J. W. (1998) J. Bacteriol. 180, 2409-2417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Guzmán-Verri C., Manterola, L., Sola-Landa, A., Parra, A., Cloeckaert, A., Garin, J., Gorvel, J.-P., Moriyón, I., Moreno, E. & López-Goñi, I. (2002) Proc. Natl. Acad. Sci. USA 99, 12375-12380. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.