Abstract
The Brucella BvrR/BvrS two-component regulatory system is homologous to the ChvI/ChvG systems of Sinorhizobium meliloti and Agrobacterium tumefaciens necessary for endosymbiosis and pathogenicity in plants. BvrR/BvrS controls cell invasion and intracellular survival. Probing the surface of bvrR and bvrS transposon mutants with monoclonal antibodies showed all described major outer membrane proteins (Omps) but Omp25, a protein known to be involved in Brucella virulence. Absence of Omp25 expression was confirmed by two-dimensional electrophoresis of envelope fractions and by gene reporter studies. The electrophoretic analysis also revealed reduction or absence in the mutants of a second set of protein spots that by matrix-assisted laser desorption ionization MS and peptide mass mapping were identified as a non-previously described Omp (Omp3b). Because bvrR and bvrS mutants are also altered in cell-surface hydrophobicity, permeability, and sensitivity to surface-targeted bactericidal peptides, it is proposed that BvrR/BvrS controls cell envelope changes necessary to transit between extracellular and intracellular environments. A genomic search revealed that Omp25 (Omp3a) and Omp3b belong to a family of Omps of plant and animal cell-associated α-Proteobacteria, which includes Rhizobium leguminosarum RopB and A. tumefaciens AopB. Previous work has shown that RopB is not expressed in bacteroids, that AopB is involved in tumorigenesis, and that dysfunction of A. tumefaciens ChvI/ChvG alters surface properties. It is thus proposed that the BvrR/BvrS and Omp3 homologues of the cell-associated α-Proteobacteria play a role in bacterial surface control and host cell interactions.
The brucellae are facultative intracellular parasites of animals and humans causing a disease of worldwide importance. These bacteria are phylogenetically entwined with animal and plant cell-associated Proteobacteria of the α subclass, such as Rhizobium, Sinorhizobium, Mesorhizobium, Agrobacterium, Bartonella, and Rickettsia species. Similar to other facultative intracellular parasites, Brucella organisms survive outside cells, but they must infect and replicate intracellularly in animals to perpetuate. The brucellae are extremely well adapted to the intracellular niche and, accordingly, they should be described as facultatively extracellular intracellular parasites (1).
Brucella organisms have to deal with two very different environments during their life cycle. On one hand, the extracellular milieu confronts the bacteria with bactericidal substances such as antibodies, antibiotics, complement, and leukocyte discharges. On the other, the bacteria must invade cells and resist the various cellular strategies aimed to eliminate parasites. It is thus predicted that some genetic systems are turned on and off to achieve the adjustments in metabolism and structure necessary for a successful extracellular/intracellular life transition. So far, only one such system, the two-component regulatory system BvrR/BvrS, has been conclusively implicated in Brucella virulence (2). BvrR/BvrS mutants are avirulent in mice, show reduced invasiveness in cells, and are unable to inhibit lysosome fusion and to replicate intracellularly (2). Dysfunction of bvrR/bvrS diminishes the characteristic resistance of Brucella to bactericidal polycations and increases its permeability to surfactants. Because these properties relate to the structure of the Brucella outer membrane (3–5), we reasoned that some of its molecular features should be under the control of BvrR/BvrS. This membrane has overall properties that depart from those of many Gram-negative bacteria, including a complex outer membrane protein (Omp) profile (reviewed in ref. 5). Brucella Omps were originally grouped by their mobility in SDS/PAGE as group 1 (94 or 89 kDa), group 2 (38–36 kDa), and group 3 (31–25 kDa). Group 2 Omps are porins, and Omp25, of group 3, has been recently shown to be involved in virulence (6, 7). In addition, three lipoproteins (10–19 kDa) and a peptidoglycan bound lipoprotein have been identified (5).
The BvrR/BvrS system is highly homologous to some two-component systems of cell associated α-2 Proteobacteria (2): BvrR has 86–76% similarity to Mesorhizobium loti ChvI, Bartonella bacilliformis BatR, Sinorhizobium meliloti ChvI, and Agrobacterium tumefaciens ChvI, and BvrS has 68–59% similarity to M. loti ExoS, S. meliloti ExoS, B. bacilliformis BatS, and A. tumefaciens ChvG. It has been demonstrated that the ChvI/ExoS system of S. meliloti controls the succinoglycan production necessary for endosymbiosis (8). Also, A. tumefaciens ChvI/ChvG mutants are not tumorigenic in plants, are comparatively sensitive to detergents, antibiotics, and acid pH, and have altered cell envelope permeability (9, 10). However, little is known on the molecular determinants controlled by these regulatory systems. In the present study, we analyzed the expression of Omps in wild type and bvrR and bvrS mutants. We report that BvrR/BvrS regulates the expression of at least two Omps, one not described previously and the other known to be involved in Brucella virulence. Genomic comparisons showed that homologous proteins are present in animal and plant cell associated α-Proteobacteria, and perusal of the literature suggests that at least some of them play a critical role in bacterial–host cell interactions.
Experimental Procedures
Bacterial Strains, Plasmids, and Growth Conditions.
The strains and plasmids used are described in Table 1. E. coli was grown in LB and Brucella in tryptic soy medium. For isolation of bacterial fractions, cells were propagated as described (11). When needed, nalidixic acid at 25 μg/ml, ampicillin at 100 μg/ml, kanamycin at 50 μg/ml, or gentamicin at 20 μg/ml was added to the medium.
Table 1.
Bacterial strains and plasmids
| Strain or plasmid | Relevant characteristics | Source |
|---|---|---|
| B. abortus | ||
| 2308 Nalr | Wild type, virulent, smooth-LPS, Nalr spontaneous mutant of strain 2308 | Ref. 40 |
| 2.13 | 2308 Nalr, bvrS:Tn5, Kanr, avirulent, smooth LPS | Ref. 2 |
| 65.21 | 2308 Nalr, bvrR:Tn5, Kanr, avirulent, smooth LPS | Ref. 2 |
| 65.21p | Reconstituted bvrR strain, 65.21 carrying plasmid pBBR1MCS-4 with a 6.0-kb fragment encoding bvrR | Ref. 2 |
| E. coli | ||
| SM10 (λ pir) | recA:RP4-2-Tet:Mu, Kmr, thi leu thr supE λ pir | Ref. 41 |
| Plasmids | ||
| pCR2.1 | Cloning vector, pUC ori, Ampr, Kmr | Invitrogen |
| pEGZ | Suicide plasmid used to generate chromosomal lacZ fusions, Ampr, Gmr | Ref. 42 |
| pEGZ-omp3a | Promoterless omp3a 550 bp fragment cloned upstream the promoterless lacZ into vector pEGZ | This study |
| pEGZ-omp3b | Promoterless omp3b 473 bp fragment cloned upstream the promoterless lacZ into vector pEGZ | This study |
Bacterial Fractionation.
Cell envelopes and cytosolic fractions were obtained and characterized as described (12). The outer membrane fragments released by exponentially growing brucellae were obtained by ultracentrifugation of spent broth. This fraction is devoid of cytoplasmic or inner membrane markers and is enriched in Brucella group 3 Omps (13). LPS extracts rich in tightly bound, protease-resistant group 3 Omps were obtained from live bacteria with SDS and proteinase K as described (14).
Analysis of Omps by PAGE.
Standard SDS/PAGE was performed according to Laemmli (15). Proteins were detected by the Coomassie G or silver staining methods (16). Total cell lysates for two-dimensional gel electrophoresis (2DGE) were obtained from overnight cultures. Bacteria were collected by centrifugation, washed twice with PBS, resuspended in sample buffer without SDS or β-mercaptoethanol (15), and boiled for 10 min. To analyze cell envelopes or outer membrane fragments by 2DGE, 300 μg of these materials were precipitated with 80% acetone, and aliquots were first run by SDS/PAGE to normalize total protein content. First dimensions were performed on Immobiline DryStrip gels (Pharmacia Biotech) according to the manufacturer's instructions. After second-dimension separation by SDS/PAGE and silver staining (16), gels were dried, scanned, and compared using Z3 software (Compugen, Jamesburg, NJ). For comparative purposes, at least five independent 2DGE analyses were performed with each material.
Matrix-Assisted Laser Desorption Ionization (MALDI)-MS and Peptide Mass Mapping.
One milligram of outer membrane fragments from the different strains was precipitated with acetone for 20 min and resuspended in 60 μl of isoelectrofocusing lysis buffer, and 2DGE was performed as described above. After Coomassie staining, the proteins were localized by using reference spots from the same gel, from silver stained gels, and from Western blots run in parallel by using the same sample. Protein spots cut out from the gels were analyzed by tandem MS and MALDI-MS followed by peptide mass mapping at the Laboratoire de Chimie des Proteines (CEA, Grenoble, France) or at Bruker Daltonik (Bremen, Germany).
Immunochemical Methods.
For dot-blot analysis, 5 μl of cell envelope suspensions (5 mg protein/ml in water) was dispensed onto poly(vinylidene difluoride) membranes (Schleicher & Schuell), which were then incubated overnight in a humid atmosphere, washed with 0.1% Tween 20 in PBS, and blocked overnight with 3% skim milk in the same solution. Amido black staining before blotting showed no quantitative differences in adsorption of proteins obtained from mutants or wild-type bacteria. Membranes were incubated with monoclonal antibodies (mAbs), washed (see above), and revealed with peroxidase-conjugated goat anti-mouse antibodies (Nordic, Tilburg, The Netherlands) and 4-chloro-1-naphthol (Merck). For Western blot, the same mAbs were used and revealed either with peroxidase-conjugated protein G and 4-chloro-1-naphthol or with a peroxidase-labeled secondary antibody and a chemiluminescence kit (Roche). mAbs to the following Omps were used (17–19): Omp10 (A68/07G11/C10), Omp16 (A68/08C03/G03), Omp19 (A76/05C10/A08, A76/18B02/D06), Omp25 (A18/13D02/F05, A19/12B10/F04, A59/05F01/C09, A70/06B05/A07, A76/02C12/C11, A68/04B10/F05, A68/07D11/B03, A68/28G06/C07), Omp2b (Omp36) (A63/08D08/C07, A63/05A07/A08, A63/03H02/B01, A63/11E05/D11, A63/13G02/C04, A63/04D11/G01, A68/25G05/A05), and Omp1 (Omp89) (A53/10B02/A01). Anti-Omp25 mAbs A18/13D02/F05, A19/12B10/F04, and A59/05F01/C09 recognize a linear epitope close to the amino-terminal section (amino acids 21–40). A mAb against the C/Y epitope was conjugated with peroxidase (20), and was used for LPS detection.
Chromosomal omp3a:lacZ and omp3b:lacZ Transcriptional Fusions and β-Galactosidase Assays.
To construct a lacZ under the transcriptional control of the omp3a promoter, a promoterless omp3a 550-bp DNA fragment was synthesized by PCR using oligonucleotides annealing at positions 255 (5′-TGCGCTGCTGCCGTTCTCTG-3′) and 776 (5′-GGATCCGGCCAGATCATAGTTCTTGT-3′; where the italics mark the terminal extension containing a BamHI restriction site) of Brucella abortus 544 omp3a (omp25) sequence (GenBank accession no. X79284). The PCR product was cloned into vector pCR2.1 (Invitrogen), and then subcloned into the promoter probe vector pEGZ (Table 1; ref. 21). The suicide plasmid generated, pEGZ-omp3a, encoding an omp3a:lacZ fusion, was delivered into the appropriate B. abortus strain by mating with E. coli SM10 λ-pir, resulting in the construction of 2308:pEGZ-omp3a, 2.13:pEGZ-omp3a, 65.21:pEGZ-omp3a, and 65.21p:pEGZ- omp3a. Correct integration of pEGZ-omp3a was confirmed by Southern blot and PCR. Construction of strains 2308:pEGZ-omp3b, 2.13:pEGZ-omp3b, 65.21:pEGZ-omp3b, and 65.21p:pEGZ-omp3b was performed likewise, but using a promoterless omp3b 473-bp DNA fragment synthesized by PCR using oligonucleotides annealing at positions 379 (5′-GCGCGCAGGTTGGTGGTT-3′) and 827 (5′-GGATCCGCCGGCCTTGATCGAATG-3′) of the omp3b sequence. The β-galactosidase activity was assessed as described (22), and specific activity was expressed as nmol of o-nitrophenol produced/min × mg protein.
Cloning and Nucleotide Sequencing of B. abortus 2308 omp3b.
Partial amino acid sequences obtained by MALDI-MS and peptide mass mapping from the Omp3b spots resolved by 2DGE were compared against preliminary B. suis complete genome sequence data (see below). Based on this, the following oligonucleotides were designed: (5′-CCCGGCTGTTACATATGCTG-3′) and (5′-CGCGCTGATATCGACATGAC-3′). A PCR was performed using Pfu DNA polymerase (Stratagene) and the 1.1-kbp product was subsequently cloned into pCR2.1 (Invitrogen). Sequencing was performed with universal M13 primers by Sistemas Genómicos (Valencia, Spain). DNA sequences were assembled and analyzed using the GENEWORK V.2.45N (IntelliGenetics) program.
Nucleotide and Protein Sequence Analyses.
Comparison of sequences, search for homologies, and analyses of protein characteristics (hydrophobicity profiles, localization site and signal sequence, and secondary and tertiary structure) were obtained using standard software (www.tigr.org; www.expasy.org; http://psort.nibb.ac.jp/form.html; www.bmm.icnet.uk/∼3dpssm/; http://bibiserv.techfak.uni-bielefeld.de/dialign/). For the construction of the neighbor-joining tree, protein sequences were aligned using the CLUSTAL W program and a matrix of Dayhoff was generated (www.infobiogen.fr/services/menuserv.html).
Results
Dysfunction of bvrR and bvrS Alters the Profile of B. abortus Group 3 Omps.
As pointed out in the Introduction, indirect evidence suggests that transposon mutants in either bvrR or bvrS have an altered cell envelope. Thus, as a first approach to identify changes in outer membrane components, we probed the cell surface of the wild-type (parental) and mutant strains by dot blot with a collection of mAbs recognizing all hitherto described B. abortus Omps. In contrast to the wild-type and bvrR reconstituted 65.21p strain, Omp25 was not or barely detected on the BvrR/BvrS mutants despite the several epitope specificities of the anti-Omp25 mAbs used (data not shown). All other Omps described so far in B. abortus (Omp10, Omp16, Omp19, Omp2b, and Omp1) were similarly immunodetected on both mutants, the wild-type and the reconstituted bvrR strain (data not shown). It is distinctive of Brucella group 3 Omps (to which Omp25 belongs) to be tightly bound to LPS in the outer membrane in a form partially resistant to protease digestion, and to coextract with this molecule (13, 23). Thus, SDS-proteinase K extracts and outer membrane fragments were examined by SDS/PAGE, and these analyses showed that the intensity of the band corresponding to group 3 proteins was weaker in the bvrR and bvrS mutants than in the wild type (Fig. 1). Consistent with the dot-blot results, Omp25 was not detected by Western blotting in SDS-proteinase K extracts of the mutants, but traces of this protein were detected in the outer membrane fragments of the bvrS mutant (Fig. 1).
Fig 1.
Envelope fractions of B. abortus 2308 wild type and BvrR/BvrS mutants contain different amounts of group 3 Omps. SDS-proteinase K extracts or outer membrane fragments were analyzed on SDS/12% PAGE gels and either silver stained for proteins or electroblotted and developed with anti-Omp25 mAbs. The arrow marks the position of the group 3 Omps band.
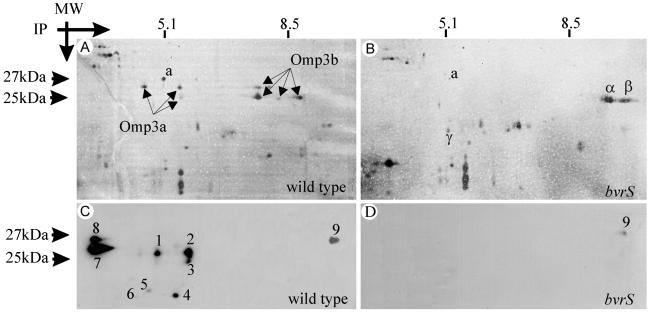
To clarify the small discrepancies observed in the above-described analyses, and also to explore other possible differences in Omp profiles, the outer membrane fractions were examined by 2DGE. This method confirmed and extended the previous findings because it revealed that two sets of proteins were either considerably reduced or beyond detection in the mutants. The first set consisted of three protein spots [all marked similarly as Omp3a (see below) in Fig. 2A] of about 25 kDa, with an intermediate isoelectric point of 5.4, and the second set consisted of five protein spots [all marked similarly as Omp3b (see below) in Fig. 2A] of about 27 kDa, with an intermediate isoelectric point of 8.2. As compared with the wild type (Fig. 2A), spots marked as Omp3b were considerably diminished in the bvrR mutant (not shown) and beyond detection in the bvrS mutant (Fig. 2B). These differences were observed consistently in all preparations, and were further confirmed by 2DGE analysis of the whole cell envelope fraction (not shown). Comparative lack, reduction, or presence of other spots was also evident in outer membrane fragments of the mutants. One spot, marked as “a” (Fig. 2 A and B) was considerably reduced in the bvrS mutant and not detected in the bvrR mutant (not shown). On the other hand, spots marked as α, β, and γ were detected in outer membrane fragments of the bvrS mutant but not in the wild-type or the bvrR mutant (Fig. 2 A and B and data not shown).
Fig 2.
Dysfunction of bvrR and bvrS alters the profile of B. abortus group 3 Omps. 2DGE (A and B) and the corresponding Western blot analysis (C and D) of outer membrane fragments of B. abortus 2308 wild type and BvrS mutant. The arrows indicate the silver-stained Omp3a and Omp3b spot groups. Numbers indicate spots immunodetected with anti-Omp25 mAbs. Notice that spots marked as Omp3a in A are the same as spots 1, 2, and 3 in C, and that spot “a” is detected in the wild type but only barely in the mutant. On the contrary, spots α, β, and γ present in the mutant were not detected in the wild type. Trace amounts of spot 9 were detected in the mutant.
To establish the correspondence between the dot-blot, SDS/PAGE, and 2DGE results, an anti-Omp25 mAb reacting against a linear epitope was used in Western blots of two-dimensional gels. At least nine major spots (including the three Omp3a spots) in the wild type and one discrete spot in the bvrS mutant were detected (Fig. 2 C and D). Therefore, the Omp3a spots (Fig. 2A) corresponded to Omp25. The resolution of Omp3a into several spots by 2DGE could result from several phenomena. Proteolytic degradation could account for large differences (for example, spots 4, 5, and 6 in Fig. 2C), secondary modifications for small differences in molecular weight (for example, between spots 2 and 3 in Fig. 2C), and interactions with charged molecules could generate shifts in isoelectric points without large changes in molecular weight (for example, spots 7–9 in Fig. 2C). In this regard, strong interactions between group 3 Omps and LPS have been shown (refs. 23 and 24; see also below). Because Brucella Omps clustering around 25 kDa were adequately designated as group 3 Omps in early studies (25), the name Omp3a should be preferred over Omp25. This is consistent with the accepted use of the names Omp2a and Omp2b (26) for the two porin proteins of Brucella, which were assigned to group 2 in the same studies (25).
Of the additional spots recognized by the same anti-Omp25 (Omp3a) mAb, two major ones (spots 7 and 8 in Fig. 2C) were resistant to proteolytic digestion and, therefore, not suitable for regular MALDI-MS sequence analysis or peptide map comparison. These spots had molecular weights between 27 and 25 kDa, and it seemed possible that they could be associated to LPS (see above). To investigate this, a two-step Western blot of outer membrane fragments was performed by probing the membrane first with the anti-Omp25 (Omp3a) and then with the anti-LPS mAbs. The results were registered after each step, and the reactions were superposed using standard software. This analysis demonstrated that spots 7 and 8 colocalized with the lower section of the LPS complexes that migrated in the acidic region of the 2DGE (data not shown).
It has been observed that the expression and apparent molecular weight of Omp3a in SDS/PAGE changes with time under regular culture conditions (27). To examine whether the heterogeneity of the group 3 Omps found by 2DGE analysis related to this observation, we analyzed the outer membrane fractions of the bvrR and bvrS mutants obtained at early and late bacterial growth. The results (not shown) demonstrated that the protein pattern was the same at the two stages of growth.
To look for further differences in protein expression, total bacterial SDS extracts and cytosols from the wild-type strain and the bvrR and bvrS mutants were subjected to 2DGE analysis. However, no significant qualitative or quantitative differences were observed in these materials.
Omp3b Is a Hitherto Unnoticed B. abortus Omp That Is Absent in bvrR and bvrS Mutants.
MALDI-MS and peptide mass mapping revealed that the five Omp3b spots in the wild-type strain had the same amino acid sequence (data not shown), and possible explanations for its apparent heterogeneity are the same as those proposed for Omp3a. Partial amino acid sequences were compared against preliminary B. suis genome sequence data, and PCR primers were designed to clone and sequence B. abortus omp3b. The whole sequence (GenBank accession no. AJ313014) contained an ORF with a potential ribosome-binding site located 5 bp upstream of the initiation codon. The omp3b ORF encoded a protein of 212 amino acid residues with a calculated mass of 22 kDa and a pI of 9.1. The analysis of the deduced amino acid sequence predicted an outer membrane localization of the protein and revealed homology (E value = 1.077e-39) with the OmpA group of bacterial Omps (28). A database search showed that Omp3b had 29% identity (47% similarity) to RopB from the plant endosymbiont Rhizobium leguminosarum, 29% identity (45% similarity) to Omp31 from Brucella melitensis, and 26% identity (45% similarity) to Omp3a (Omp25) from different Brucella species. The sequence presents a putative signal peptide of approximately 23 amino acids and a phenylalanine residue as a C-terminal amino acid, required for efficient translocation of Omps (29). Secondary structure prediction suggested a predominant extended conformation, with eight putative β-sheets, five of them hydrophobic. However, the overall hydrophobicity was only moderate (see Fig. 5, which is published as supporting information on the PNAS web site, www.pnas.org).
Omp3a and Omp3b Expression Is Transcriptionally Regulated by BvrR/BvrS.
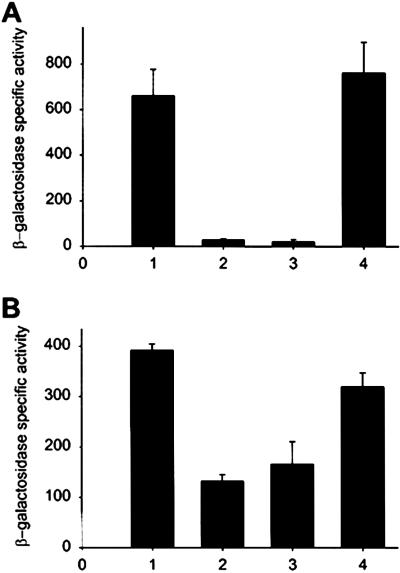
Omp3a was not detected by Western blot analysis of total soluble and purified cytoplasmic extracts of the bvrS mutant (not shown), strongly suggesting that this Omp was not accumulated in the bacterial cytoplasm and that its absence in the envelope could not be due to lack of translocation. Thus, we considered the hypothesis that expression of these proteins could be transcriptionally regulated and, accordingly, the β-galactosidase activity of omp3a:lacZ and omp3b:lacZ transcriptional fusions was investigated (Fig. 3). The results showed that the activity of the omp3a and omp3b promoters was decreased in both bvrR and bvrS mutants as compared with that in the wild type or in the reconstituted bvrR 65.21p. These results demonstrate that omp3a and omp3b are under the transcriptional control of BvrR/BvrS. Future DNA binding experiments will clarify whether this is the result of direct control by BvrR or is mediated through an intermediate regulatory system.
Fig 3.
Omp3a and Omp3b expression is transcriptionally regulated by BvrR/BvrS. The β-galactosidase activity of omp3a:lacZ (A) and omp3b:lacZ (B) transcriptional fusions was investigated in four genetic backgrounds: (1) B. abortus wild-type 2308; (2) bvrS mutant 2.13; (3) bvrR mutant 65.21; and (4) reconstituted bvrR 65.21p. The values are the mean and the standard error of three independent assays.
Omp3a and Omp3b Homologues Are Present in Members of the Rhizobiaceae.
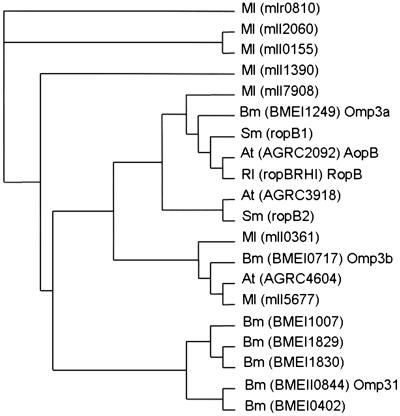
Using the B. abortus Omp3a and Omp3b sequences, we searched for homologous genes in the available genomes of B. melitensis and phylogenetically related bacteria (A. tumefaciens, S. meliloti, and M. loti). The search yielded a surprising number of homologues: seven in B. melitensis and in M. loti, three in A. tumefaciens, and two in S. meliloti. The corresponding protein sequences were aligned with the CLUSTAL W program, and a neighbor-joining tree derived from a distance-matrix was obtained (Fig. 4). According to this analysis, these proteins can be divided into four main groups: (i) sequences closely similar to Omp3a, including RopB and AopB; (ii) sequences closely similar to Omp3b; (iii) sequences only present in M. loti; and (iv) sequences closely similar to Omp31 only present in Brucella.
Fig 4.
Neighbor-joining tree derived from distance-matrix analysis of 22–31-kDa Omps sequences of M. loti (Ml), B. melitensis (Bm), S. meliloti (Sm), R. leguminosarum (Rl), and A. tumefaciens (At). The names of the corresponding genes (as they appear in GenBank) are in brackets.
Discussion
Brucella lacks classical virulence factors such as toxins or adhesins, and its virulence seems ascribed to type IV secretion systems and to the peculiar properties of the cell envelope (1). In this respect, the results presented here stress the role of the cell envelope in Brucella pathogenicity because they demonstrate that BvrR/BvrS regulates the expression of B. abortus Omps. Two major sets of proteins, Omp3a (formerly Omp25) and Omp3b, related to B. melitensis Omp31 (absent in B. abortus), to RopB of R. leguminosarum, and to AopB of A. tumefaciens, were identified as regulated by BvrR/BvrS.
The Omp3b protein described here has features reported for other bacterial Omps such as the characteristic eight membrane spanning amphipathic β-sheets, no high overall hydrophobicity, presence at the amino-terminal region of a signal peptide, and phenylalanine as the carboxyl-terminal residue (29). Thus, it is concluded that Omp3b is a hitherto nondescribed Brucella Omp.
The facts that Omp3a and Omp3b are severely diminished or absent, and that the promoter activities of omp3a and omp3b are also reduced in the bvrR and bvrS mutants, indicate that the synthesis of these proteins is under the control of the BvrR/BvrS two-component system. Although experimental evidence is lacking, it might be that this system also regulates the expression of other Omps because we observed a decrease of protein spot “a” and an increase of protein spots α, β, and γ. It is also possible that the increase in some protein spots and concomitant absence or reduction of Omp3a and Omp3b in the bvrR and bvrS mutants result from a compensatory effect to maintain the homeostasis of the outer membrane. An alternative possibility is suggested by the relatively high number of omp3 genes present in Brucella, particularly intriguing when considering that only Omp3a and Omp3b were identified. It is conceivable that some could code for the protein spots (such as α, β, and γ) that could not be analyzed by MALDI-MS for technical reasons, even though it is also possible that some of these genes are not expressed under laboratory conditions. If so, these omp genes would not be an exception because B. abortus carries two porin genes (omp2a and omp2b), only one of which is expressed in vitro (30).
In a previous study (2), we found that BvrR/BvrS mutants are attenuated in the mouse model, and Edmonds et al. (6, 7) have reported that omp3a-deficient mutants, which keep intact the BvrR/BvrS system, are attenuated in mice, cattle, and pregnant goats. The comparison of the results obtained in mice shows that, whereas the BvrR/BvrS mutants are cleared from spleens by the second week, the omp3a mutants last longer than 4 weeks (more than 18 weeks for the parental strain). Thus, although both kinds of mutants are attenuated, attenuation is more intense in the former. This is consistent with the characteristic control of multiple elements by two-component regulatory systems. Indeed, it is necessary to study omp3b and double omp3a-omp3b mutants to determine whether the attenuation of BvrR/BvrS mutants is due only to down-regulation of Omp3 proteins or whether additional factors are also involved. The same mutants should be examined for those outer membrane defects characteristic of BvrR/BvrS (i.e., polycation sensitivity, altered outer membrane hydrophobicity and permeability, etc.).
B. suis Omp3a has been shown to be involved in the negative regulation of TNF-α production after infection of human macrophages, supporting a role for this protein in Brucella capability to survive within cells (31). Furthermore, we have found recently that bvrS mutants do not stimulate the generation of active forms of small GTPases of the Rho family on cell contact (32), indicating that this two-component system is also necessary for establishing an adequate cross talk with the host cell. In contrast to wild-type bacteria, BvrR/BvrS mutants are poor cell invaders and, when ingested by professional and nonprofessional phagocytes, they are directed to lysosomes and destroyed (2). Taken together, these observations and the results of the present study lead to the hypothesis that BvrR/BvrS regulates the expression of Omps involved in cell invasion and also possibly in intracellular survival, perhaps through a direct or indirect control of intracellular trafficking. This could be due to a role of group 3 proteins in the interaction with cell surface receptors, but also with internal receptors aimed to detect intracellular parasite molecules (33). However, because BvrR/BvrS mutants have properties indicative of overall outer membrane alteration, it cannot be excluded that deficient internalization relates to this rather than to the Omp3 deficiency by itself. It also possible that an outer membrane structural stability is necessary for the correct assembly and functioning of the VirB membrane components known to be involved in the control of intracellular trafficking (34). Although these hypotheses and observations concern the role of Omp3 in intracellular survival, they do not exclude the contribution of other outer membrane components, such as LPS, in virulence (1), or their regulation by BvrR/BvrS. This aspect is also currently under investigation.
The similarity of the Brucella BvrR/BvrS system with some of the chromosomally encoded two-component regulatory systems present in plant endosymbionts and pathogens of the α-2 Proteobacteria is considerably higher than with other known two-component systems involved in bacterial virulence (2). S. meliloti ChvI/ExoS regulates production of polysaccharides like succinoglycans (8), but these kinds of molecules have not been described in Brucella, and we have evidence that production of Brucella native hapten polysaccharides and periplasmic cyclic glucans is not affected by mutation of bvrR and bvrS (unpublished results). However, the fact that the ChvI/ExoS and ChvI/ChvG of S. meliloti and A. tumefaciens are also critical for symbiosis and parasitism strongly argues in favor of the hypothesis that these systems are crucial for the adaptation to eukaryotic pericellular or intracellular habitats. From this perspective, the high homology displayed by Omp3a, Omp3b, and their Rhizobiaceae counterparts becomes very significant in the light of the data presented here and in previous works. In R. leguminosarum, RopB (Fig. 4), which is expressed in explanted bacterial cells (35), is severely decreased during bacteroid formation (36). This is also true of R. leguminosarum group III Omps such as RopA (37). Moreover, A. tumefaciens AopB, also highly homologous to Omp3a (Fig. 4), is involved in tumorigenesis (38). Obvious comparisons between the lifestyle of Brucella and rhizobiae emerge (39) and, on this basis, it can be predicted that expression of RopB, RopA, AopB, and other proteins homologous to Brucella group 3 Omps are both under the control of systems homologous to BvrR/BvrS and critical to establish prokaryotic–eukaryotic cell interactions. At least with regard to AopB, this prediction is confirmed by the results presented in the accompanying paper by Li et al (43).
Supplementary Material
Acknowledgments
We are indebted to E. Chaves for his helpful discussions. C.G.-V. was a recipient of a grant from the Karolinska International Research Training Program. Fellowship support to L.M. from Ministerio de Ciencia y Tecnología (Spain) and to A.S.-L. from Fundación R. Areces is also gratefully acknowledged. This work was supported by European Community Grant ICA4-CT-1999-10001 and the RTD project Noveltargetvacccines, Consejo Nacional de Ciencia y Tecnología (Costa Rica), Ministerio de Ciencia y Tecnología (Spain) Grant AGL2000-0305-C02-01, Programa de Cooperación Científica con Iberoamérica (Agencia Española de Cooperación Iberoamericana), and Plan de Investigación de la Universidad de Navarra.
Abbreviations
2DGE, two-dimensional gel electrophoresis
LPS, lipopolysaccharide
Omp, outer membrane protein
MALDI, matrix-assisted laser desorption ionization
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AJ313014).
References
- 1.Moreno E. & Moriyón, I. (2002) Proc. Natl. Acad. Sci. USA 99, 1-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sola-Landa A., Pizarro-Cerdá, J., Grilló, M. J., Moreno, E., Moriyón, I., Blasco, J. M., Gorvel, J. P. & López-Goñi, I. (1998) Mol. Microbiol. 29, 125-138. [DOI] [PubMed] [Google Scholar]
- 3.Freer E., Moreno, E., Moriyón, I., Pizarro-Cerdá, J., Weintraub, A. & Gorvel, J. P. (1996) J. Bacteriol. 178, 5867-5876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Martínez de Tejada G., Pizarro-Cerdá, J., Moreno, E. & Moriyón, I. (1995) Infect. Immun. 63, 3054-3061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Moriyón I. & López-Goñi, I. (1998) Int. Microbiol. 1, 19-26. [PubMed] [Google Scholar]
- 6.Edmonds M. D., Cloeckaert, A., Booth, N. J., Fulton, W. T., Hagius, S. D., Walker, J. V. & Elzer, P. H. (2001) Am. J. Vet. Res. 62, 1461-1466. [DOI] [PubMed] [Google Scholar]
- 7.Edmonds M. D., Cloeckaert, A., Hagius, S. D., Samartino, L. E., Fulton, W. T., Walker, J. V., Enright, F. M., Booth, N. J. & Elzer, P. H. (2002) Res. Vet. Sci. 72, 235-239. [DOI] [PubMed] [Google Scholar]
- 8.Cheng H. P. & Walker, G. C. (1998) J. Bacteriol. 180, 20-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Charles T. C. & Nester, E. W. (1993) J. Bacteriol. 175, 6614-6625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mantis N. J. & Winans, S. C. (1993) J. Bacteriol. 175, 6626-6636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Aragón V., Díaz, R., Moreno, E. & Moriyón, I. (1996) J. Bacteriol. 178, 1070-1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Moriyón I. & Berman, D. T. (1982) J. Bacteriol. 152, 822-828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gamazo C. & Moriyón, I. (1987) Infect. Immun. 55, 609-615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Garín-Bastuji B., Bowden, R. A., Dubray, G. & Limet, J. N. (1990) J. Clin. Microbiol. 28, 2169-2174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Laemmli U. K. (1970) Nature (London) 227, 680-685. [DOI] [PubMed] [Google Scholar]
- 16.Coligan J. E., Dunn, B. M., Ploegh, H. L., Speicher, D. W. & Wingfield, P. T., (1995) Current Protocols in Protein Science (Wiley, New York).
- 17.Bowden R. A., Cloeckaert, A., Zygmunt, M. S., Bernard, S. & Dubray, G. (1995) Infect. Immun. 63, 3945-3952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cloeckaert A., de Wergifosse, P., Dubray, G. & Limet, J. N. (1990) Infect. Immun. 58, 3980-3987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cloeckaert A., Jacques, I., Bosseray, N., Limet, J. N., Bowden, R., Dubray, G. & Plommet, M. (1991) J. Med. Microbiol. 34, 175-180. [DOI] [PubMed] [Google Scholar]
- 20.Rojas N., Freer, E., Weintraub, A., Ramírez, M., Lind, S. & Moreno, E. (1994) Clin. Diagn. Lab. Immunol. 1, 206-213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sambrook J., Fritsch, E. F. & Maniatis, T., (1989) Molecular Cloning: A Laboratory Manual (Cold Spring Harbor Lab. Press, Plainview, NY).
- 22.Miller J. F., (1972) Experiments in Molecular Genetics (Cold Spring Harbor Lab. Press, Plainview, NY).
- 23.Gamazo C., Vitas, A. I., Moriyón, I., López-Goñi, I. & Díaz, R. (1993) FEMS Microbiol. Lett. 112, 141-146. [DOI] [PubMed] [Google Scholar]
- 24.Moriyón I., Gamazo, C. & Díaz, R. (1987) Ann. Ins. Pasteur Microbiol. 138, 89-91. [DOI] [PubMed] [Google Scholar]
- 25.Verstreate D. R., Creasy, N. T., Caveney, N. T., Baldwin, C. L., Blab, M. W. & Winter, A. J. (1982) Infect. Immun. 35, 979-989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cloeckaert A., Verger, J. M., Grayon, M. & Vizcaíno, N. (1996) FEMS Microbiol. Lett. 145, 1-8. [DOI] [PubMed] [Google Scholar]
- 27.Gallot-Lavallee T., Zygmunt, M. S., Cloeckaert, A., Bezard, G. & Dubray, G. (1995) Res. Microbiol. 146, 227-236. [DOI] [PubMed] [Google Scholar]
- 28.MacCallum R. M., Kelley, L. A. & Sternberg, M. J. E. (2000) Bioinformatics 16, 125-129. [DOI] [PubMed] [Google Scholar]
- 29.Struyvé M., Moons, M. & Tommassen, J. (1991) J. Mol. Biol. 218, 141-148. [DOI] [PubMed] [Google Scholar]
- 30.Marquis H. & Ficht, T. A. (1993) Infect. Immun. 61, 3785-3790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jubier-Maurin V., Boigegrain, R. A., Cloeckaert, A., Gross, A., Alvarez-Martínez, M. T., Terraza, A., Liautard, J., Köhler, S., Rouot, B., Dornand, J. & Liautard, J. P. (2001) Infect. Immun. 69, 4823-4830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Guzmán-Verri C., Chaves-Olarte, E., von Eichel-Streiber, C., López-Goñi, I., Thelestam, M., Arvidson, S., Gorvel, J. P. & Moreno, E. (2001) J. Biol. Chem. 276, 44435-44443. [DOI] [PubMed] [Google Scholar]
- 33.Girardin S. E., Sansonetti, P. J. & Philpott, D. J. (2002) Trends Microbiol. 10, 193-199. [DOI] [PubMed] [Google Scholar]
- 34.Comerci D. J., Martínez-Lorenzo, M. J., Sieira, R., Gorvel, J. P. & Ugalde, R. A. (2001) Cell. Microbiol. 3, 159-168. [DOI] [PubMed] [Google Scholar]
- 35.Roest H. P., Mulders, I. H., Wijffelman, C. A. & Lugtenberg, B. J. (1995) Mol. Plant–Microbe Interact. 8, 576-583. [PubMed] [Google Scholar]
- 36.de Maagd R. A., de Rijk, R., Mulders, I. H. M. & Lugtenberg, B. J. J. (1989) J. Bacteriol. 171, 1136-1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.de Maagd R. A., Yang, W. C., Goosen-de Roo, L., Mulders, I. H. M., Roest, H. P., Spaink, H. P., Bisseling, T. & Lugtenberg, B. J. J. (1994) Mol. Plant–Microbe Interact. 7, 276-281. [Google Scholar]
- 38.Jia Y. H., Li, L. P., Hou, Q. M. & Pan, S. Q. (2002) Gene 284, 113-124. [DOI] [PubMed] [Google Scholar]
- 39.Moreno E., Stackebrandt, E., Dorsch, M., Wolters, J., Busch, M. & Mayer, H. (1990) J. Bacteriol. 172, 3569-3576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sangari F. & Agüero, J. (1991) Microb. Pathog. 11, 443-446. [DOI] [PubMed] [Google Scholar]
- 41.Simons R. W., Priefer, U. & Puhler, A. (1983) Bio/Technology 1, 784-791. [Google Scholar]
- 42.Martínez de Tejada G., Miller, J. F. & Cotter, P. A. (1996) Mol. Microbiol. 22, 895-908. [DOI] [PubMed] [Google Scholar]
- 43.Li L., Jia, Y., Hou, Q., Charles, T. C., Nester, E. W. & Pan, S. Q. (2002) Proc. Natl. Acad. Sci. USA 99, 12369-12374. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.