Abstract
Adeno-associated virus (AAV) undergoes site-specific integration into human chromosome 19 through a deletion-substitution mechanism at the well characterized AAVS1 site. We have shown previously that a cis element within the left end of the AAV genome enhances the efficiency of Rep-mediated site-specific integration into chromosome 19 when present in inverted terminal repeat-containing recombinant AAV (rAAV) plasmids. We now demonstrate that a 138-bp cis element, the p5 integration efficiency element (p5IEE), mediates efficient integration. The p5IEE is not only required for efficient site-specific integration, it is also sufficient. Integration mediated by the p5IEE occurs in the absence of the AAV inverted terminal-repeat elements. The data presented in this study demonstrate that the p5IEE is a multifunctional element, serving as the highly regulatable Rep promoter and the primary substrate for targeted integration.
Adeno-associated virus (AAV) is a single-stranded parvovirus with a 4.7-kb genome (1). The DNA contains two ORFs, rep and cap, that are flanked by two inverted terminal repeats (ITRs; refs. 2–4). AAV is the only known virus that site-specifically integrates into the human genome, and this targeted integration of the AAV genome occurs at the AAVS1 site on chromosome 19q13.3-qter (5–8).
The Rep protein (specifically Rep 68/78) is essential in mediating recombination between the AAV genome and the AAVS1 chromosomal target (9–13). Rep 68/78 functions by binding to the Rep-binding elements (RBEs) situated in both the AAV genome and at the AAVS1 site (7, 14, 15). Through poorly understood interactions, the Rep protein/AAV–DNA complex localizes to the AAVS1 site, and a nonhomologous deletion-insertion recombination event occurs, resulting in integration of the AAV genome (16–22). It has been shown that head-to-tail concatemers of the wild-type (wt)AAV genome are able to site-specifically integrate in this manner (23).
Rep-mediated integration of recombinant AAV (rAAV)(ITR+) plasmids [such as pTRUF2 (24) or p2ITRLacZ (10)] is extremely inefficient, with between 0.1 and 1% of transduced cells demonstrating rAAV genome persistence after 6 weeks (25). In contrast, plasmids that carry the entire AAV genome integrate at efficiencies of greater than 10%. This difference between rAAV plasmids and wtAAV plasmids led to the discovery of a previously unknown cis sequence domain present in the left end of the AAV genome that enhances integration efficiency of rAAV(ITR+) constructs by 10–100-fold (25). The current study extends the characterization of the left-end cis element in mediating site-specific integration. We have discovered a 138-bp AAV integration efficiency element (p5IEE) that is not only necessary for efficient site-specific integration but is also sufficient. Data presented in this study clearly demonstrate efficient Rep-mediated integration of p5IEE-containing plasmids that do not contain AAV ITR elements. Furthermore, in nearly 100% of cases the integration event was targeted to the AAVS1 site of chromosome 19.
Materials and Methods
Plasmid Constructs.
Plasmid constructs pAAV/Ad [pRepCap(itr−) (26)] and pSub201 [pRepCap(itr+) (27)] are well established AAV constructs. Plasmids pGFPCap and pT7-Rep were constructed as described previously (25). The pAd-p5CAT plasmid was generated by replacement of the cytomegalovirus (CMV) promoter in pAd-chloramphenicol acetyltransferase (CAT) (28) with a 138-bp PCR fragment of the p5 promoter element (corresponding to AAV nucleotides 151–289).
Southern Blot Analysis.
Whole-cell DNA was isolated from HeLa cell lines by using a standard salting-out protocol (29). EcoRI was used to digest 7.5 μg of DNA from each clone, and digested DNA was separated on 1% agarose gels. After transferring DNA fragments to nylon membranes, hybridization was carried out by using 32P-labeled probes at a concentration of 3 million cpm/ml Sigma prehybridization solution according to manufacturer instructions. The following DNA fragments were generated for DNA probes: the 800-bp Rep PCR fragment was generated from oligos gatcgaagcttccgcgtctgacgtcgatgg and ggaccaggcctcatacatctccttcaatgc; the AAVS1 1-kb PCR product was obtained by using oligos gaactctgccctctaacgctgc and caccagataaggaatctgcc (5, 30); the NotI digest of pTRUF2 to generate 700-bp GFP fragment; and the 1.2-kb plasmid backbone sequence was generated by AseI-digesting pRepCap(itr−). DNA probes were 32P-labeled by using Rediprime kit (Amersham Pharmacia) according to manufacturer instructions. Bands were visualized by autoradiography.
Plasmid Integration Assay.
HeLa cells were electroporated with plasmid(s), and 48 h posttransfection, GFP+ cells were isolated by fluorescence-activated cell sorter (FACS) sorting using a Beckman Coulter Altra cell sorter. Cells were plated at 1 cell per well into 96-well plates, and clonal cell lines were grown for 6 weeks in Dulbecco's modified Eagle's medium containing 5% calf serum and 5% FCS. Whole-cell DNA was harvested, EcoRI-digested, and separated on 1% agarose gels. The DNA was transferred to nylon membranes and hybridized to 32P-labeled probes (using the Southern blot protocol described above).
Results
pRepCap(itr−) Integration into AAVS1.
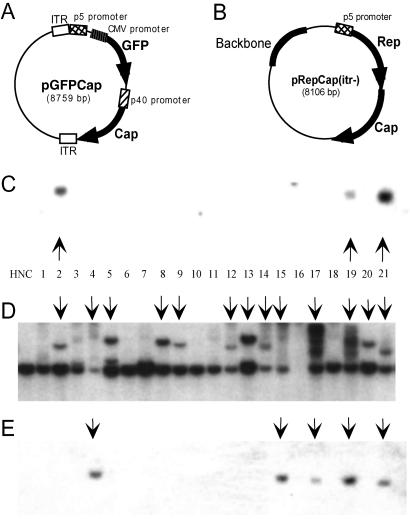
We have used a simple transfection assay to characterize the integration efficiency of ITR+ substrate plasmids in HeLa cells (25). The transfection assay includes three elements: an AAV integration substrate, a Rep-expressing plasmid to mediate the integration event, and a GFP plasmid for FACS-transduced cells. Briefly, HeLa cells are cotransfected with plasmids containing the three elements, and 48 h posttransfection, GFP+ cells are FACS-sorted and plated into 96-well dishes to establish clonal cell lines over a period of 6–8 weeks. Cell lines were established from a cotransfection of the rAAV plasmid pGFPCap (Fig. 1A), which contains the first 7% of the AAV genome followed by a GFP transgene and AAV Cap sequence, and the Rep-expressing plasmid pRepCap(itr−) (Fig. 1B) [pAAV/Ad (26)], which contains wtAAV sequences with both flanking ITR elements deleted. Clones were screened by Southern analysis for persistence of pGFPCap DNA and disruption of the AAVS1 genomic locus. It is found commonly that when AAV integrates into HeLa AAVS1 sites, resulting disruptions are variable in AAVS1 restriction fragment length and fragment-band intensity. Several factors may influence the character of AAVS1 integrants: HeLa cells are aneuploid; the AAVS1 integration site is instable, particularly in the presence of rep; and the deletion-insertion mechanism of Rep-mediated AAVS1 integrations imprecisely alter the sequence of the AAVS1 site. In these studies, we are looking for evidence of AAVS1 disruption and persistence of the target substrate DNA as well as comigration of plasmid DNA with an AAVS1 disruption band as indicating a site-specific integration event. In agreement with previous results, ≈6% of the 78 cell lines tested contained integrated pGFPCap DNA (Fig. 1C). When cell lines were screened for genomic alterations of the AAVS1 site of chromosome 19, 54% of the clones were found to have AAVS1 site disruptions (Fig. 1D). Based on these data, it seems that AAVS1 disruptions occur at a much higher frequency than pGFPCap integration events.
Fig 1.
Southern blot analysis of pGFPCap and pRepCap(itr−)-cotransfected HeLa cell lines: Rep integration. Schematic representations of AAV plasmid constructs pGFPCap (A) and pRepCap(itr−) (B). HeLa cells were transfected with pGFPCap and pRepCap(itr−), 48-h posttransfection cells were FACS-sorted, and clonal cell lines were grown for 6 weeks. Whole-cell DNA was harvested, EcoRI-digested, and characterized by Southern blot analysis by probing with 32P-labeled GFP (C), AAVS1 (D), or Rep (E) probes. HNC shows negative-control HeLa cell DNA. Vertical arrows indicate GFP-containing clones, S1 disruptions, and rep-containing clones (C–E, respectively).
Several explanations may account for the above observation: Rep-mediated disruption of AAVS1 sites can occur in the absence of integration; integration events are occurring at AAVS1 sites, but the integrated DNA is unstable; or DNA elements other than pGFPCap are integrating at the AAVS1 site. The pGFPCap and pRepCap(itr−) plasmids (Fig. 1 A and B, respectively) have significant homology at both the 5′ and 3′ ends of their respective AAV regions. Therefore an overlap recombination between the two plasmids theoretically could occur and result in generation of a wtAAV genome. To confirm that none of the cell lines had taken up a wt rearrangement between pGFPCap and pRepCap(itr−), genomic DNA from the clones was examined for the presence of integrated Rep DNA. Surprisingly, we found that 14% of the cell lines contained Rep DNA that in most cases was integrated site-specifically (Fig. 1 E and D, respectively). To determine whether a recombination event between pRepCap(itr−) and pGFPCap had occurred, we carried out PCR analysis of genomic DNA by using one primer complementary to the pRepCap(itr−) Rep sequence and another complementary to the pGFPCap ITR sequence. A wt recombination event would yield a 700-bp PCR product, whereas pGFPCap or pRepCap(itr−) would not serve as a substrate for the PCR primer pair. As positive and negative controls, the primer pair was used to amplify from pRepCap(itr+) [pSub201 (27)] and pRepCap(itr−), respectively. A PCR product was not obtained from any of the pGFPCap-RepCap(itr−)-cotransfected cell lines (data not shown). This result suggested that recombination between the pRepCap(itr−) and pGFPCap(itr+) to form a wtAAV plasmid was unlikely to have accounted for the Rep integrants identified in Fig. 1E and raised the possibility that pRepCap(itr−) could be an independent substrate for Rep-mediated site-specific integration into AAVS1.
AAV-ITR Elements Are Not Required for AAV Site-Specific Integration.
Based on the results in Fig. 1, we hypothesized that pRepCap(itr−) contains all the cis and trans factors necessary for site-specific integration despite the fact that it does not contain ITR elements. To test this hypothesis, HeLa cells were transfected with the pRepCap(itr−) plasmid and a GFP-expressing plasmid, pCMV-GFP, that does not contain AAV sequence. In parallel, we carried out a transfection of pRepCap(itr+) (Fig. 2A) and pCMV-GFP. After plating GFP+ FACS-sorted single cells into 96-well plates, clonal cell lines were grown for 6 weeks, and then genomic DNA was harvested.
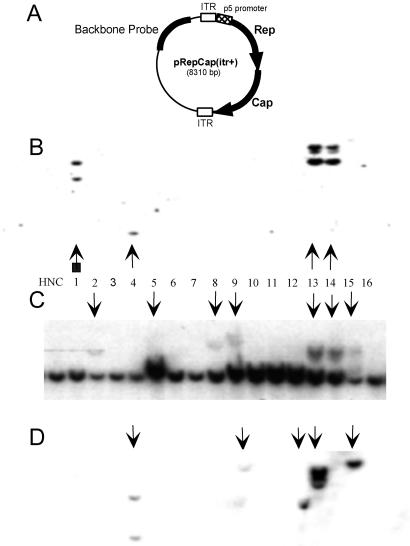
Fig 2.
AAV-ITR elements are potential substrate-integration boundaries when using wt plasmid pRepCap(itr+). (A) Schematic representation of wtAAV plasmid construct pRepCap(itr+). HeLa cells were cotransfected with pRepCap(itr+) and a GFP-expressing plasmid. Clonal cell lines were grown for 6 weeks and processed as described previously. Southern blots were probed with 32P-labeled Rep (B), AAVS1 (C), or backbone (D) DNA. HNC shows negative-control HeLa cell DNA. Vertical arrows indicate S1 disruptions and rep-containing or backbone-containing clones (C, B, and D, respectively). The arrow with a square indicates rep band comigrating with the 8-kb AAVS1 band; therefore, disruption to the S1 site is not visible.
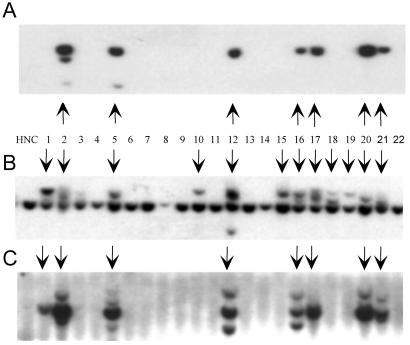
The DNA from pRepCap(itr+) cell lines was digested with EcoRI, and Southern blotting was used to detect integration of the rep transgene and also to visualize disruptions in the AAVS1 site. In agreement with earlier results, the wt ITR-containing plasmid pRepCap(itr+) had a Rep-integration efficiency of 12% (Fig. 2B and Table 1). Consistent with previous observations, AAVS1 disruptions were found in a higher proportion of cell lines (33%) than those that retained rep DNA sequence (compare 2C with 2B). DNA from cell lines transfected with pRepCap(itr−) was characterized by Southern analysis for Rep integration. Rep was found in 22% of cell lines tested, and in most cases Rep was integrated into AAVS1 (Fig. 3A and Table 1). A large percentage of pRepCap(itr−)-transfected cell lines (45%) had AAVS1 genomic fragment disruptions (Fig. 3B). We can make two conclusions from these results: first, plasmid constructs that lack the AAV ITR elements can serve as substrate DNA for rep-mediated site-specific integration into human chromosome 19 at the AAVS1 site (Table 1); and second, higher levels of AAVS1 disruption than substrate DNA integrations are found in cell lines established from either ITR+ or ITR− substrate plasmids.
Table 1.
Site-specific integration efficiency of plasmid vectors
| Plasmid | No. of clones tested | % with Rep (∼) or CAT (*) | % with backbone | % with AAVS1 disruption | % site specificity |
|---|---|---|---|---|---|
| pRepCap(itr+) | 48 | 12∼ | 23 | 33 | 90 |
| pRepCap(itr−) | 49 | 22∼ | 24 | 45 | 91 |
| pT7-Rep + pAd-p5CAT | 45 | 22* | nd | 29 | 100 |
| pAd-p5CAT | 48 | 0* | nd | 0 | 0 |
HeLa cells were transfected with plasmid vectors, and clonal cell lines were grown for 6 weeks. Whole-cell DNA was harvested, digested with EcoRI, and separated on 1% agarose gels. DNA was transferred to nylon membranes and hybridized to 32P-labeled transgene probes. nd indicates that Southern blots were not done.
Fig 3.
Site-specific integration of pRepCap(itr−) in the absence of additional cis elements (ITRs). HeLa cells were transfected with pRepCap(itr−) and a GFP-expressing plasmid, then 48 h posttransfection, cells were FACS-sorted, and GFP+ clonal cell lines were grown for 6 weeks. Whole-cell DNA was harvested, EcoRI-digested, and characterized by Southern blot analysis by probing with 32P-labeled Rep (A), AAVS1 (B), or plasmid backbone (C) probes. HNC shows negative-control HeLa cell DNA. Vertical arrows indicate disruptions of the chromosome 19 AAVS1 8-kb EcoRI genomic fragment (B) and rep- or backbone-containing clones (A and C, respectively).
ITR Elements Influence Boundaries of Integration Substrates.
Results from the previous section demonstrate that not only does pRepCap(itr−) contain all the elements necessary to mediate targeted integration, but it seems to be a more effective integration substrate than wt pRepCap(itr+) [compare 22 with 12% Rep(+) integrants, respectively; Table 1]. Because a major difference between these constructs is the presence of the AAV ITR elements in pRepCap(itr+), we sought to determine how the ITR may be influencing integration efficiency. The terminal hairpin structure of the AAV ITR serves as the viral origin of replication. In a double-stranded plasmid substrate such as in pRepCap(itr+), duplex cruciform structures can be generated at each ITR, with each cruciform being a substrate for Rep binding and nicking. If nicking at an ITR cruciform occurs, it may result in defining the segment of the pRepCap(itr+) element that is able to undergo site-specific integration. If this occurs, we can envision at least three distinct integration substrates: RepCap flanked by ITRs; plasmid backbone flanked by ITRs; or both RepCap and plasmid backbone flanked by ITRs. In contrast, pRepCap(itr−) lacks the complexity and function of the ITR elements, and in theory all integrants of pRepCap(itr−) should include the entire pRepCap(itr−) DNA sequence.
To determine if ITR elements function to define integration boundaries, we probed genomic DNA isolated from the pRepCap(itr−) and the pRepCap(itr+)-transfected clones with 1.2-kb AseI-digested pRepCap(itr−) backbone sequence (Figs. 3C and 2D, respectively). In agreement with our prediction, the plasmid backbone Southern blot of pRepCap(itr−) shows that 24% of the cell lines contained plasmid backbone (Table 1). Furthermore, 91% of the Rep+ clones are also positive for the plasmid backbone. Based on the data from this experiment, it seems that the entire plasmid sequence is the substrate for AAVS1 integration in cell lines derived from a pRepCap(itr−) transduction of HeLa cells.
In contrast to the cell lines derived from pRepCap(itr−) integration, the plasmid backbone screen of pRepCap(itr+)-transfected cell lines (Fig. 2D) resulted in a more complex pattern of AAVS1 integration. We found that 27% of cell lines tested had at least part of the pRepCap(itr+) sequence integrated. As previously indicated, rep was site-specifically integrated into 12% of the cell lines; however, when screened for plasmid backbone sequence, 23% of the cell lines were shown to contain plasmid integrants (Fig. 2 B and D, respectively, and Table 1). The distribution of the three predicted types of integration substrate was biased slightly toward the plasmid backbone, roughly half of the cell lines had only backbone integrants, approximately a third of the integrants had both backbone and RepCap sequence, and the remainder contained only RepCap sequence.
The overall integration efficiency of ITR-containing or ITR-deleted constructs (27 versus 24%) is similar. Therefore we are led to conclude that the AAV ITRs make a minimal contribution toward integration efficiency. However, when using a plasmid integration substrate, the presence of the AAV ITR elements can act to generate integration boundaries. Presumably Rep binding to the RBE and nicking at the terminal resolution site of the ITR generates potential 5′ and 3′ boundaries of the integration substrate. In a wt virus infection, boundary definition is naturally present because of the ITR hairpin structure present at the ends of the linear, single-stranded viral genome.
p5IEE Sequence Is the Only cis Element Required for Site-Specific Integration.
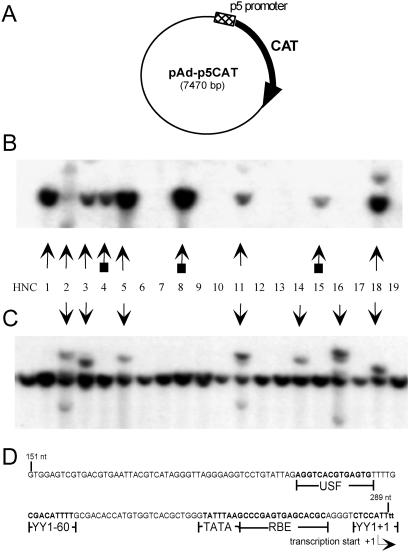
The experiments presented in this study indicate that AAV ITR elements, which have long been considered important cis elements in mediating site-specific integration, do not play a primary role in this event. We previously identified a cis element, the IEE, that is essential for efficient Rep-mediated site-specific integration (25). Based on the new observations presented in this study, the IEE may not only be a necessary cis element but also sufficient in mediating efficient Rep-dependent integration into AAVS1. The IEE region overlaps with the p5 promoter region of AAV (31, 32). To determine whether the p5 promoter was coincident with the IEE, a plasmid, pAd-p5CAT, was constructed that contains the 138-bp p5 promoter sequence (AAV nucleotide 151 to nucleotide 289) upstream of a CAT reporter gene (Fig. 4A). Promoter activity was confirmed with this construct in transient transfection assays. In HeLa cells, the p5 promoter was able to mediate expression of CAT at levels comparable to the CMV promoter and was vulnerable to trans repression by Rep (data not shown).
Fig 4.
Southern blot analysis showing rep-mediated site-specific integration of pAd-p5CAT. (A) Schematic representation of pAd-p5CAT plasmid. HeLa cells were cotransfected with pAd-p5CAT and pT7-Rep, a rep-expressing plasmid. Cell lines were grown and processed as described previously. (B) Hybridization to CAT probe. (C) Hybridization to AAVS1 probe. HNC shows negative-control HeLa cell DNA. Vertical arrows indicate CAT-containing clones (B) and disruptions of the chromosome 19 AAVS1 8-kb EcoRI genomic fragment (C). The arrows with squares indicate CAT bands comigrating with the 8-kb AAVS1 band; therefore, disruption to the S1 site is not visible. (D) Schematic representation of the 138-nt IEE showing YY1 and Rep-binding sites, a putative upstream stimulating factor (USF)-binding site, and a TATA box.
To determine the function of this construct in an integration assay, HeLa cells were cotransfected with pAd-p5CAT and the Rep-expressing plasmid, pT7-Rep (25). The pT7-Rep plasmid also expresses GFP, which allowed for FACS sorting 48 h posttransfection. Cell lines were established as described previously, and after 6 weeks each cell line was harvested for genomic DNA. Genomic DNA was digested with EcoRI, and a Southern blot analysis was performed as described previously, probing for the presence of CAT or the presence of AAVS1 disruptions (sample shown in Fig. 4 B and C, respectively, and Table 1). Of 45 cell lines tested, 29% contained AAVS1 disruptions, and CAT gene integrations were found in 22% of cell lines. In all cases, the data indicate comigration of pAd-p5CAT DNA with the AAVS1 probe. As a negative control, HeLa cells were transfected with pAd-p5CAT alone. In the absence of a Rep-expressing plasmid, the CAT transgene was unable to integrate, which confirms that the targeted integration event is Rep-dependent (Table 1). The results of this experiment indicate that the 138-bp p5 promoter region (p5IEE) is the only AAV element required in cis to mediate site-specific integration of a substrate DNA through Rep-dependent integration into AAVS1.
Discussion
The data presented in this study offer several important new insights into the molecular mechanisms that mediate site-specific integration of the AAV genome into human chromosome 19. It has generally been thought that AAV replication and its replication origins (the ITR elements) are involved in AAV-targeted integration. Although unexpected, our data present a strong argument against a direct role for the AAV ITR elements in mediating the integration event. We have found that when using plasmid integration substrates, a 138-bp element, the p5IEE, is the only cis element required to target a substrate DNA for site-specific integration to the AAVS1 site on chromosome 19.
In previous studies, the characterization of ITR-containing rAAV integrants had relied on drug selection to isolate cell lines that had stably integrated the target DNA substrate. Under these conditions, site-specific integration into chromosome 19 occurs in ≈30% of selected cell lines, with the remaining cell lines containing random integrants (33, 34). This high percentage of cell lines undergoing ITR-mediated site-specific integration demonstrates that the presence of the RBE in the AAV ITR can function to facilitate localization of the rAAV genome to the target integration site. However, recent studies have separated the replication function of the AAV origin from the potential integration function of the ITR elements (22). Therefore we are left to conclude that under these conditions the RBE may be the sole cis integration element functioning in the ITR. In comparison to ITR-mediated integration, the p5IEE is 10–100-fold more effective at inducing integration than the AAV ITR. Furthermore, the p5IEE is able to mediate nearly 100% localization to the AAVS1 site. The enhanced integration specificity and efficiency of p5IEE integration enables us to conclude that the p5IEE serves as a unique platform for the formation of an AAV/Rep “integration complex.”
The p5IEE has been extensively characterized as a highly regulated promoter of Rep 68/78 (refs. 31, 32, 35, and 36; Fig. 4D). After presentation of a double-stranded template for transcription, the p5 promoter is activated through binding of cellular transcription factors, resulting in Rep-68/78 production. Autoregulation is a key feature of the p5 promoter. Low-level Rep is sufficient to allow Rep binding to RBEs in the p5 promoter (35) as well as in the ITR (14). The binding of Rep to the p5 promoter causes down-regulation of p5 transcription by RNA polymerase II and a subsequent reduction in Rep 68 and 78 expression (37). The rationale for p5/Rep autorepression is not well understood. The presence of helper virus induces high-level Rep expression, which is associated with AAV rescue, replication, and cytotoxicity; hence AAV enters the lytic cycle. In contrast, AAV latency occurs in the absence of helper virus (6, 38). The autoregulation of Rep leads to low-level Rep expression, which is associated with integration and generation of the latent state of AAV infection. Our view of p5 promoter down-regulation is that it represents a transition from a low-activity transcription complex to a high-activity integration complex. The presence of an array of transcription factors (31, 32, 39) and/or preinitiation complexes (35), which are present before Rep binding, offer a potential platform for the formation of a unique integration structure that mediates the specialized localization of the p5IEE to the AAVS1 site. Further studies may be carried out to characterize this integration complex.
Acknowledgments
We thank Christopher Colon for the FACS sorting. This work was supported by National Institutes of Health Grants P50 HL59312 and EY13101.
Abbreviations
AAV, adeno-associated virus
ITR, inverted terminal repeat
RBE, Rep-binding element
wt, wild type
rAAV, recombinant AAV
IEE, integration efficiency element
FACS, fluorescence-activated cell sorter
CMV, cytomegalovirus
CAT, chloramphenicol acetyltransferase
References
- 1.Berns K. I. & Linden, R. M. (1995) BioEssays 17, 237-245. [DOI] [PubMed] [Google Scholar]
- 2.Srivastava A., Lusby, E. W. & Berns, K. I. (1983) J. Virol. 45, 555-564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kotin R. M. (1994) Hum. Gene Ther. 5, 793-801. [DOI] [PubMed] [Google Scholar]
- 4.Srivastava A. (1987) Intervirology 27, 138-147. [DOI] [PubMed] [Google Scholar]
- 5.Kotin R. M., Siniscalco, M., Samulski, R. J., Zhu, X. D., Hunter, L., Laughlin, C. A., McLaughlin, S., Muzyczka, N., Rocchi, M. & Berns, K. I. (1990) Proc. Natl. Acad. Sci. USA 87, 2211-2215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Samulski R. J., Zhu, X., Xiao, X., Brook, J. D., Housman, D. E., Epstein, N. & Hunter, L. A. (1991) EMBO J. 10, 3941-3950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Giraud C., Winocour, E. & Berns, K. I. (1994) Proc. Natl. Acad. Sci. USA 91, 10039-10043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kearns W. G., Afione, S. A., Fulmer, S. B., Pang, M. C., Erikson, D., Egan, M., Landrum, M. J., Flotte, T. R. & Cutting, G. R. (1996) Gene Ther. 3, 748-755. [PubMed] [Google Scholar]
- 9.Dyall J., Szabo, P. & Berns, K. I. (1999) Proc. Natl. Acad. Sci. USA 96, 12849-12854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Surosky R. T., Urabe, M., Godwin, S. G., McQuiston, S. A., Kurtzman, G. J., Ozawa, K. & Natsoulis, G. (1997) J. Virol. 71, 7951-7959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lamartina S., Roscilli, G., Rinaudo, D., Delmastro, P. & Toniatti, C. (1998) J. Virol. 72, 7653-7658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xiao X., Li, J. & Samulski, R. J. (1996) J. Virol. 70, 8098-8108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Walsh C. E., Liu, J. M., Xiao, X., Young, N. S., Nienhuis, A. W. & Samulski, R. J. (1992) Proc. Natl. Acad. Sci. USA 89, 7257-7261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Weitzman M. D., Kyostio, S. R., Kotin, R. M. & Owens, R. A. (1994) Proc. Natl. Acad. Sci. USA 91, 5808-5812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chiorini J. A., Wendtner, C. M., Urcelay, E., Safer, B., Hallek, M. & Kotin, R. M. (1995) Hum. Gene Ther. 6, 1531-1541. [DOI] [PubMed] [Google Scholar]
- 16.Dyall J. & Berns, K. I. (1998) J. Virol. 72, 6195-6198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Linden R. M., Ward, P., Giraud, C., Winocour, E. & Berns, K. I. (1996) Proc. Natl. Acad. Sci. USA 93, 11288-11294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Linden R. M., Winocour, E. & Berns, K. I. (1996) Proc. Natl. Acad. Sci. USA 93, 7966-7972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang C. C., Xiao, X., Zhu, X., Ansardi, D. C., Epstein, N. D., Frey, M. R., Matera, A. G. & Samulski, R. J. (1997) J. Virol. 71, 9231-9247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Urcelay E., Ward, P., Wiener, S. M., Safer, B. & Kotin, R. M. (1995) J. Virol. 69, 2038-2046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Young S. M., McCarty, D. M., Degtyareva, N. & Samulski, R. J. (2000) J. Virol. 74, 3953-3966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Young S. M. & Samulski, R. J. (2001) Proc. Natl. Acad. Sci. USA 98, 13525-13530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Giraud C., Winocour, E. & Berns, K. I. (1995) J. Virol. 69, 6917-6924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zolotukhin S., Potter, M., Hauswirth, W. W., Guy, J. & Muzyczka, N. (1996) J. Virol. 70, 4646-4654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Philpott N. J., Giraud-Wali, C., Dupuis, C., Gomos, J., Hamilton, H., Berns, K. I. & Falck-Pedersen, E. (2002) J. Virol. 76, 5411-5421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Samulski R. J., Chang, L. S. & Shenk, T. (1989) J. Virol. 63, 3822-3828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Samulski R. J., Chang, L. S. & Shenk, T. (1987) J. Virol. 61, 3096-3101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gall J., Kass-Eisler, A., Leinwand, L. & Falck-Pedersen, E. (1996) J. Virol. 70, 2116-2123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Miller S. A., Dykes, D. D. & Polesky, H. F. (1988) Nucleic Acids Res. 16, 1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tsunoda H., Hayakawa, T., Sakuragawa, N. & Koyama, H. (2000) Virology 268, 391-401. [DOI] [PubMed] [Google Scholar]
- 31.Shi Y., Seto, E., Chang, L. S. & Shenk, T. (1991) Cell 67, 377-388. [DOI] [PubMed] [Google Scholar]
- 32.Chang L. S., Shi, Y. & Shenk, T. (1989) J. Virol. 63, 3479-3488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Palombo F., Monciotti, A., Recchia, A., Cortese, R., Ciliberto, G. & La Monica, N. (1998) J. Virol. 72, 5025-5034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pieroni L., Fipaldini, C., Monciotti, A., Cimini, D., Sgura, A., Fattori, E., Epifano, O., Cortese, R., Palombo, F. & La Monica, N. (1998) Virology 249, 249-259. [DOI] [PubMed] [Google Scholar]
- 35.Kyostio S. R., Wonderling, R. S. & Owens, R. A. (1995) J. Virol. 69, 6787-6796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McCarty D. M., Pereira, D. J., Zolotukhin, I., Zhou, X., Ryan, J. H. & Muzyczka, N. (1994) J. Virol. 68, 4988-4997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Beaton A., Palumbo, P. & Berns, K. I. (1989) J. Virol. 63, 4450-4454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kotin R. M., Menninger, J. C., Ward, D. C. & Berns, K. I. (1991) Genomics 10, 831-834. [DOI] [PubMed] [Google Scholar]
- 39.Seto E., Shi, Y. & Shenk, T. (1991) Nature (London) 354, 241-245. [DOI] [PubMed] [Google Scholar]