Abstract
The 2,160,267 bp genome sequence of Streptococcus agalactiae, the leading cause of bacterial sepsis, pneumonia, and meningitis in neonates in the U.S. and Europe, is predicted to encode 2,175 genes. Genome comparisons among S. agalactiae, Streptococcus pneumoniae, Streptococcus pyogenes, and the other completely sequenced genomes identified genes specific to the streptococci and to S. agalactiae. These in silico analyses, combined with comparative genome hybridization experiments between the sequenced serotype V strain 2603 V/R and 19 S. agalactiae strains from several serotypes using whole-genome microarrays, revealed the genetic heterogeneity among S. agalactiae strains, even of the same serotype, and provided insights into the evolution of virulence mechanisms.
Streptococcus agalactiae, or group B Streptococcus, is the leading cause of bacterial sepsis, pneumonia, and meningitis in neonates in the U.S. and Europe. Although S. agalactiae usually behaves as a commensal organism that colonizes the gastrointestinal or genital tract of 25–40% of healthy women, it can cause life-threatening invasive infection in susceptible hosts: newborn infants, pregnant women, and nonpregnant adults with underlying chronic illnesses (1, 2). Since guidelines recommending intrapartum antibiotic prophylaxis for high-risk or colonized women were issued in 1996 (3), the incidence of neonatal infections has decreased, and invasive S. agalactiae infections in immunocompromised adults have become more common. Adult disease now accounts for the majority of serious S. agalactiae infections. First recognized as a pathogen in bovine mastitis, S. agalactiae is distinguished from other pathogenic streptococci by the cell wall-associated group B carbohydrate.
Another polysaccharide constitutes the organism's capsule, an important S. agalactiae virulence determinant. S. agalactiae strains of capsular serotype V were rarely isolated before the mid-1980s but now account for approximately one-third of clinical isolates in the U.S. (4–6). Type V is the most common capsular serotype associated with invasive infection in nonpregnant adults, and the emergence of type V strains over the past decade has been temporally linked to an increase in S. agalactiae disease in this population (7). As a species, S. agalactiae shares certain features with other pathogenic streptococci; however, the precise repertoire of shared and unique attributes that account for the emergence of S. agalactiae as a major pathogen for specific human populations remains undefined. To elucidate the molecular basis for S. agalactiae virulence, we determined the complete genome sequence (8) of the recent clinical type V isolate 2603 V/R (www.tigr.org) and performed comparative analyses with other S. agalactiae strains and with other species of pathogenic streptococci.
Methods
ORF Prediction and Gene Identification.
ORFs were predicted by glimmer (9, 10) trained with ORFs larger than 600 bp from the genomic sequence and S. agalactiae genes available in GenBank. All predicted proteins larger than 30 aa were searched against a nonredundant protein database (11). Frame-shifts and point mutations were detected and corrected where appropriate; those remaining were annotated as “authentic frame-shift” or “authentic point mutation.” Protein membrane-spanning domains were identified by toppred (12). Candidate lipoprotein signal peptides (13) were flagged by N-terminal exact matches to the pattern {DERK} (6)-[LIVMFWSTAG] (2)-[LIVMFYSTAGCQ]-[AGS]-C. Putative signal peptides were identified by using signalp (14). Two sets of hidden Markov models were used to determine ORF membership in families and superfamilies: pfam version 5.5 (15) and tigrfams 1.0 (16). Domain-based paralogous families were built by performing all-versus-all searches on the protein sequences by using a modified version of a previously described method (17). Potential lineage-specific gene duplications were estimated by identification of ORFs more similar to other ORFs within the S. agalactiae genome than to ORFs from other complete genomes. All ORFs were searched with FASTA3 (18) against all ORFs from the complete genomes and matches with a fasta P value of 10−15 were considered significant.
Western Blot.
S. agalactiae 2603 V/R strain cells were grown in Todd–Hewitt broth (Difco) to OD600 nm = 0.5. The culture was centrifuged for 20 min at 3,000 × g. The supernatant was discarded, and bacteria were washed once with PBS, resuspended in 2 ml of 50 mM Tris⋅HCl pH 6.8, containing 400 units of Mutanolysin (Sigma), and incubated 2 h at 37°C. After three cycles of freeze and thaw, cellular debris was removed by centrifugation at 13,000 × g for 10 min, and the protein concentration of the supernatant was measured by the Bio-Rad Protein assay, with BSA as a standard. Purified recombinant proteins (50 ng) (see purification method in ref. 19), and total cell extracts (25 μg) derived from S. agalactiae serotype V 2603 V/R strain were separated by SDS/PAGE and electroblotted onto nitrocellulose membranes for 1 hr at 100 V. The membranes were saturated by overnight incubation at 4°C in 5% skimmed milk and 0.1% Tween 20 in PBS and incubated for 1 hr at room temperature with sera from immunized mice diluted 1:500–1:1,000 in saturation buffer. To reduce background due to antibodies raised against contaminating Escherichia coli proteins, sera were preincubated with E. coli protein extracts absorbed on nitrocellulose strips. The membranes were washed twice in 3% skimmed milk and 0.1% Tween 20 in PBS and incubated for 1 hr with a 1:1,000 dilution of horseradish peroxidase-conjugated anti-mouse Ig (DAKO). After washing with 0.1% Tween 20 in PBS, the membranes were developed with the Opti-4CN Substrate kit (Bio-Rad).
Fluorescence-Activated Cell Sorter (FACS).
S. agalactiae 2603 V/R strain cells were grown in Todd–Hewitt broth (Difco) to OD600 nm = 0.5. The culture was centrifuged for 20 min at 3,000 × g, and bacteria were washed once with PBS, resuspended in PBS containing 0.05% paraformaldehyde, and incubated for 1 hr at 37°C and then overnight at 4°C. Fifty microliters of fixed bacteria (OD600 nm 0.1) was washed once with PBS, resuspended in 20 μl of newborn calf serum (Sigma), and incubated for 20 min at room temperature. The cells were then incubated for 1 hr at 4°C in 100 μl of preimmune or immune sera and diluted 1:200 in dilution buffer (PBS, 20% newborn calf serum, 0.1% BSA). After centrifugation and washing with 200 μl of washing buffer (0.1% BSA in PBS), samples were incubated for 1 hr at 4°C with 50 μl of R-phycoerythrin-conjugated F(ab)2 goat anti-mouse IgG (Jackson ImmunoResearch) diluted 1:100 in dilution buffer. Cells were washed with 200 μl of washing buffer and resuspended in 200 μl of PBS. Samples were analyzed by using a FACS calibur apparatus (Becton Dickinson), and data were analyzed by using cell quest (Becton Dickinson). A shift in mean fluorescence intensity of >75 channels compared with preimmune sera from the same mice was considered positive. This cutoff was determined from the mean plus two standard deviations of shifts obtained with control sera raised against mock purified recombinant proteins from cultures of E. coli carrying the empty expression vector and included in every experiment. Artifacts due to bacterial lysis were excluded by using antisera raised against six different known cytoplasmic proteins, all of which gave negative results.
Regions of Atypical Nucleotide Composition.
These regions were identified by the χ2 analysis: the distribution of all 64 trinucleotides (3 mers) was computed for the complete genome in all six reading frames, followed by the 3-mer distribution in 2,000 bp windows. Windows overlapped by 1,000 bp. For each window, the χ2 statistic on the difference between its 3-mer content, and that of the whole genome was computed.
In Silico Genome Comparisons.
The protein sets of S. agalactiae, Streptococcus pneumoniae, and Streptococcus pyogenes were compared by using fasta3 (18). Shared genes were defined by using a fasta3 P value cutoff of 10−15. These shared genes and genes that S. agalactiae did not share with the other streptococci using this cutoff were subsequently searched against all completely sequenced genomes, and genes were defined as unique to streptococci or S. agalactiae when they did not share similarity with any other gene sets with a fasta3 P value of 10−5 or lower. The use of two cutoffs provides for a more stringent analysis of shared or unique genes.
Synteny.
Regions of conservation of gene synteny were computed as windows of 10 kb spanning at least three genes whose order was conserved in the other species. Regions were merged if they were less than 20 kb apart. The number of genes within each broad region was then calculated.
Comparative Genome Hybridizations (CGH).
Predicted genes from strain 2603 V/R were amplified by PCR and arrayed on glass microscope slides (20). Genomic DNA was labeled according to protocols provided by J. DeRisi (www.microarrays.org/pdfs/GenomicDNALabel_B.pdf), except that the DNA was not digested or sheared before labeling. Arrays were scanned with a genepix 4000B scanner (Axon Instruments, Foster City, CA), and individual hybridization signals were quantitated with tigr spotfinder (21). Cy3/Cy5 (2603 V/R signal/test strain) ratio cutoffs were defined arbitrarily as Cy3/Cy5 = 1.0–3.0, gene present in test strain; 3.0–10.0, ambiguous result; >10.0, gene absent. For ambiguous results, the gene may be divergent in the test strain relative to 2603 V/R, or the gene may be absent in the test strain but still produces a hybridization signal because the 2603 V/R gene is part of a paralogous gene family or a repetitive element. Although cutoffs are arbitrary, they fit nicely the results for the variation of the capsule locus in the strains tested (see region 9 on Fig. 1) where most genes are slightly divergent and only a few are completely different.
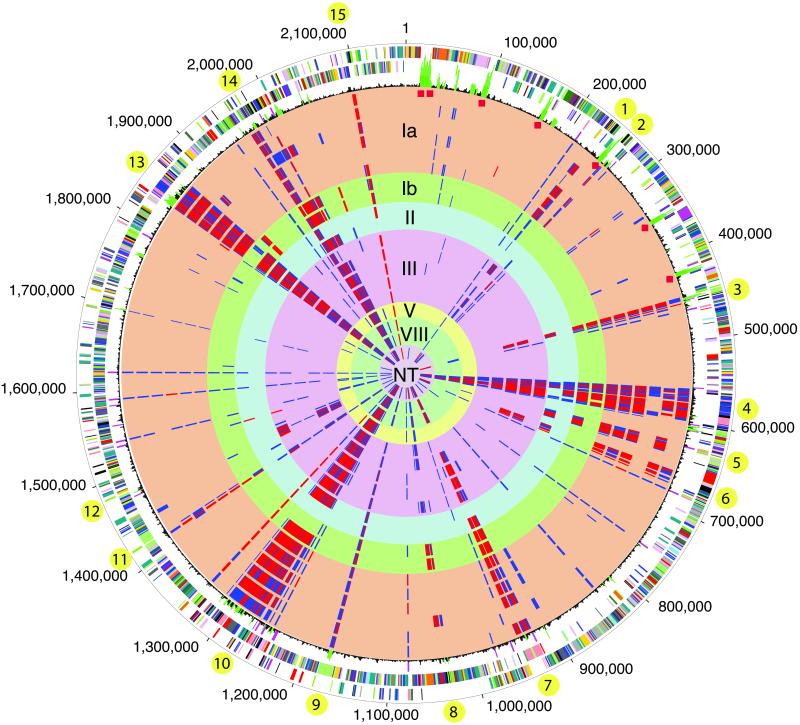
Fig 1.
Circular representation of the S. agalactiae genome and comparative genome hybridizations using microarrays. Outer circle: predicted coding regions on the plus strand color coded by role categories: violet, amino acid biosynthesis; light blue, biosynthesis of cofactors, prosthetic groups, and carriers; light green, cell envelope; red, cellular processes; brown, central intermediary metabolism; yellow, DNA metabolism; light gray, energy metabolism; magenta, fatty acid and phospholipid metabolism; pink, protein synthesis and fate; orange, purines, pyrimidines, nucleosides, and nucleotides; olive, regulatory functions and signal transduction; dark green, transcription; teal, transport and binding proteins; gray, unknown function; salmon, other categories; blue, hypothetical proteins. Second circle: predicted coding regions on the minus strand. Third circle: black, atypical nucleotide composition curve; green, most atypical regions; magenta, insertion elements; red diamonds indicate rRNAs. Circles 4–22: comparative genome hybridizations of strain 2603 V/R with 19 S. agalactiae strains. Cy3/Cy5 (2603 V/R signal/test strain) ratio cutoffs were defined arbitrarily as Cy3/Cy5 = 1.0–3.0, gene present in test strain (not displayed); 3.0–10.0, ambiguous result (blue); >10.0, gene absent in test strain (red). Circles 4–9: type Ia strains 090, 515, A909, Davis, DK1, DK8; 10–11: type Ib (S7) 7357b and H36B; 12–13: type II 18RS21 and DK21; 14–18: type III COH1, COH31, D136C, M732 and M781; 19: type V strain CJB111; 20–21: type VIII strains SMU014 and JM9130013; 22: nontypable (NT) strain CJB110. Varying regions of five or more consecutive genes are indicated by yellow bullets.
Profile Clustering.
The information on presence and absence of genes based on the CGH results was used to group genes based on their distribution patterns. The analysis used was essentially identical to that used for phylogenetic profile analysis (22). Each gene was assigned a binary profile based on its presence/absence across the different strains, with presence determined by a Cy3/Cy5 ratio <3.0 and absence ≥3.0. The gene profiles were then clustered by using the single-linkage clustering algorithm with column weighting (all with default settings) of cluster (http://rana.lbl.gov). The cluster program also groups the strains (columns) based on similarity of gene profiles. Clusters of genes and strains were viewed by using treeview (http://rana.lbl.gov).
Phylogeny.
The sequences of genes from the different strains were aligned by using clustalw (23) and trimmed to remove ambiguously aligned regions. Phylogenetic trees of individual genes and of concatenated alignments of multiple genes were inferred by using maximum likelihood methods of paup* 4.0 b10 (Sinauer, Sunderland, MA). Bootstrap analysis was carried out by using paup* as well. The possibility of recombination among strains was examined by using analysis of sequence variation using simplot (http://sray.med.som.jhmi.edu/RaySoft/SimPlot/) and analysis of phylogenetic heterogeneity by using macclade (Sinauer).
Results
General Genome Features.
The genome consists of a circular chromosome of 2,160,267 bp with a G+C content of 35.7%. Base pair one of the chromosome was assigned within the putative origin of replication. The genome contains 80 tRNAs, 7 rRNAs, and 3 sRNAs. Approximately 78% of the 2,175 predicted genes (Table 1, which is published as supporting information on the PNAS web site, www.pnas.org) are transcribed in the same direction as that of DNA replication, a feature also observed in S. pneumoniae and other low-GC Gram-positive organisms (8) (Fig. 1 and Fig. 4, which is published as supporting information on the PNAS web site). Biological roles were assigned to 1,333 (61%) of the predicted proteins according to the classification scheme adapted from Riley (24). Another 623 predicted proteins (29%) matched proteins of unknown function, and the remaining 219 (10%) had no database match. The expression of 50 of these hypothetical proteins was confirmed by Western blot analysis, and the proteins were annotated as “proteins of unknown function.” A total of 339 paralogous protein families were identified in strain 2603, containing 941 predicted proteins (43% of the total).
Polysaccharides.
S. agalactiae produces two surface polysaccharides: the cell wall-associated group B carbohydrate common to all S. agalactiae strains and the type-specific capsular polysaccharide. A cluster of 13 adjacent genes (SAG1410-SAG1424) was tentatively identified as encoding enzymes required for synthesis of the group B carbohydrate, a complex multiantennary structure of rhamnose, glucitol phosphate, N-acetylglucosamine, and galactose. Predicted proteins encoded within this cluster include seven putative glycosyltransferases, four of which are similar to rhamnosyltransferases in other streptococcal species; a putative dTDP-l-rhamnose synthase; and proteins involved in glucitol synthesis. All nine recognized S. agalactiae capsular polysaccharide types contain sialic acid residues as part of their repeating unit structure, a feature that contributes to virulence by inhibiting activation of the alternative complement pathway (25). The type V capsular polysaccharide gene cluster consists of 18 genes. A region of glycosyltransferases and related proteins (SAG1162-SAG1170) that direct the synthesis of the type V polysaccharide repeat unit is flanked on either side by genes that are conserved in all known S. agalactiae capsule serotypes. Downstream of this region are genes that encode enzymes for the biosynthesis and activation of sialic acid (SAG1158-SAG1161). Upstream of the serotype-specific region are genes (SAG1171-SAG1175) found not only in all nine S. agalactiae capsular serotypes but also in a variety of other polysaccharide-producing streptococci. The sequence of the capsule gene cluster in strain 2603 V/R is consistent with that in type V strain CNCTC 1/82 previously deposited in GenBank (AF349539).
Surface Proteins.
Several genes identified as encoding S. agalactiae surface proteins and secreted products are potential virulence factors or targets of protective immunity: Sip (SAG0032) (26), CAMP factor (SAG2043), R5 protein (SAG1331), streptococcal enolase (SAG0628), hyaluronidase (SAG1197) (27) and hemolysin/cytolysin (cylE, SAG0669) (28). Two genes, lmb encoding laminin-binding protein (SAG1234) and scpB encoding C5a peptidase (SAG1236), are adjacent to a group II intron as previously described (29, 30), but the peptidase is frame-shifted near its C terminus in strain 2603 V/R. A putative protease similar to the C5a peptidase (SAG0416) is located elsewhere in this genome. Surface protein Rib (SAG0433), a member of the tandem-repeat-containing family of S. agalactiae surface proteins (31), was identified in this genome. Each of the 14 tandem repeats in the gene here encodes 67 amino acids, 12 fewer than those in the published sequence. The Rib protein has previously been detected predominantly in S. agalactiae strains of serotypes II and III, whereas type V S. agalactiae strains generally express a related member of the protein family, Alp3 (32).
Prediction algorithms (see Methods and ref. 19) estimate that approximately 650 proteins are membrane-associated or secreted, some of which are potential adhesion factors or virulence-associated determinants. These include 24 proteins predicted to be anchored on the cell wall through their LPxTG motif, 51 lipoproteins, and 177 proteins carrying a signal peptide (Table 2, which is published as supporting information on the PNAS web site). Two novel enzymes are related to metabolism of host sugars or hexosamines, a putative pullulanase (SAG1216) and a neuraminidase-related protein (SAG1932).
The expression of the 650 predicted surface-exposed proteins was tested experimentally. The 291 recombinant proteins successfully expressed were used to immunize mice and the corresponding antibodies were used in Western blot analysis against the total protein extract of the homologous type V strain 2603 V/R. One hundred thirty-nine sera revealed a predominant protein of the expected molecular weight, and all but seven of the proteins were present in the heterologous type III strain COH1 with no detectable difference in electrophoretic mobility. The 139 sera were also tested by fluorescence-activated cell sorter analysis with whole S. agalactiae cells and revealed 55 proteins clearly exposed on the cell surface (Table 2). The failure to detect surface expression with the remaining 84 sera may reflect masking of the proteins by the cell wall and capsule or overestimation of the surface prediction programs. One of the surface-exposed proteins is adenylate kinase (SAG0079), a ubiquitous enzyme generally intracellular, secreted by both Vibrio cholerae (33) and pathogenic strains of Pseudomonas aeruginosa, where it acts as a virulence factor through its role in macrophage cell death (34).
SAG2063 is a novel protein of 630 amino acids predicted to be cell-wall anchored. It contains 43 imperfect repeats of the amino acid motif PE(D)A(V)K at its C terminus, a structure similar to that of known virulence factors of Gram-positive bacteria. SAG1206 is predicted to be secreted, shares similarity with a metalloprotease from Clostridium acetobutylicum, and contains several repeats of a GW amino acid motif necessary for binding to lipoteichoic acids in Listeria (35). Finally, two genes (SAG0832 and SAG0833) in a region of very low G+C content (30%) are predicted to encode secreted proteins.
Comparative Genomics.
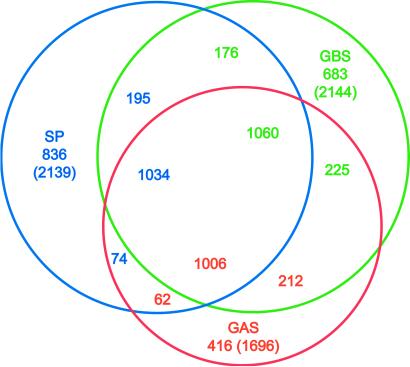
The complete genome sequence is available for two other important human pathogens of the genus Streptococcus, S. pneumoniae (8) and S. pyogenes (36). Although these species colonize different regions of the body, they are all capable of causing severe invasive disease. Hence, it is likely that these species will share some virulence factors, whereas other factors will determine specific colonization, invasion, and disease characteristics. One thousand sixty genes (50%) have homologs in all three genomes (Fig. 2). S. agalactiae shares 176 genes with S. pneumoniae and 225 genes with S. pyogenes, whereas S. pneumoniae and S. pyogenes share only 74 genes. Finally, 683 genes are unique to S. agalactiae. Conservation of gene synteny is more pronounced between S. agalactiae and S. pyogenes (828 genes in 35 regions spanning ≈1,104 kb) than between S. agalactiae and S. pneumoniae (128 genes in 9 regions spanning ≈131 kb).
Fig 2.
In silico comparisons between streptococci. The protein sets of S. agalactiae [group B Streptococcus (GBS)], S. pneumoniae (SP), and S. pyogenes [group A Streptococcus (GAS)] were compared by using FASTA3. Numbers under the species name indicate genes that are not shared with the other species; values in parentheses are the number of proteins in each species (excluding frame-shifted and degenerated genes). Numbers in the intersections indicate genes shared by two or three species. These are displayed in the color corresponding to the species used as the query (GBS, green; SP, blue; GAS, red). Numbers in any given intersection are slightly different due to gene duplications in some species.
To place the variation in a wider context, the 1,060 genes shared by the three streptococcal species were compared with the gene complements of all of the completely sequenced genomes. Using a more stringent cutoff, 12 Streptococcus-specific genes were identified (Table 3, which is published as supporting information on the PNAS web site). Using the same approach, 315 of the 683 genes that S. agalactiae did not share with the other two streptococci were not similar to genes from other completely sequenced genomes. These include six proteins predicted to be anchored on the cell wall (SAG0677, SAG0771, SAG1052, SAG1331, SAG1473, and SAG1996), three of the capsule-related genes (SAG1163, SAG1167, and SAG1168), six transcriptional regulators, and four genes of the cyl operon (SAG0663-SAG0673) essential for S. agalactiae hemolytic activity and production of pigment (28). The rest of the 315 proteins include 219 hypothetical proteins with no similarity to other proteins in databases.
Metabolic differences between the streptococci inferred from genome analysis may reflect the availability of compounds in their respective microenvironments in the human host. S. agalactiae has genes required for the synthesis of arginine, aspartate, and citrulline, whereas it apparently lacks the systems the other two streptococci have for the metabolism of fucose, lactose, mannitol, raffinose, lysine, and threonine. S. pneumoniae was notable for its large array of sugar phosphoenolpyruvate-dependent phosphotransferase system (PTS) transporters, which may be important for utilization of host-derived carbohydrates and for recycling of capsule constituents (8). Although S. agalactiae has uptake and metabolism systems for ribose and gluconate and a complete operon for the metabolism of glucuronic acid, it lacks the uptake systems for lactose, cellobiose, mannitol, and the putative fucose-related system present in S. pneumoniae, and it also lacks a raffinose ABC transporter. Instead, S. agalactiae has a galactitol PTS, a ribose ABC transporter, a gluconate transporter, and a sodium ion-driven glucuronide uptake system, correlating with the novel pathways present in S. agalactiae. S. agalactiae also possesses two additional peptide ABC uptake systems not present in S. pneumoniae, which together with the presence of many secreted peptidases may imply that host proteins/peptides are an important nutrient source for this organism. S. agalactiae lacks two of the ABC ferric chelate transporters present in S. pneumoniae, but instead encodes two Nramp transporters for manganese or iron uptake, suggesting distinct mechanisms for acquisition of these ions that may be important in virulence. These metabolic and transport systems specific to S. agalactiae probably relate to adaptation to distinct niches in its human and animal hosts.
Seventeen two-component signaling systems are identified in the S. agalactiae genome, one of which (SAG2127-SAG2128) is absent from the other two streptococci. Another one (SAG1921-SAG1922) is shared with S. pyogenes only.
Many of the 315 genes specific to S. agalactiae are located in regions likely to constitute mobile genetic elements. Two of these regions resemble prophages (SAG0545-SAG0610 and SAG1835-SAG1885) displaying a mosaic structure with segments most similar to different bacteriophages, a pattern that suggests frequent recombination events. PblA and PblB are adhesins from a Streptococcus mitis prophage where they contribute to endocarditis by binding to human platelets (37, 38). Their orthologs in S. agalactiae are located on separate prophages and display a different protein structure. Another region (SAG1247-SAG1299) encodes a putative conjugative transposon that carries genes for cadmium efflux and mercury resistance.
Finally, 130 genes in S. agalactiae (Table 4, which is published as supporting information on the PNAS web site) were probably duplicated after the divergence of this species from other lineages for which complete genomes are available. Such lineage-specific gene duplications may reveal species-specific adaptations because gene duplication is frequently accompanied by functional diversification and divergence. These duplications include glycosyl transferases, sortases, proteins anchored on the cell wall, β lactam resistance factors, and many hypothetical proteins.
CGH and Phylogeny.
Little is known of the genome differences between the serotypes or of the genome variation within serotypes. To shed light on this variation, CGH (Fig. 1) using DNA microarrays were performed between the sequenced type V strain 2603 V/R and 19 other S. agalactiae strains of multiple serotypes (Table 5, which is published as supporting information on the PNAS web site). The CGH detected 1,698 genes in all of the strains, whereas 401 genes from strain 2603 V/R (18% of the gene complement) were not detected in at least one other strain, suggesting that they are absent or significantly divergent in those strains. Two hundred sixty (38%) of the 683 genes specific to S. agalactiae when compared with the other two streptococci (Fig. 2), including virulence determinants and surface proteins, vary among S. agalactiae strains, whereas only 47 (4%) of the genes common to all three streptococcal species, including 5 of the 6 sortases identified in the genome, vary among strains. Thus, the in silico analysis of genes shared by the streptococci that are not expected to vary among this genus is consistent with the CGH analysis. Forty-four (25%) of the genes shared by S. agalactiae and S. pneumoniae and 44 (20%) of those shared by S. agalactiae and S. pyogenes vary in the CGH analysis. The first set contains many glycosyl transferases and proteins carrying a cell-wall anchor, whereas the second set displays many phage-related genes. One hundred thirty-six of the 315 genes unique to S. agalactiae when compared with all sequenced genomes vary among strains. These include R5, three capsular genes, two cell wall-anchored proteins, and three transcriptional regulators.
Three hundred sixty-four (91%) of the 401 varying genes correspond to 15 regions containing 5 or more contiguous genes. Ten of these regions display an atypical nucleotide composition in strain 2603 V/R (Fig. 1), consistent with the possibility that they were laterally transferred into this strain. Two of the largest regions (region 4, a prophage and region 7, similar to Tn916 from Enterococcus faecalis) are flanked by insertion sequence elements. The 15 regions contain many proteins predicted to be anchored on the cell wall or surface exposed, including Rib (region 3), sortases, glycosyl transferases, the capsule locus (region 9, divergent in all strains but the other type V strain CJB111), and phage-related genes. Region 14 is unique to S. agalactiae and spans 33 genes (SAG1989-SAG2021), including 25 proteins of unknown function, some of which carry a cell-wall anchor. It is flanked by an ISL3 transposase and displays an atypical nucleotide composition. Region 1, unique to S. agalactiae, is a possible plasmid or remnant of a phage (SAG0218-SAG0238), contains mostly hypothetical proteins, and is flanked by a site-specific recombinase. Region 8 is specific to S. agalactiae, comprises 20 proteins of unknown function (SAG1018-SAG1037), most of which are predicted to be membrane associated or secreted, and displays an atypical nucleotide composition.
The CGH results were analyzed by profile clustering where genes are grouped based on their distribution patterns (Fig. 5, which is published as supporting information on the PNAS web site). Sixteen clusters of five or more contiguous and noncontiguous genes comprising a total of 300 genes were identified (Table 6, which is published as supporting information on the PNAS web site). Several clusters correspond to regions of contiguous genes described above. Some clusters of genes that do not share sequence similarity and are located at different loci in the genome display an identical profile. For instance, a cluster of genes containing a surface antigen (SAG0674-SAG0681) follows the same distribution as another cluster containing only hypothetical proteins (SAG0247-SAG0249). A putative pathogenicity protein (SAG2063) also clusters with a region containing several glycosyl transferases and Sec proteins (SAG1447-SAG1462).
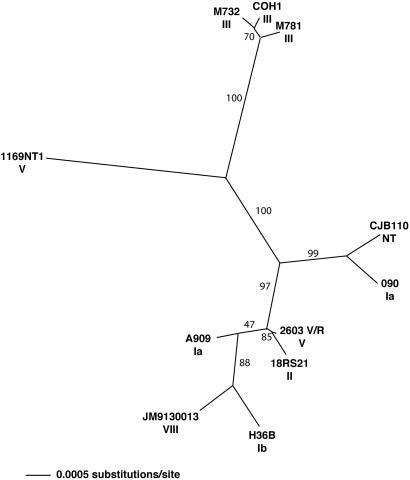
Profile clustering was also used to group strains based on similarity of gene content (Fig. 5). This method is not ideal for inferring relationships among strains, because the genes specific to strains other than 2603 V/R were not represented on the microarray and were therefore not interrogated. To generate a more accurate picture of the strain relationships, the sequences of 19 genes from each of 11 S. agalactiae strains were determined after PCR amplification and used for phylogenetic analyses. The set comprised 8 housekeeping genes and 11 genes coding for proteins predicted to be surface-exposed (Table 7, which is published as supporting information on the PNAS web site). Phylogenetic trees were inferred for the complete set of 19 genes and for the subsets of housekeeping and surface-exposed genes. Because the branching patterns in all three trees were identical, only the tree of the 19 genes is shown in Fig. 3. The degree of polymorphism of the housekeeping and the surface-exposed genes is similar (≈1 variable site among all of the strains per 100 bp). Analysis of this variation showed no evidence for major recombination events between the strains. There were no long stretches of polymorphic sites that strongly supported other trees (analysis with macclade), and there were no significant crossover events in plots of sequence similarity between strains (analysis with simplot).
Fig 3.
Phylogenetic tree of S. agalactiae strains based on PCR sequences. The sequences of 19 genes (Table 7) from each of 11 S. agalactiae strains were aligned and trimmed to remove ambiguously aligned regions, and phylogenetic trees were inferred. Strain names are indicated in bold, and serotypes are indicated under the strain names. Bootstrap values are indicated on the branches.
Some strain groupings (clades) generated by phylogenetic analysis were similar to clusters from the profile analysis (type III strains M781, M732 and COH1; type Ia strain 090 and nontypable strain CJB110), whereas others were different, possibly because of the aforementioned problems with the profile clustering. In both the phylogenetic analysis and the profile clustering, there is serotype-dependent and -independent clustering (Figs. 3 and 5). The presence of strains of the same serotype in different clades or clusters could be due to lateral gene transfer. Previous studies showed that a single gene in the capsule gene locus converts serotype Ia to III (39). Serotype switching during natural infection was shown in S. pneumoniae (40–42) and in Neisseria meningitidis (43–45), which may result from selective pressure of host immunity and could lead to convergent or parallel evolution. Serotype-independent clustering supports the possibility of capsule switching, although such events have not yet been demonstrated to occur spontaneously in S. agalactiae.
Conclusion
The S. agalactiae genome reveals substantial similarity with those of the related human pathogens S. pyogenes and S. pneumoniae. S. agalactiae differs from the other streptococci in several metabolic pathways and related membrane transport systems that probably relate to adaptation to distinct niches in its human and animal hosts. Comparison with all characterized bacterial genomes identified genes unique to S. agalactiae that are expected to play a role in colonization or disease: surface proteins, capsule synthesis genes, hemolysin, and several transcriptional regulators. Many of these are associated with mobile elements, including bacteriophages, transposons, and insertion sequences, an observation that supports acquisition of virulence traits from other species. The presence of more than 100 genes probably duplicated recently suggests evolution of additional species-specific functions. Genetic heterogeneity among S. agalactiae strains, even of the same serotype, provides evidence that these mechanisms of acquisition, duplication, and reassortment have produced the genetic diversity within the species that has permitted S. agalactiae to adapt to new environmental niches and to emerge as a major human pathogen.
Supplementary Material
Acknowledgments
We thank O. White, S. Angiuoli, L. Umayam, J. Peterson, M. Wu, M. Heaney, S. Lo, M. Holmes, B. Lee, R. Karamchedu, and V. Sapiro for informatics support at The Institute for Genomic Research (TIGR) and the TIGR faculty and sequencing core for expert advice and assistance. We also thank G. Orefici and M. Oggioni for isolation and characterization of the sequenced strain. This work was supported by Chiron Corporation and National Institutes of Health Grants AI23339, AI38424, and AI42940, and Contract AI75326.
Abbreviations
CGH, comparative genome hybridizations
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AE009948).
References
- 1.Zangwill K. M., Schuchat, A. & Wenger, J. D. (1992) Mor. Mortal Wkly. Rep. CDC Surveill. Summ. 41, 25-32. [PubMed] [Google Scholar]
- 2.Schuchat A. & Wenger, J. D. (1994) Epidemiol. Rev. 16, 374-402. [DOI] [PubMed] [Google Scholar]
- 3.Anonymous (1996) MMWR Recomm. Rep. 45, 1-24. [Google Scholar]
- 4.Harrison L. H., Elliott, J. A., Dwyer, D. M., Libonati, J. P., Ferrieri, P., Billmann, L. & Schuchat, A. (1998) J. Infect. Dis. 177, 998-1002. [DOI] [PubMed] [Google Scholar]
- 5.Lin F. Y., Clemens, J. D., Azimi, P. H., Regan, J. A., Weisman, L. E., Philips, J. B., III, Rhoads, G. G., Clark, P., Brenner, R. A. & Ferrieri, P. (1998) J. Infect. Dis. 177, 790-792. [DOI] [PubMed] [Google Scholar]
- 6.Zaleznik D. F., Rench, M. A., Hillier, S., Krohn, M. A., Platt, R., Lee, M. L., Flores, A. E., Ferrieri, P. & Baker, C. J. (2000) Clin. Infect. Dis. 30, 276-281. [DOI] [PubMed] [Google Scholar]
- 7.Tyrrell G. J., Senzilet, L. D., Spika, J. S., Kertesz, D. A., Alagaratnam, M., Lovgren, M. & Talbot, J. A. (2000) J. Infect. Dis. 182, 168-173. [DOI] [PubMed] [Google Scholar]
- 8.Tettelin H., Nelson, K. E., Paulsen, I. T., Eisen, J. A., Read, T. D., Peterson, S., Heidelberg, J., DeBoy, R. T., Haft, D. H., Dodson, R. J., et al. (2001) Science 293, 498-506. [DOI] [PubMed] [Google Scholar]
- 9.Delcher A. L., Harmon, D., Kasif, S., White, O. & Salzberg, S. L. (1999) Nucleic Acids Res. 27, 4636-4641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Salzberg S. L., Delcher, A. L., Kasif, S. & White, O. (1998) Nucleic Acids Res. 26, 544-548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fleischmann R. D., Adams, M. D., White, O., Clayton, R. A., Kirkness, E. F., Kerlavage, A. R., Bult, C. J., Tomb, J. F., Dougherty, B. A., Merrick, J. M., et al. (1995) Science 269, 496-512. [DOI] [PubMed] [Google Scholar]
- 12.Claros M. G. & von Heijne, G. (1994) Comput. Appl. Biosci. 10, 685-686. [DOI] [PubMed] [Google Scholar]
- 13.Hayashi S. & Wu, H. C. (1990) J. Bioenerg. Biomembr. 22, 451-471. [DOI] [PubMed] [Google Scholar]
- 14.Nielsen H., Engelbrecht, J., Brunak, S. & von Heijne, G. (1997) Protein Eng. 10, 1-6. [DOI] [PubMed] [Google Scholar]
- 15.Bateman A., Birney, E., Durbin, R., Eddy, S. R., Howe, K. L. & Sonnhammer, E. L. (2000) Nucleic Acids Res. 28, 263-266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Haft D. H., Loftus, B. J., Richardson, D. L., Yang, F., Eisen, J. A., Paulsen, I. T. & White, O. (2001) Nucleic Acids Res. 29, 41-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nierman W. C., Feldblyum, T. V., Laub, M. T., Paulsen, I. T., Nelson, K. E., Eisen, J., Heidelberg, J. F., Alley, M. R., Ohta, N., Maddock, J. R., et al. (2001) Proc. Natl. Acad. Sci. USA 98, 4136-4141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pearson W. R. (2000) Methods Mol. Biol. 132, 185-219. [DOI] [PubMed] [Google Scholar]
- 19.Pizza M., Scarlato, V., Masignani, V., Giuliani, M. M., Arico, B., Comanducci, M., Jennings, G. T., Baldi, L., Bartolini, E., Capecchi, B., et al. (2000) Science 287, 1816-1820. [DOI] [PubMed] [Google Scholar]
- 20.Peterson S., Cline, R. T., Tettelin, H., Sharov, V. & Morrison, D. A. (2000) J. Bacteriol. 182, 6192-6202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hegde P., Qi, R., Abernathy, K., Gay, C., Dharap, S., Gaspard, R., Hughes, J. E., Snesrud, E., Lee, N. & Quackenbush, J. (2000) Biotechniques 29, 548-550., 552–554, 556 passim. [DOI] [PubMed] [Google Scholar]
- 22.Pellegrini M., Marcotte, E. M., Thompson, M. J., Eisenberg, D. & Yeates, T. O. (1999) Proc. Natl. Acad. Sci. USA 96, 4285-4288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Thompson J. D., Higgins, D. G. & Gibson, T. J. (1994) Nucleic Acids Res. 22, 4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Riley M. (1993) Microbiol. Rev. 57, 862-952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Edwards M. S., Kasper, D. L., Jennings, H. J., Baker, C. J. & Nicholson-Weller, A. (1982) J. Immunol. 128, 1278-1283. [PubMed] [Google Scholar]
- 26.Brodeur B. R., Boyer, M., Charlebois, I., Hamel, J., Couture, F., Rioux, C. R. & Martin, D. (2000) Infect. Immun. 68, 5610-5618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pritchard D. G. & Lin, B. (1993) Infect. Immun. 61, 3234-3239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pritzlaff C. A., Chang, J. C., Kuo, S. P., Tamura, G. S., Rubens, C. E. & Nizet, V. (2001) Mol. Microbiol. 39, 236-247. [DOI] [PubMed] [Google Scholar]
- 29.Franken C., Haase, G., Brandt, C., Weber-Heynemann, J., Martin, S., Lammler, C., Podbielski, A., Lutticken, R. & Spellerberg, B. (2001) Mol. Microbiol. 41, 925-935. [DOI] [PubMed] [Google Scholar]
- 30.Granlund M., Michel, F. & Norgren, M. (2001) J. Bacteriol. 183, 2560-2569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wastfelt M., Stalhammar-Carlemalm, M., Delisse, A. M., Cabezon, T. & Lindahl, G. (1996) J. Biol. Chem. 271, 18892-18897. [DOI] [PubMed] [Google Scholar]
- 32.Lachenauer C. S., Creti, R., Michel, J. L. & Madoff, L. C. (2000) Proc. Natl. Acad. Sci. USA 97, 9630-9635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Punj V., Zaborina, O., Dhiman, N., Falzari, K., Bagdasarian, M. & Chakrabarty, A. M. (2000) Infect. Immun. 68, 4930-4937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Markaryan A., Zaborina, O., Punj, V. & Chakrabarty, A. M. (2001) J. Bacteriol. 183, 3345-3352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jonquieres R., Bierne, H., Fiedler, F., Gounon, P. & Cossart, P. (1999) Mol. Microbiol. 34, 902-914. [DOI] [PubMed] [Google Scholar]
- 36.Ferretti J. J., McShan, W. M., Ajdic, D., Savic, D. J., Savic, G., Lyon, K., Primeaux, C., Sezate, S., Suvorov, A. N., Kenton, S., et al. (2001) Proc. Natl. Acad. Sci. USA 98, 4658-4663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bensing B. A., Siboo, I. R. & Sullam, P. M. (2001) Infect. Immun. 69, 6186-6192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bensing B. A., Rubens, C. E. & Sullam, P. M. (2001) Infect. Immun. 69, 1373-1380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chaffin D. O., Beres, S. B., Yim, H. Y. & Rubens, C. E. (2000) J. Bacteriol. 182, 4466-4477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Coffey T. J., Dowson, C. G., Daniels, M., Zhou, J., Martin, C., Spratt, B. G. & Musser, J. M. (1991) Mol. Microbiol. 5, 2255-2260. [DOI] [PubMed] [Google Scholar]
- 41.Barnes D. M., Whittier, S., Gilligan, P. H., Soares, S., Tomasz, A. & Henderson, F. W. (1995) J. Infect. Dis. 171, 890-896. [DOI] [PubMed] [Google Scholar]
- 42.Coffey T. J., Enright, M. C., Daniels, M., Morona, J. K., Morona, R., Hryniewicz, W., Paton, J. C. & Spratt, B. G. (1998) Mol. Microbiol. 27, 73-83. [DOI] [PubMed] [Google Scholar]
- 43.Frosch M. & Meyer, T. F. (1992) FEMS Microbiol. Lett. 79, 345-349. [DOI] [PubMed] [Google Scholar]
- 44.Swartley J. S., Marfin, A. A., Edupuganti, S., Liu, L. J., Cieslak, P., Perkins, B., Wenger, J. D. & Stephens, D. S. (1997) Proc. Natl. Acad. Sci. USA 94, 271-276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vogel U., Claus, H. & Frosch, M. (2000) N. Engl. J. Med. 342, 219-220. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.