Abstract
The view that parasites can develop cooperative symbiotic relationships with their hosts is both appealing and widely held; however, there is no molecular genetic evidence of such a transition. Here we demonstrate that a mutualistic bacterial endosymbiont of grain weevils maintains and expresses inv/spa genes encoding a type III secretion system homologous to that used for invasion by bacterial pathogens. Phylogenetic analyses indicate that inv/spa genes were present in presymbiotic ancestors of the weevil endosymbionts, occurring at least 50 million years ago. The function of inv/spa genes in maintaining symbiosis is demonstrated by the up-regulation of their expression under both in vivo and in vitro conditions that coincide with host cell invasion.
Many bacterial lineages have evolved close associations with eukaryotic hosts, with effects ranging from invasive parasitism to obligate mutualism. In many of these associations, bacteria must enter and replicate within host cells. The principal mechanism by which Gram-negative bacterial pathogens invade eukaryotic cells involves the deployment of a type III secretion system (TTSS) to deliver effector proteins that assist in host cell entry (1). Gene clusters encoding TTSSs, often situated within pathogenicity islands, have been acquired by a diverse array of horizontally transmitted plant and animal pathogens and symbionts, including species of Shigella, Salmonella, Yersinia, Pseudomonas, Xanthomonas, Erwinia, and Rhizobium (2). Recently, the insect endosymbiont, Sodalis glossinidius, was also shown to depend on a TTSS for invasion of host cells (3). These symbionts contain homologs of inv/spa genes encoding the Salmonella/Shigella TTSS, and mutants lacking a key component of type III secretion (invC) are known to be neither invasive in vitro nor symbiotic in their natural tsetse fly host.
As a vertically transmitted symbiont, S. glossinidius is not expected to greatly reduce the fitness of its host. Nonetheless, such “secondary” endosymbionts of insects resemble pathogens in that they confer no known benefits on hosts, are not essential for host growth or reproduction, invade a diverse range of host tissues, and undergo occasional horizontal transmission (4, 5). On the basis of 16S ribosomal RNA phylogenies, the closest known relatives of S. glossinidius are the primary endosymbionts of grain weevils (genus Sitophilus: Insecta: Coleoptera: Curculionidae; ref. 6). “Primary” endosymbionts differ from “secondary” symbionts in several respects: they are essential for host survival, reside only within specialized host organs (bacteriomes), and are strictly vertically transmitted. Primary symbionts show stereotyped patterns of replication and migration within specific host tissues; this movement results in their incorporation into bacteriocytes and developing eggs. On the basis of microscopic observations, these patterns have been interpreted as reflecting host-based mechanisms governing movement of the bacterial cells (5).
In the case of the primary symbionts of Sitophilus spp., eggs are infected before they are laid. The inoculating bacteria multiply in the developing embryo and are ultimately localized within a bacteriome at the foregut periphery that persists throughout the larval stages (5, 7). At metamorphosis, the larval bacteriome disappears, and the symbionts must migrate and infect a new bacteriome associated with the hindgut outer lining in the adult weevil (5). Experimental reduction and removal of bacteria have confirmed that they are required for normal growth, development, and reproduction in Sitophilus (8). The endosymbionts of Sitophilus have not yet been cultivated in vitro and currently have no official nomenclature. In the present study, we have focused on the Sitophilus zeamais primary endosymbiont (SZPE).
It has been proposed that parasites can evolve into mutualists after the establishment of a vertical mode of transmission, concomitant with the attenuation of parasite virulence (9–12). To date, there is little empirical evidence supporting this evolutionary transition. In particular, if obligate “primary” symbionts evolve from pathogens, we might expect continuity in the molecular basis of the infection mechanism, despite changes in how the association affects the host, the loss of horizontal transmission, and the inability to infect many host cell types. Until now, there have been no reported examples of vertically transmitted mutualistic symbionts that use pathogenicity determinants to facilitate their associations with hosts. We searched for TTSS-encoding genes that might enable SZPE to invade the cells of the host bacteriome (bacteriocytes), as required during the insect life cycle.
SZPE and S. glossinidius provide an excellent model for studying the evolutionary transition to mutualism, because the divergence of these two lineages from a shared ancestor has produced different symbiotic outcomes. In this study, we show that SZPE, a weevil endosymbiont, harbors TTSS-encoding genes closely related to those found in S. glossinidius, and that expression of these genes coincides with the timing of bacteriome infection within the developing weevil. We then discuss the implications of these findings for the evolution of mutualistic associations involving bacteria and animals.
Materials and Methods
PCR Amplification and Sequence Determination.
S. glossinidius was grown in liquid culture, as described previously (4). Sitophilus zeamais weevils, the host of SZPE, were maintained in maize at 27.5°C and 70% humidity. Bacteriomes (containing SZPE) were dissected from fifth-instar S. zeamais larvae, and DNA was prepared from cultured S. glossinidius and S. zeamais bacteriomes with the DNeasy tissue kit (Qiagen, Valencia, CA). To determine whether SZPE harbors inv/spa homologs, we designed degenerate oligonucleotide primers based on alignments of the inv/spa genes from S. glossinidius, Salmonella enterica, and Shigella flexneri. PCR primers used for the amplification of invA and rplB were synthesized with inosine (I) as follows (listed 5′ to 3′): invA (1,020-bp product) ATG CCI GGI AAR CAR ATG and TIG TYT CYT GDT TIC CRA A; rplB (665 bp) AAC CCT GAR YTI CAY AAR GGI AA and TTA CCY TTI GTY TGI ACI CCC CA. An invA-spaM fragment, a spaP-spaR fragment, and a fusA fragment were amplified with the following primers (listed 5′ to 3′): invA-spaM (3,323 bp) GTC AAC TGT ACG GTG CGC TTG and CCG CGT AAA ACC CTG CTG TTT CC; spaPQR (614 bp) ATG ATG ATG ATG AGC CCG and AGC CCA TGC ATA ACC CAA AA; fusA (760 bp) CAT CGG CAT CAT GGC NCA YAT HGA and CAG CAT CGG CTG CAY NCC YTT RTT. PCR reactions contained 50–100 ng of template DNA, 25 pmol of each primer, and 2.5 units of TaqDNA polymerase (Promega) in a final reaction volume of 50 μl with 3.5 mM MgCl2. Reactions proceeded with an initial denaturation step (5 min at 95°C) followed by 40 cycles of denaturation (1 min at 95°C), annealing (1 min at 50°C), and extension (1 min/kb at 72°C), followed by a final extension (5 min at 72°C) to promote A-tailing. PCR products were gel-purified, extracted with a Qiaquick PCR purification kit (Qiagen), and cloned into pGEM T-easy vector (Promega) or pTOPO-XL (Invitrogen), according to manufacturers' instructions. For each PCR product, at least six clones were sequenced in both directions to establish error-free consensus sequences.
Phylogenetic Analysis.
DNA sequences of invA were translated and the inferred amino acid sequences aligned by using pileup (Genetics Computer Group package, Madison, WI). Phylogenetic trees were obtained by both maximum parsimony and distance methods, as implemented in paup*4.0 (13). Similarly, 16S rDNA sequences were aligned with pileup, and tree topologies were ascertained with paup. The uncorrected pairwises distances in Table 1 were computed by using the gcg package (Genetics Computer Group).
Table 1.
Pairwise distances between InvA amino acid sequences
| Sz | Se | Sf | Yp | Ea | Ps | Xc | Rs | Bb | Cp | Rh | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Sg | 7.1 | 45.8 | 48.6 | 43.8 | 63.1 | 63.5 | 62.5 | 64.0 | 59.5 | 62.3 | 63.1 |
| Sz | 53.4 | 53.9 | 51.8 | 68.8 | 69.4 | 68.0 | 71.3 | 66.2 | 67.5 | 66.9 | |
| Se | 34.9 | 47.0 | 61.1 | 60.6 | 61.5 | 64.9 | 55.6 | 62.4 | 60.8 | ||
| Sf | 48.5 | 63.3 | 62.0 | 62.2 | 65.8 | 59.5 | 60.5 | 62.6 | |||
| Yp | 64.7 | 64.8 | 62.2 | 64.8 | 60.8 | 62.5 | 64.1 | ||||
| Ea | 35.8 | 62.4 | 62.9 | 57.3 | 63.2 | 63.9 | |||||
| Ps | 61.8 | 62.5 | 56.8 | 65.1 | 64.8 | ||||||
| Xc | 33.5 | 53.1 | 60.1 | 59.7 | |||||||
| Rs | 54.1 | 60.4 | 62.1 | ||||||||
| Bb | 52.6 | 55.6 | |||||||||
| Cp | 61.2 |
Sg, S. glossinidius; Sz, SZPE; Se, S. enterica; Sf, S. flexneri; Yp, Yersinia pestis; Ea, E. amylovora; Ps, P. syringae; Xc, X. campestris; Rs, R. solanacearum; Bb, B. bronchiseptica; Cp, C. pneumoniae; Rh, Rhizobium spp. NGR 234.
Estimation of Nucleotide Sequence Divergence.
Sequence divergence at synonymous and nonsynonymous sites was computed by using the method of Li (14), implemented in the GCG package (Genetics Computer Group). Analyses were performed on a 1,020-bp fragment of invA, a 760-bp fragment of fusA, a 665-bp fragment of rplB, and the complete coding sequences of invB (408 bp), invC (1,311 bp), and spaQ (261 bp). Note that Escherichia coli does not harbor inv/spa homologs, thus precluding pairwise comparisons for these genes.
In Vivo Gene Expression Assays.
To analyze inv/spa gene expression in vivo, S. zeamais development was synchronized by allowing 250 adults to mate and oviposit overnight in maize at 27.5°C and 70% relative humidity. Progeny were extracted from maize grains at 3-day intervals from day 6 after oviposition through day 33. After each collection, insects were dissected on ice in saline (0.85% wt/vol), and bacteriomes were removed and frozen in a dry ice/ethanol bath. RNA was prepared with the RNeasy minikit (Qiagen) with on-column DNase treatment. Relative transcript levels were obtained by real-time quantitative RT-PCR in a LightCycler (Roche Molecular Biochemicals). Reactions consisted of 10 ng of RNA, 0.3 μM of primers, and 2.75 mM (for invA only) or 3.25 mM Mn(OAc)2 in 1× Hot Start LightCycler-RNA Master SYBR Green I (Roche Molecular Biochemicals). The following primers (listed 5′ to 3′) were designed to amplify 220 bp of invA (TCA AGA AAC GAC GTG AAG TAC and ACA AGT GGG TAT AAA CGG TAA G), 280 bp of spaPQ (AAT TTG TTG TTT GCC GGT AAC and AGA GGC GAG ATC ATC AGT CAG) and 220 bp of fusA (GTC CAT TTT GTT TAC GAA CGC and CAT CAA TAT CAT CGA CAC CCC). RT-PCR cycling parameters followed the manufacturer's recommendations, with a 10-sec annealing step at 57°C for all reactions. The integrity of reactions was confirmed by analyzing reaction product melting curves; no nonspecific products were observed. All RNA samples were free of DNA contamination, tested in standard LightCycler PCR reactions with no reverse transcription step. Relative expression levels of each gene were obtained through comparisons to external standard curves generated for each primer set with serial dilutions of template RNA.
In Vitro Gene Expression Assays.
To analyze inv/spa gene expression in vitro, bacteriomes were removed from 14-day-old S. zeamais larvae under sterile conditions and homogenized in 0.85% (wt/vol) saline by using a Dounce tissue homogenizer (Kontes). The homogenate was filtered through a 5-μm poly(vinylidene difluoride) membrane, and the filtrate was centrifuged (6,000 × g, 10 min, 25°C) to pellet bacterial symbionts. Symbionts were purified away from cellular debris by three successive washes in sterile 0.85% (wt/vol) saline and equilibrated for assay by three additional washes in minimal medium (20 mM NaHPO4/0.1% casamino acids/25 mM glucose). Bacteria were resuspended in minimal medium at a concentration of 107 cells/ml. The bacterial suspension was combined with solutions of minimal medium containing either 2 mM CaCl2, 2 mM MgCl2, or 2 mM CuCl2 to provide a range of samples of equal volume and cell densities having metal cation concentrations ranging from 20 μM to 1 mM. Bacterial suspensions were then maintained under uniform conditions at 15 and 27.5°C for 15 h. After incubation, an aliquot of each suspension was plated on LB agar to ensure that samples were free of contamination. After pelleting bacteria from each culture, RNA was extracted by using the RNeasy minikit (Qiagen). Relative transcript levels of invA, spaPQ, and fusA were determined by LightCycler RT-PCR, as described above.
Results
Cloning and Sequencing of SZPE inv/spa Homologues.
By using degenerate PCR primers, we amplified and sequenced fragments of appropriate lengths from template DNA extracted from the bacteriomes of S. zeamais. The SZPE inv/spa sequences are 86.5, 81.9, 73.8, and 83.7% identical to invA, invB, invC, and spaPQR of S. glossinidius, respectively (3), and have intact ORFs with no mutations resulting in frameshifts or stop codons. The conceptual translations of invA and invC contain conserved functional protein domains corresponding to an inner cytoplasmic membrane component (invA) and a cytoplasmic ATP synthase nucleotide-binding domain (invC). The base compositions of the inv/spa genes (G+C: invA, 47.7%; invB, 45.1%; invC, 54.6%; spaPQR, 54.9%) are in agreement with the estimated genomic base composition of the primary endosymbiont of rice weevils (6) and are almost identical to those reported for the inv/spa genes of S. glossinidius (3).
Phylogenetic Analysis.
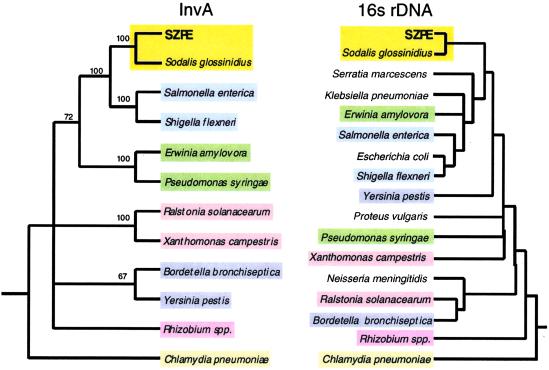
We determined the ancestry of the SZPE inv/spa genes through phylogenetic reconstructions with homologs from S. glossinidius and several plant and animal pathogens. For comparison, we also constructed a tree based on the sequences of 16S rDNA from the same bacterial species. Results are shown for the phylogeny generated by parsimony (Fig. 1, Table 1), with bootstrap support based on 1,000 replicates, and identical tree topologies were obtained by using maximum parsimony and distance methods. In both the 16S rDNA and InvA trees, the SZPE and S. glossinidius sequences form a single robust clade distinct from the enteric pathogens, indicative of a single acquisition of the invA homologs in the common ancestor of these symbionts.
Fig 1.
Phylogenetic trees based on the InvA and 16S rDNA sequences from SZPE, S. glossinidius, and selected plant and animal pathogens. For the InvA tree, only clades supported by bootstrap values greater than 60% are shown as resolved. Accession numbers of InvA sequences are as follows: SZPE (AF467290), S. glossinidius (AF306649), Yersinis pestis (NC001972), S. enterica (U43239), S. flexneri (NC002698), P. syringae (AAG33877), Ralstonia solanacearum (P35656), Chlamydia pneumoniae (NC000922), Erwinia amylovora (X99768), Rhizobium sp. NGR234 (P55726), Xanthomonas campestris (P80150). The invA homolog from Bordetella bronchiseptica was obtained from a dataset produced by the Bordetella bronchiseptica Sequencing Group at the Sanger Center, and can be obtained from ftp://ftp.sanger.ac.uk/pub/pathogens/bb/.
Nucleotide Sequence Divergence Estimates.
Because our phylogenetic analyses supported the presence of invA in the common ancestor to SZPE and S. glossinidius, the inv/spa gene cluster should follow the same evolutionary history as other genes ancestral to these species. To test this hypothesis, we compared the extent of sequence divergence of several inv/spa genes to that of two informational genes, fusA and rplB. Informational genes that are involved in transcription, translation, and related processes provide a robust standard for evolutionary comparisons, because these genes are rarely subject to horizontal transfer (15, 16).
For all genes, frequencies of both synonymous and nonsynonymous substitutions (Ks and Ka, respectively) are lower for S. glossinidius—SZPE than for SZPE—S. enterica and S. glossinidius—S. enterica, supporting the occurrence of all six genes in the shared ancestor of S. glossinidius and SZPE (Table 2). In addition, pair-wise comparisons yielded similar Ka values for each gene between SZPE—S. enterica and S. glossinidius—S. enterica, as expected if inheritance of each gene has been strictly vertical since the divergence of S. glossinidius and SZPE. Estimates of Ks for S. glossinidius—SZPE are similar between the inv/spa genes but substantially higher than Ks values for both fusA and rplB, reflecting the strong codon bias in fusA and rplB within this group of bacteria. For example, fusA and rplB have high codon adaptation indices in E. coli (0.75 and 0.72, respectively), and thus these genes are expected to show atypically low Ks values for the E. coli—S. enterica comparison (17, 18).
Table 2.
Divergence among E. coli, Salmonella, Sodalis, and SZPE homologs
| Synonymous substitutions per site, Ks | ||||||
|---|---|---|---|---|---|---|
| invA | invB | invC | spaQ | fusA | rplB | |
| SZPE–S. glossinidius | 0.57 | 0.70 | 0.87 | 0.58 | 0.19 | 0.13 |
| SZPE–S. enterica | >2 | >2 | >2 | >2 | 0.66 | 0.57 |
| S. glossinidius–S. enterica | >2 | >2 | >2 | >2 | 0.70 | 0.61 |
| S. enterica–E. coli | — | — | — | — | 0.19 | 0.11 |
| Nonsynonymous substitutions per site, Ka | ||||||
| invA | invB | invC | spaQ | fusA | rplB | |
| SZPE–S. glossinidius | 0.04 | 0.09 | 0.21 | 0.04 | 0.002 | 0.002 |
| SZPE–S. enterica | 0.53 | 0.90 | 0.46 | 0.24 | 0.070 | 0.047 |
| S. glossinidius–S. enterica | 0.53 | 0.90 | 0.42 | 0.23 | 0.065 | 0.042 |
| S. enterica–E. coli | — | — | — | — | 0.017 | 0.003 |
In Vivo Gene Expression Assays.
Because SZPE has not been cultivated in vitro, we were unable to use gene knockouts to examine the function of the inv/spa genes in the SZPE–weevil symbiosis. Instead, we examined inv/spa gene expression at different points in the host life cycle to determine whether transcript levels change when SZPE invades bacteriomes during metamorphosis of the insect host (5). Expression patterns of invA and spaPQ were assayed by quantitative RT-PCR by using RNA extracted from the bacteriomes of weevils throughout the 33-day period of weevil development.
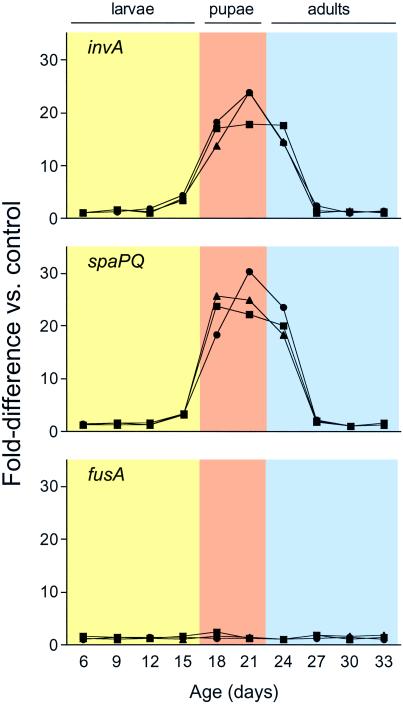
As anticipated from the polycistronic organization of the inv/spa genes (2, 3), the expression patterns of invA and spaPQ were very similar (Fig. 2). Both invA and spaPQ exhibited a 20-fold increase in expression coincident with the onset of metamorphosis, when symbionts must infect new bacteriomes. We observed no significant changes in the number of fusA transcripts in weevil bacteriomes throughout the entire period of weevil development. Because fusA is known to encode a ribosomal translocase (elongation factor G) expressed consistently during translation in bacteria, these results demonstrate that up-regulation of inv/spa expression occurs independently of any global changes in SZPE transcription and translation during insect metamorphosis. The integrity of the experimental approach was confirmed by testing RNA extracted from three individual cohorts of animals, maintained under identical conditions.
Fig 2.
Quantitative RT-PCR analysis of gene expression in weevil bacteriomes at different developmental stages. Expression of invA, spaPQ, and fusA was analyzed for three individual cohorts of reproductively synchronized animals at 3-day intervals through the course of their development into adulthood. To calculate relative transcript numbers, standard curves were generated for each gene by serially diluting RNA templates with the highest transcript numbers for each gene. “Fold-difference” values were determined by comparing transcript numbers in each sample to a control sample, defined within each cohort as the sample with the lowest number of transcripts for a given gene. Peak levels of the invA and spaPQ transcripts are detected in weevils undertaking pupation and metamorphosis, whereas levels of the housekeeping gene (fusA) transcripts remain constant throughout weevil development.
In Vitro Gene Expression Assays.
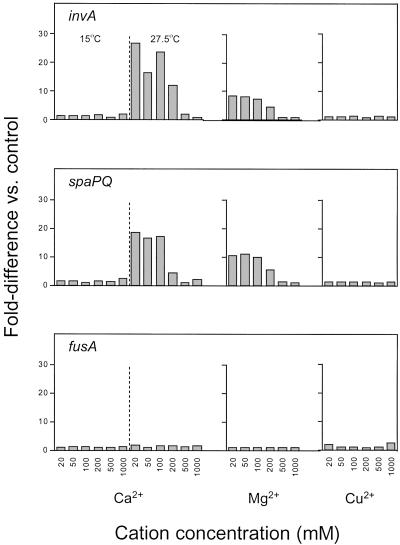
Because the regulation of the inv/spa genes in S. enterica is governed by the PhoP/PhoQ signal transduction system and is modulated by environmental Mg2+ and Ca2+ (19, 20), we tested whether inv/spa expression in SZPE is similarly controlled by external concentrations of divalent cations. For these assays, symbionts were purified directly from bacteriocytes of S. zeamais and resuspended in aliquots of minimal medium containing a series (20 μM to 1 mM) of three different divalent cations. After 15-h exposure, the relative levels of the symbiont invA, spaPQ, and fusA transcripts were measured by quantitative RT-PCR. In these in vitro assays, we observed at least a 10-fold increase in the relative expression of both invA and spaPQ—but not of fusA—as the extracellular concentrations of Ca2+ or Mg2+ decreased (Fig. 3), showing that the inv/spa genes in SPZE are regulated by the same cues as their Salmonella homologs.
Fig 3.
Effect of divalent cations and temperature on invA, spaPQ, and fusA gene expression in SZPE. To calculate relative transcript numbers, standard curves for each gene were generated by serially diluting RNA templates with the highest transcript numbers for each gene. “Fold-difference” values were determined by comparing transcript numbers in each sample to a control sample, defined within each experiment as the sample with the lowest number of transcripts for a given gene. At high concentrations of Ca2+ or Mg2+ (≥200 μM), but not Cu2+, invA and spaPQ transcription is repressed. Transcription of the housekeeping gene, fusA, is not affected by any of the metal cations tested. At a reduced assay temperature (15°C), inv/spa expression is completely repressed regardless of Ca2+ availability.
To test whether the up-regulation of inv/spa was due simply to a change in the ionic strength of the medium, we measured transcript levels in symbionts exposed to varying concentrations of Cu2+. The relative expression levels of the inv/spa genes, as well as the fusA controls, were unaffected by Cu2+ concentration in the medium (Fig. 3). To test the effect of temperature, which is known to affect inv/spa expression in Shigella, we set up a duplicate series of Ca2+ assays that were maintained at 15°C instead of 27.5°C. At the reduced incubation temperature, we detected only a 2-fold reduction in the numbers of fusA transcripts in these samples, indicating that global transcription and translation processes are still active in SZPE at 15°C. We were unable to detect any invA or spaPQ transcripts in RNA extracted from symbionts maintained at 15°C, suggesting that the processes of cell invasion are repressed in SZPE at reduced temperature.
Discussion
Many intracellular bacterial pathogens of animals and plants harbor a TTSS that mediates their uptake and entry into eukaryotic cells (21). In contrast, the transmission of primary endosymbionts of insects has traditionally been viewed as the result of host-directed mechanisms (5), a view that was recently corroborated by analysis of the full genome sequence of Buchnera aphidicola, the primary bacteriome-associated endosymbiont of aphids. Inspection of the highly reduced gene inventory of Buchnera suggested that this symbiont is a passive captive of its host (22); in particular, Buchnera was found to lack a TTSS. For these reasons, symbionts have not been expected to exploit protein translocation systems to actively invade host cells. In the present study, we have established that the infection of new host bacteriomes by certain endosymbionts is facilitated by a TTSS homologous to those identified in enteric pathogens.
Our findings indicate that the weevil endosymbiont, SZPE, contains a tandem array of genes (inv/spa) that share high levels of sequence identity with genes encoding components of the TTSS molecular syringe in other bacteria. The SZPE genes have intact ORFs and share an identical spatial and cistronic organization with their homologs in S. glossinidius, S. enterica, and S. flexneri. The lineage comprising SZPE and S. glossinidius is of interest, because these endosymbionts diverged from a common ancestor and have evolved in symbiotic relationships with different insect hosts. On the basis of sequence analyses of inv/spa and other genes, it appears that the TTSS was acquired by the common ancestor of SZPE and S. glossinidius. Subsequently, the TTSS was retained in both lineages as an essential element in the establishment and maintenance of infection.
Comparisons of the extent of nucleotide sequence divergence in the inv/spa genes with that of the housekeeping genes provide additional evidence for the origin of the inv/spa genes in the common ancestor of SZPE and S. glossinidius. Furthermore, nonsynonymous nucleotide sequence divergence values are nearly identical for the S. glossinidius—SZPE comparisons and for the corresponding E. coli—S. enterica comparisons. If these genes evolve at similar rates in all four lineages, an assumption supported by sequence comparisons of E. coli and endosymbionts (23), then S. glossinidius and SZPE diverged at the same time as E. coli and S. enterica, approximately 100 million years ago (24). This date is consistent with estimates derived from independent analyses indicating that Sitophilus and its symbionts have coevolved through strict vertical transmission for only 50–100 million years (6). The recent establishment of the weevil–bacterial endosymbiosis may account for the observation that the genomes of the weevil endosymbionts do not display the typical A+T bias and extreme reduction in genome size, as observed in more ancient obligate bacterial endosymbionts such as B. aphidicola and Wigglesworthia glossinidia (8, 22, 25).
Although SZPE resides exclusively in bacteriomes and provides its weevil host with vitamins and amino acids essential for growth and reproduction (8), this endosymbiont also displays certain characteristics that are typical of facultative “secondary” endosymbionts. This includes a requirement for SZPE to invade new host cells during weevil metamorphosis, as indicated by the 20-fold increase in inv/spa transcript numbers in SZPE during metamorphosis relative to the entire period of larval development. Because there is no global change in SZPE transcription over the host life cycle, it appears that SZPE is responding to host metamorphosis by specifically up-regulating transcription of the inv/spa genes, promoting cell invasion.
Many bacteria have evolved mechanisms enabling them to modulate gene expression in response to environment cues (26, 27). For example, Salmonella species are able to sense the nature of their immediate environment by measuring Ca2+ and Mg2+ availability through the PhoP/PhoQ signal transduction system (20). In conditions of low Ca2+ and Mg2+ availability, as encountered intracellularly, PhoP/PhoQ modulates expression of Salmonella invasion genes, including inv/spa (28). By using a transient in vitro culture system and RT-PCR assays, we determined that SZPE is also able to modulate expression of inv/spa genes in response to Ca2+ and Mg2+ availability and temperature. Under conditions of low Ca2+ and Mg2+ availabilities (<100 μM), we observed an order of magnitude increase in the expression of the inv/spa genes, whereas expression of housekeeping genes was largely unchanged. We postulate that SZPE is responding to differences in the concentrations of these cations to coordinate the timing of cell invasion, in the same way as Salmonella spp. An alternative possibility is that SZPE retains, but no longer utilizes, a regulatory system that served to coordinate opportunistic host cell invasion by a free-living ancestor. However, the divergence at neutral nucleotide sites between S. glossinidius and SZPE is sufficiently high (Table 2) that we would expect functionless genes to have been inactivated by mutation.
We have demonstrated that two distinct members of an endosymbiotic clade have evolved in different hosts with dependence on a specialized cell invasion apparatus that is also commonly used by pathogens. It has been suggested that parasites, through a coevolutionary process, can develop commensal or mutualistic relationships, and that the molecular mechanisms of symbiosis and pathogenesis might be similar (29, 30). Our findings support these notions, indicating that a TTSS has been adapted in the context of mutualism to maintain a permanent intracellular association between symbiotic bacteria and their hosts.
Acknowledgments
We thank Serap Aksoy (Yale University) for providing S. glossinidius cultures and Jim Baker (U.S. Department of Agriculture, Manhattan, KS) for providing S. zeamais. This work was supported by National Institutes of Health Grant GM56120 (to H.O.) and National Science Foundation Grant DEB-9978518 (to N.A.M.).
Abbreviations
TTSS, type III secretion system
SZPE, S. zeamais primary endosymbiont
References
- 1.Galan J. E. & Collmer, A. (1999) Science 284 1322-1328. [DOI] [PubMed] [Google Scholar]
- 2.Hueck C. J. (1998) Microbiol. Mol. Biol. Rev. 62 379-433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dale C., Young, S. A., Haydon, D. T. & Welburn, S. C. (2001) Proc. Natl. Acad. Sci. USA 98 1883-1888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dale C. & Maudlin, I. (1999) Int. J. Syst. Bacteriol. 49 267-275. [DOI] [PubMed] [Google Scholar]
- 5.Buchner P., (1965) Endosymbiosis of Animals with Plant Microorganisms (Interscience, New York), pp. 160–190.
- 6.Heddi A., Charles, H., Khatchadourian, C., Bonnot, G. & Nardon, P. (1998) J. Mol. Evol. 47 52-61. [DOI] [PubMed] [Google Scholar]
- 7.Nardon P. & Grenier, A. M. (1988) in Cell to Cell Signals in Plant, Animal and Microbial Symbiosis, eds. Scannerini, S., Smith, D. C., Bonfante-Fasolo, P. & Gianinazzi-Pearson, V. (Springer, Berlin), pp. 255–270.
- 8.Heddi A., Grenier, A. M., Khatchadourian, C., Charles, H. & Nardon, P. (1999) Proc. Natl. Acad. Sci. USA 96 6814-6819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ewald P. (1987) Ann. N.Y. Acad. Sci. 503 295-306. [DOI] [PubMed] [Google Scholar]
- 10.Dubos R., (1965) Man Adapting (Yale Univ. Press, New Haven).
- 11.Axelrod R. & Hamilton, W. D. (1981) Science 211 1390-1396. [DOI] [PubMed] [Google Scholar]
- 12.Frank S. A. (1995) J. Theor. Biol. 176 403-410. [DOI] [PubMed] [Google Scholar]
- 13.Swofford D., (2000) paup* 4.0 (Sinauer Associates, Sunderland, MA).
- 14.Li W.-H. (1993) J. Mol. Evol. 36 96-99. [DOI] [PubMed] [Google Scholar]
- 15.Jain R., Rivera, M. C. & Lake, J. A. (1999) Proc. Natl. Acad. Sci. USA 96 3801-3806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lawrence J. G. (1999) Curr. Opin. Microbiol. 2 519-523. [DOI] [PubMed] [Google Scholar]
- 17.Sharp P. M. & Li, W.-H. (1987) Mol. Biol. Evol. 4 222-230. [DOI] [PubMed] [Google Scholar]
- 18.Lawrence J. G. & Ochman, H. (1997) J. Mol. Evol. 44 383-397. [DOI] [PubMed] [Google Scholar]
- 19.Groisman E. A. (1998) BioEssays 20 96-101. [DOI] [PubMed] [Google Scholar]
- 20.Vescovi E. G., Ayala, Y. M., Di Cera, E. & Groisman, E. A. (1997) J. Biol. Chem. 17 1440-1443. [DOI] [PubMed] [Google Scholar]
- 21.Marcus S. L., Brumell, J. H., Pfeifer, C. G. & Finlay, B. B. (2000) Microbes Infect. 2 145-156. [DOI] [PubMed] [Google Scholar]
- 22.Shigenobu S., Watanabe, H., Hattori, M., Sakaki, Y & Ishikawa, H. (2000) Nature (London) 407 81-86. [DOI] [PubMed] [Google Scholar]
- 23.Ochman H., Elwyn, S. & Moran, N. A. (1999) Proc. Natl. Acad. Sci. USA 96 12638-12643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ochman H. & Wilson, A. C. (1987) J. Mol. Evol. 26 74-86. [DOI] [PubMed] [Google Scholar]
- 25.Akman L. & Aksoy, S. (2001) Proc. Natl. Acad. Sci. USA 98 7546-7551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mekalanos J. J. (1992) J. Bacteriol. 174 1-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Smirnova A., Li, H., Weingart, H., Aufhammer, S., Burse, A., Finis, K., Schenk, A. & Ullrich, M. S. (2001) Arch. Microbiol. 176 393-399. [DOI] [PubMed] [Google Scholar]
- 28.Groisman E. A. (2001) J. Bacteriol. 183 1835-1842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Goebel W. & Gross, R. (2001) Trends Microbiol. 9 267-273. [DOI] [PubMed] [Google Scholar]
- 30.Hentschel U. & Steinert, M. (2001) Trends Microbiol. 9 585. (lett.). [DOI] [PubMed] [Google Scholar]