Abstract
Xylella fastidiosa (Xf) causes wilt disease in plants and is responsible for major economic and crop losses globally. Owing to the public importance of this phytopathogen we embarked on a comparative analysis of the complete genome of Xf pv citrus and the partial genomes of two recently sequenced strains of this species: Xf pv almond and Xf pv oleander, which cause leaf scorch in almond and oleander plants, respectively. We report a reanalysis of the previously sequenced Xf 9a5c (CVC, citrus) strain and the two “gapped” Xf genomes revealing ORFs encoding critical functions in pathogenicity and conjugative transfer. Second, a detailed whole-genome functional comparison was based on the three sequenced Xf strains, identifying the unique genes present in each strain, in addition to those shared between strains. Third, an “in silico” cellular reconstruction of these organisms was made, based on a comparison of their core functional subsystems that led to a characterization of their conjugative transfer machinery, identification of potential differences in their adhesion mechanisms, and highlighting of the absence of a classical quorum-sensing mechanism. This study demonstrates the effectiveness of comparative analysis strategies in the interpretation of genomes that are closely related.
It is believed that globally a fifth of potential crop yields are lost each year because of disease (1), and a significant proportion of these diseases are of bacterial origin. The xylem-inhabiting Gram-negative bacterium Xylella fastidiosa (Xf) is an important phytopathogen causing Pierce's disease of grapevine, citrus variegated chlorosis in citrus, and leaf scorch disease in numerous other plants (2). Based on microscopy (3) and cross-inoculation experiments (4), the causative agent of this disease group was demonstrated to be the same bacterium. Insects, including the glassy-winged sharpshooter (5), are the vectors for transmitting these diseases. Xf can be differentiated into subspecies or pathovars depending on such criteria as plant host specificity and pathogenicity (2, 6–11). We will refer throughout to the citrus, almond, and oleander strains as: Xf pv citrus (XFA), Xf pv almond (XFX), and Xf pv oleander (XFY).
The genome sequence of the Xf (strain 9a5c), which causes citrus variegated chlorosis (12), and the gapped-genome sequencing of XFX (Dixon strain) and XFY (Ann1 strain) causing almond and oleander leaf scorch, respectively, have been reported (32). Here, we present a functional reconstruction of this phytopathogenic microbe based on genomic sequences, as well as the analysis of a three-way genome comparison that shows the shared and unique functions present in each strain. The results of the comparison of the two draft Xylella genome sequences with the completed citrus strain provides significant advancement in the understanding of the physiology of these organisms, as well as insights into their pathogenicity and host-range specificity.
Materials and Methods
ORF Prediction.
ORFs were predicted with a proprietary ORF calling software system developed at Integrated Genomics. The system combines statistically predicted ORFs with ORFs derived from external sources (if available) and blast and fasta similarities. ORFs were predicted by using this approach for all Xf genomes including the previously published XFA (12) extracted from GenBank. The ORF calling algorithm was not run on contigs with <20 reads because of low coverage and poor sequence quality.
Annotation and Functional Reconstruction.
We used the ergo bioinformatics suite (www.integratedgenomics.com) for the genome analyses. ergo currently contains an integration of more than 470 genomes (including complete and partial, public and proprietary from all kingdoms), with a manually curated set of functional annotations and more than 5,300 cellular pathways. The analysis of the three Xf strains was performed as described (13–16). A complete set of genomic data for the Xylella strains used in this study, including ORF coordinates, annotations, etc. has been made available (www.integratedgenomics.com/genomereleases.html). Sequences of the XFX, Dixon strain (NC_002723), and XFY, Ann-1 strain (NC_002722), have been deposited at GenBank. Other aspects of the cellular reconstruction are published as supporting information on the PNAS web site, www.pnas.org.
Whole-Genome Comparative Analysis.
A clustering algorithm (workbench; Integrated Genomics), part of the ergo bioinformatics suite was used to calculate the protein clusters among all three sequenced Xf strains. The total set of clusters determined from this three-way comparison was then partitioned into seven composite subsets comprising clusters that were either unique to each organism (three sets), those shared between two specific Xf species (three pair-wise sets), and finally the set of clusters present in all Xf strains (the workbench data files are available at http://ergo.integratedgenomics.com/Genomes/Xyllela/Cluster). The minimum similarity threshold used to calculate the clusters had an E value of 10−5. The global genome clustering data sets together with the “in silico” functional reconstructions of the Xf genomes were subsequently analyzed to determine the respective similarities and differences between each organism. Comparative analysis of the universally shared Xf clusters used ORFs of high DNA sequence quality. A gene sequence was considered high quality if the average predicted base error probability was <1/10,000 and no single nucleotide has an error of >1/100. The error probabilities were determined by inspection of the Phrap quality scores for the draft genomes. The sequence of the citrus genome (12) by definition meets this criterion.
Results and Discussion
Reanalysis of the XFA Genome.
We re-examined the complete genome of XFA to add as much value to the raw sequence data as possible by using the ergo bioinformatics suite. First, we reassessed the predicted ORFs by merging those from the published data (12) with those generated from our own ORF prediction algorithm. Intergenic regions were then analyzed with a blastt search against all ORFs in the ergo database (currently more than 106 ORFs) to generate a third set of predicted ORFs, which was then postprocessed and merged with the initial data set described above. This version of the XFA genome was then used for all subsequent analyses.
Our methods predicted a total of 2,985 ORFs [in comparison to the 2,782 previously reported (12)], of which 58% had functional assignments (versus 46%). A total of 131 potential additional ORFs, not previously reported, were identified, including cases that were truncated or had frame-shifted gene sequences; 35% (41 ORFS) of these ORFs could be annotated. Table 1 shows those ORFs that appear to encode a putative full-length version of the respective gene. From this list we found several interesting functions including six additional ORFs on the plasmid from the XFA genome, two of which have a predicted function: a type IV secretion system component, VirB7 protein (RXFA03058) and a conjugal transfer protein TrbE (RXFA02884). There are also numerous phage-related proteins and several DNA recombinase-type proteins that may be associated with regions of DNA mobility. We also identified a toxin-like protein (RXFA02868) with 48% identity to a Pseudomonas fluorescens toxin protein (tr|Q9F7Y3).
Table 1.
Identification of ORFs with functional assignment from XFA not identified previously
| XFA ID | Functional assignment | DNA coordinates |
|---|---|---|
| Phage proteins | ||
| RXFA02870 | Phage DNA packaging protein GP2 (terminase large subunit) | main_652111_652584 |
| RXFA02874 | Phage protein with ZN-finger domain | main_1602437_1603396 |
| RXFA02919 | Phage-related protein | main_682557_683123 |
| RXFA03000 | Phage-related ATP/GTP binding protein | main_1046293_1046508 |
| RXFA03028 | Phage DNA packaging protein GP2 (terminase large subunit) | main_1193843_1194085 |
| RXFA03087 | Phage-related protein | main_309199_308984 |
| RXFA03216 | Phage protein with ZN-finger domain | main_1194248_1194472 |
| Informational processing: DNA replication, recombination, modification, and repair | ||
| RXFA02885 | Transposase | main_341961_342965 |
| RXFA02886 | DNA-invertase | main_1965741_1965505 |
| RXFA02868 | Toxin-like protein | main_1621091_1620816 |
| RXFA02920 | Transposase | main_1834786_1835250 |
| RXFA02922 | Resolvase | main_1833819_1834220 |
| RXFA02936 | DNA-invertase | main_1966040_1965708 |
| RXFA02986 | Transposase | main_1834527_1834144 |
| RXFA03001 | Transposase | main_2030735_2030974 |
| Informational processing: Transcription and translation | ||
| RXFA02933 | Ribosomal protein S6 modification protein | main_2406479_2406835 |
| RXFA03064 | Transcriptional regulator | main_1685281_1684697 |
| Pathogenesis | ||
| RXFA02856 | Hemolysin precursor, truncated/partial | main_2660751_2660212 |
| RXFA02857 | Hemolysin precursor, truncated/partial | main_2423973_2423761 |
| RXFA03058 | Channel protein VIRB7 homolog | pXF51_8930_9169 |
| Conjugal transfer | ||
| RXFA02884 | Conjugal transfer protein TRBE (IS element) | pXF51_35914_33764 |
| Miscellaneous enzymatic functions | ||
| RXFA02887 | Acetyltransferase (EC 2.3.1.-) | main_2435902_2435273 |
| RXFA02895 | MG2+ transporter MGTE | main_1351993_1351661 |
| RXFA02906 | Transglutaminase-like enzymes | main_1726349_1724298 |
| RXFA02998 | HICA | main_1603449_1603703 |
| RXFA03085 | Zinc metallopeptidase | main_309199_308984 |
ORFs are shown organized into functional subsystem categories.
Some previously identified ORFs (12) displayed no sequence similarity to any known proteins. To our surprise, we found additional putative ORFs in exactly the same DNA region, but encoded on the opposite strand, showing sequence similarity to other known proteins in the public databases (Table 4, which is published as supporting information on the PNAS web site). Examples of ORFs with functional assignments that were identified as fragments (putative frame shifts) are also noted (Table 5, which is published as supporting information on the PNAS web site). These ORFs include several important functions including type I restriction and modification subunits and conjugal transfer proteins TrbJ and TrbE. A ribosomal modification protein was also found, which is orthologous to the Escherichia coli RimK protein that modifies the C terminus of ribosomal protein S6. There appear to be two copies of aconitate hydratase representing the aconitase 1 family (RXFA03019, RXFA02859; Table 5) and the aconitase 2 family (RXFA00292) proteins. A reverse transcriptase (RT)-type protein was identified that is likely to be a candidate for horizontal gene transfer (HGT) (see below).
One of the hypothetical proteins on the oleander plasmid, RXFY02526, resides in the type IV secretion system cluster, located between ORFs for VirB2 and VirB4. This is likely to be the VirB3 protein (RXFY02526). This ORF (RXFY02526) is nonorthologous to the XFA VirB3 protein (RXFA02788) but is similar to an ORF in Brucella melitensis biovar suis (B. suis, AF141604; B. abortus, AF226278) and is 29% identical to VirB3 from Agrobacterium tumefaciens (gi|10955511|NP_065363.1).
Global Comparative Analysis of Xf Genomes.
The draft genome sequences of XFX and XFY have been reported (32). To identify the signature features of each Xf genome, we compared the three genomes and characterized the protein clusters and ORFs that are unique to each genome. In instances where ORFs were identified in both XFX and XFY but were missing from XFA, we reasoned that these are likely to be bona fide differences, because the former are gapped genomes and the latter is complete. On the other hand, cases of putative ORFs missing from either of the first two strains might still be found in the nonsequenced gaps. Clusters of similar ORFs from all three Xf genomes were generated as described (see Materials and Methods). We calculated clusters of protein families from the three genomes based on a cut-off E value of 10−5. Fig. 1 summarizes the results from the global comparison of the three genomes and Table 2 shows the detailed organism-specific partitioning of the ORFs present within the protein cluster sets shown in Fig. 1 (Table 6, which is published as supporting information on the PNAS web site, documents the ORFs with functional assignments in the unique clusters for both XFX and XFY strains, as well as those shared between them but absent from XFA). Subsequently for each of those protein clusters, we identified the subsets that contain unique Xylella-specific proteins, i.e., they have no similarities to any other genomes with an E value better that 10−2 (Fig. 1, Table 2 in parentheses).
Fig 1.
Whole-genome comparison of three Xylella strains. A comparative analysis of the three Xf genomes was performed by using protein clustering algorithms. The numerals represent the number of clusters of related protein families.
Table 2.
Statistics of whole-genome protein cluster analysis for the three Xf strains
| Genomes
|
Clusters | ORFs | |||
|---|---|---|---|---|---|
| No. of clusters | ORFs in clusters | XFA | XFX | XFY | |
| XFA + XFX + XFY | 1,705 (38) | 7,002 (130) | 2,277 (42) | 2,339 (41) | 2,386 (47) |
| XFA + XFX | 114 (44) | 239 (90) | 123 (44) | 116 (46) | — |
| XFA + XFY | 183 (37) | 398 (78) | 196 (38) | — | 202 (40) |
| XFX + XFY | 85 (17) | 187 (35) | — | 93 (17) | 94 (18) |
| XFA | 375 (205) | 389 (211) | 389 (211) | — | — |
| XFX | 132 (77) | 133 (78) | — | 133 (78) | — |
| XFY | 180 (77) | 188 (81) | — | — | 188 (81) |
| Total | 2,774 (495) | 8,536 (703) | 2,985 (335) | 2,681 (182) | 2,870 (186) |
Protein clusters were calculated for the XFA, XFX, and XFY genomes (see Materials and Methods). The number of ORFs for each appropriate Xf strain comprising each protein cluster set (from Fig. 1) is indicated. In parenthesis are the corresponding clusters/ORFs with no similarity hits better than 10−2, against any other organism.
Common Xf Protein Clusters.
A total of 1,705 conserved protein clusters were identified (with the parameters described above), with each cluster having at least one ORF from each of the three strains. These clusters comprise 7,002 Xf ORFs, representing 82% of the total number of 8,536 ORFs identified in the genomes of the three Xf strains (Table 2). A total of 130 of the 7,002 ORFs (2% of the total number of the ORFs in common families) form 38 protein clusters that have no homologs in any other sequenced organism and therefore constitute the Xylella species signature. The remaining 6,872 ORFs have a homolog in other genomes and generally bear a functional assignment. An analysis of the functional subsystems comprising the common Xf protein clusters was performed to discern common aspects of Xylella biology, and some of the results of this analysis are outlined below.
Pathogenesis-Related Factors.
Xf displays polar attachment by producing fimbriae (17–21) (H.F. and A.P., unpublished observations). Fimbrial genes (e.g., fimA) have been implicated in virulence of the plant pathogenic bacteria (22). FimA and other fimbrial genes in this pathway are conserved among the Xf strains. Xf contains only one classical chaperone/usher-dependent fimbrial operon (RXFA00077-83), compared with enteric bacteria that contain 10–15 such operons. XFA displays a standard gene organization: a major subunit (RXFA00083) followed by a chaperone/usher pair that directs translocation (RXFA00082 and RXFA00081).
Xf also contains three afimbrial adhesins that may have arisen by gene duplication. RXFA01516 and RXFA01529 are separated by the 11-component operon (12 kb) of the general secretory pathway. RXFA01981 is homologous to RXFA01529, suggesting that this gene may have arisen by duplication. Furthermore, several outer membrane proteins have been identified in the Xf strains as potential afimbrial adhesins, displaying major differences in part of their sequences. In particular, in a cell surface protein (RXFA01981) homologous to the Hia gene product encoding the major adhesin of Haemophilus influenzae, we noted a similarity of this protein in the XFA and XFY strains with a truncation in the XFX ortholog. This and other examples of differences in cell surface proteins are noted (Fig. 4, which is published as supporting information on the PNAS web site). Predicted differences among the outer membrane potential adhesin gene sequences for the three Xf genomes suggest that there are organism and host-specific adhesin(s).
Quorum Sensing?
Cell–cell communication via small molecules enable bacteria to coordinate changes in gene expression in response to cell density, a process called quorum sensing. Xf does not have the Gram-negative homoserine lactone (N-AHL) signaling system. However, there is evidence that Xf may share a system of non-N-AHL diffusible signaling molecules with Xanthomonas campestris (Xc). The Xc system is regulated by a gene cluster designated rpf (regulation of pathogenicity factors) that affects a number of important phenotypes that may play a role in disease, such as extracellular polysaccharide production (1, 23, 24). Initial studies in Xc suggest that such signaling molecules may comprise fatty-acid derivatives and play a role in disease (1, 23). Orthologs of the rpf system have been found in XFA (and both other Xf strains) for a number of the key functional roles including rpfB (long-chain fatty-acid CoA ligase, RXFA00287) and rpfF (enoyl-CoA hydratase, RXFA01115). Direct experimentation will be required to determine whether Xf uses a similar mechanism.
Lipid Modifications.
Covalent modifications of lipid A have been implicated in virulence in Salmonella typhimurium (25) possibly by mediating adherance to host-cell surfaces. Orthologs of the S. typhimurium lpxO gene, encoding the enzyme lipid A-myristate β-hydroxylase that produces a 2-hydroxymyristate-containing lipid, have been found in all Xf strains (RXFA02100, RXFY01206, RXFX02032). This form of the lipid has been implicated as an important pathogenicity factor (25, 26). In Salmonella it is thought that inside phagolysosomes the 2-hydroxymyristoyl chain of lipid A is released by acyloxyacyl hydrolase, transported to the cytosol, and converted to 2-hydroxymyristoyl-CoA, which is a potent inhibitor of myristoyl-CoA:protein N-myristoyltransferase (26). Inhibition of the latter enzyme might interfere with host cell signaling functions and/or vesicular trafficking resulting from improper localization of numerous proteins that use myristoyl chains as membrane anchors.
Conjugal Transfer.
Xf has both the IncP (Trb) and the IncN (Tra) type of conjugation transfer system. In XFA there are two copies of the IncP conjugation system, one each on the plasmid (pXF51) and chromosome (Fig. 2). Both copies are present on putative mobile elements. In XFX and XFY there is only one set of the IncP transfer system, chromosomally located. The IncP genes are in an operon consisting of 12 ORFs (trbBCDEFGHIJKLN) in both XFA and XFX and nine ORFs (trbBCDEFIJLN) in XFY. The operon higAB, which is involved in maintaining cell growth, is present in all three genomes. HigB (RXFA00720), also referred to as host-addiction system protein, is toxic to segregants that do not maintain the plasmid. All of the ORFs in the IncN operon are found only in XFX, although XFA has a traBCEFJ operon of the IncN type but lacks the traKLMN, incC, and korB genes. None of the three genomes has the TracI helicase, suggesting that the conjugation system might be different from that of E. coli.
Fig 2.
Visualization of orthologous chromosomal regions encoding the IncP conjugal transfer gene cluster centered around RXFA02052: 1-trbB; 2-trbC; 3-trbD; 4-trbE; 5-trbF; 6-trbG; 7-trbH; 8-trbI; 9-trbJ; 10-trbK; 11-trbL; 12-trbN. Dotted lines denote that the region is part of a putative mobile element (e.g., insertion element). The chromosomal copy of the XFA IncP cluster is shown here. The orthologous ORFs are represented by similar color shading.
Carbohydrate Metabolism.
The chemical composition of the plant xylem within which this phytopathogen resides contains a dilute solution of organic acids, amides, glutamine, asparagine, and salts. However, analysis of the Xf genomes suggests an inability to degrade any organic acid, such as d- or l-lactate, malonate, propionate, butanoate, tartrate, glucarate, galactarate, glycolate, oxalate, or uronic acids. The exceptions to this rule are those organic acids that enter the tricarboxylic acid cycle (e.g., malate, oxaloacetate, citrate, fumarate). Additionally, Xf seems to be unable to degrade polysaccharides other than cellulose and galacturonans (e.g., xylan, rhamnan, arabinan). Bacterial polygalacturonases (PGAs) are thought to be responsible for degradation of plant tissue. PGAs have been identified in the XFA (RXFA02896, RXFA02916), XFX (RXFX01560) and XFY (RXFY01728) pathovars. Homologs of these PGAs from Erwinia carotovora and Pseudomonas solanacearum have been implicated in wilt disease (27, 28), suggesting a common mechanism of host tissue degradation. Interestingly, all Xf strains lack the flagellar biosynthetic machinery (flagellar class I, II, and III pathways are absent), therefore making them independently nonmotile. Consequently, any movement of the bacterium within the plant is likely to be controlled by fluid hydrodynamics within the xylem vessel. Further details of other aspects of the Xylella functional reconstruction will be presented elsewhere (32).
Strain-Specific Xf Protein Clusters.
A total of 133 ORFs (in 132 clusters) of the XFX strain appear to be absent from the other two strains. This represents 5% of the ORFs of the XFX genome. These ORFs include functions of the IncN conjugal transfer system (e.g., TraBHIJKMN) as well as two type II restriction endonucleases (SphI-like; RXFX00998) and NgoMI (RXFX01590). The SphI-type restriction subunit is chromosomally clustered with two ORFs on the opposite strand (RXFX00999, RXFX01000) that have methyltransferase motifs and are likely to be parts of a methylation subunit of this restriction-modification system. Similarly to the NgoMI-like type II system (e.g., Neisseria gonorrhoeae), the restriction (RXFX01590) and modification (RXFX01588) subunit genes are chromosomally clustered. Seventy-eight of these 133 XFX ORFs (59%) are unique (i.e., no homologs were identified in any other organism) hypothetical proteins and constitute the XFX signature. The remaining 55 XFA ORFs not in XFX or XFY appear to have homologs in other organisms and are candidates for HGT (see below).
A total of 188 ORFs (7%) of the XFY strain (in 180 clusters) did not show any detectable similarity to the XFA and XFX strains. These ORFs include a number of phage-related proteins. Plasmid maintenance proteins PemI (RXFY03510) and PemK (RXFY00993), identified as unique to XFY, are chromosomally clustered and are responsible for episomal stabilization during cell division. Although there are two PemK-like proteins in the XFA genome, RXFA02809 (51-kb plasmid-encoded) and RXFA01862 (chromosomal), these are nonorthologous to the XFY PemK protein. Moreover, neither of the XFA PemK-like ORFs resides in an operon with an identified PemI ORF. Interestingly, the XFY PemIK operon resides on a 30-kb plasmid, suggesting a role in maintenance of the plasmid that bears the pathogenicity island discussed above. Eighty-one of the 188 XFY ORFs (43%) are hypothetical proteins unique to that strain alone (XFY signature ORFs). Among the remaining 107 XFY ORFs there are potential candidates of HGT (see below).
From the XFA strain, 389 ORFs (in 375 clusters) did not show any similarity to the predicted ORFs of XFX and XFY. They represent 13% of the XFA ORFs, which seems much higher than the average of 6% observed in the other two strains. However, given that both of the latter genomes are incomplete, it is expected that some of those XFA ORFs will eventually merge to some of the other categories. Overall, based on the statistics of the other two strains, we predict that ≈6–7% of the XFA ORFs will eventually be identified in both the oleander and almond strains. Some of those genes include those that encode, for example, tyrosyl tRNA synthetase, the ribosomal proteins L3P, L1E, and L23P, S10P, and others. Additional XFA ORFs absent from the other strains encode transcription factors and transporters. Of particular interest is a predicted resistance protein (RXFA01765), which confers resistance to the epoxide antibiotic methylenomycin A, which appears to be absent from XFX and XFY. Finally, 211 of the 389 XFA ORFs (54%) are not seen in any other genome. Functional differences between XFA and XFX-XFY strains can be exploited to identify aspects of potential biological control. Thus, further analysis of the XFX and XFY strains should reveal whether the methylenomycin resistance protein is indeed absent from those genomes.
We identified 85 clusters containing 187 ORFs that are shared between XFX and XFY but absent from XFA. They represent a mere 2% of all of the ORFs found in all three Xf strains. Most of these ORFs are ≤ 100 aa in length with hypothetical functions. Additionally, within these pair-wise shared clusters, we identified conjugal transfer proteins TraL (RXFX00803), TraN (RXFY00996), a methylation subunit of a type II restriction-modification system (RXFX01588), a DNA cytosine (C5) methyltransferase (RXFY00809), and sensory transduction histidine kinases (RXFX02605, RXFY02878). Seventeen of those 85 clusters, containing 35 ORFs, have no homologs in any other genomes.
Although the percentage of ORFs shared between XFX and XFY is not expected to decrease (given that the citrus genome is complete), the same is not true for the other two comparisons (XFA-XFX and XFA-XFY), which currently represent 8% (239 + 396/8536) of the total number of Xf ORFs, akin to the argument presented above for the 389 XFA ORFs.
Relatedness of the Xf Strains.
As mentioned above, 82% of the Xf ORFs reside in protein clusters with at least one ORF from each strain in every cluster. To examine how this relatively high conservation compares with other closely related organisms, we applied the same clustering methodology to groups of three completely sequenced strains of Chlamydia pneumoniae, E. coli, and Neisseria meningitidis (29). As shown in Table 3, 97.7% of the ORFs in the three Chlamydia strains (CHLPN) belong to common protein families, as well as 87% of the E. coli ORFs (ECOLI), and 85% of the ORFs in the Neisseria strains (NEIME). As argued earlier, a significant number of additional common protein clusters between the three Xf strains is anticipated with the completion of the sequence of the two draft genomes. Therefore, these Xf strains display a similar pattern of genomic conservation as those observed within the E. coli and the Neisseria strains, using the same methodological approach.
Table 3.
Comparison of gene conservation within different groups of organisms
| Species group | CHLPN | ECOLI | NEIME | XYLFA |
|---|---|---|---|---|
| Level of conservation | 97.7% | 86.8% | 85% | 82.2% |
Each bacterial group represents one species and comprise three closely related strains which have a completely sequenced genome available (29). CHLPN: C. pneumoniae strains AR39, CWL029, J138; ECOLI: E. coli strains K12, O157:H7, O157:Sakai; NEIME: N. meningitidis serotype A (strain Z2491), serotype B (strain MC58), serotype C (strain FAM18); XYLFA: Xf strains CVC, almond (Dixon), and oleander (Ann1).
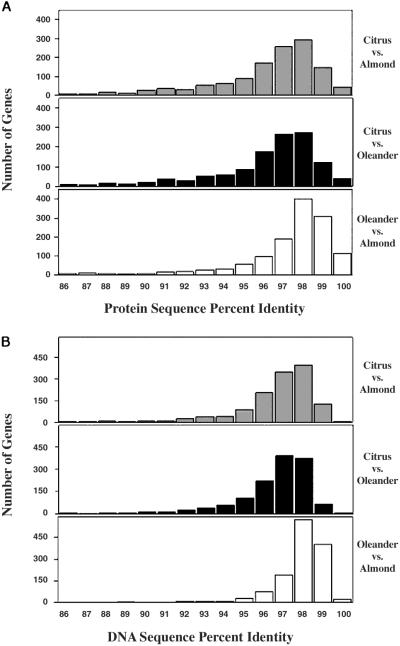
To examine the extent of inter-relatedness of the three Xf strains, we used the sequence identities of genes in the Xf clusters shared among all three genomes. Specifically, the genes from each of the 1,705 common Xf protein clusters were used for each possible pair-wise comparison. The percent DNA and protein sequence identities for these clusters were determined as follows: pair-wise alignments with fasta formats of respective DNA and proteins were performed with clustalw, and percent identities were determined algorithmically. The result of these comparisons is shown in the histograms in Fig. 3. Clearly, on the basis of both DNA and protein sequence conservation, the almond and oleander strains are more closely related to one another than either is to the citrus strain. This observation was further supported by the number of ORFs shared between XFX and XFY as opposed to either of the two with XFA. Thus, 88.5% of the ORFs from the XFX-XFY genomes are in common protein families, as compared with 87% between XFA and XFY and 86% between XFA and XFX. Therefore, the hierarchy of relative similarities between strains is thus: XFX-XFY > XFA-XFY > XFA-XFX.
Fig 3.
XFX and XFY strains are more closely similar to each other than to the XFA strain. Pair-wise comparisons based on (A) protein and (B) DNA sequence identity of the ORFs from each of the Xf protein clusters shared between all three strains.
HGT Versus Conserved Ancestry.
It had been noted previously that parts of the XFA genome might have been derived from non-Xylella origins (12). We investigated this possibility by determining which genes, if any, in any of the sequenced Xylella genomes resulted from potential HGT events from non-Xylella bacteria. Specifically, we identified all ORFs in the Xf group (XFA, XFX, XFY) that displayed stronger sequence similarity to a gene in an organism outside this group. Stronger sequence similarity here is defined as having a stronger hit (E value 10−10) with an ORF outside the Xf group than with any ORFs within the group. This analysis should be considered as an approximation to identify putative candidates for HGT. Members of the type IV secretion pathway, for example VirB1, VirB3, VirB4VirB5, VirB10, VirD2, VirD4 (BU) and the putative transcriptional regulator of this operon, comprising some of the virulence factors, appear as putative HGT candidates derived from a variety of soil-inhabiting organisms including Burkholderia fungorum (LB400), Sinorhizobium meliloti, and Agrobacterium tumefaciens. These genes reside on the 51-kb XFA plasmid and the 30-kb XFY plasmid (32).
We note here that XFA has an ADP/ATP carrier protein (30). This ORF (XFA01738) is absent in XFX and XFY, although homologs exist in intracellular bacterial pathogens (e.g., Chlamydia trachomatis, sp|O84068; C. pneumoniae, sp|Q9Z7U0; Rickettsia prowazekii, sp|P19568), phytopathogens (e.g., Xanthomonas campestris, X. axonopodis) (31), and plastid forms of plant homologs (e.g., Solanum tuberosum, tr|O24381). It is thought that in Rickettsia this ADP/ATP translocase provides the bacterial cell with host ATP in exchange for bacterial ADP, resulting in energy acquisition from the host (23). We speculate that such types of bacterial-host interaction may also exist in Xylella, which availed itself through a transfer from an organism in a similar ecological niche.
The ORFs for RT-like proteins (RXFA02955, RXFA02961), noted in Table 2, appear to have a foreign origin. In this case the best similarity hit is to a pathogenic enteric bacterium, E. coli O157:H7 (RECS05399; tr|082894). Although present in the XFA genome as two contiguous ORFs with this function, this putative RT is likely to be a single gene because both ORFs have a best-hit similarity to a single orthologous gene in numerous genomes. In particular, this ORF (RXFA02961) has strong similarity to two plant pathogen-encoded RTs from Ralstonia metallidurans (RREU02079, 1.97e-118; RREU01827, 7.98e-116) as well as to an RT (RPR00226, 4.04e-37) and a putative group II intron-encoded endonuclease (RPR00225, 1.64e-34) from the mitochondrial genome of the red alga, Porphyria purpleus. These results suggest that the RT is likely to be associated with an active mobile genetic element that is moving through the biosphere.
Another example of an entire operon likely to be derived from a non-Xylella source is the case of the IncN conjugal transfer region of XFX. ORFs from this genome (RXFX00238, RXFX00239, RXFX03105, RXFX00808, RXFX00805, RXFX00803, RXFX03106, RXFX02654, RXFX00802, RXFX03501, RXFX00801) for the IncN-type conjugal transfer system (IncC protein, KorB regulator, TraBNCLKJH and TrbB) bear a strong similarity to proteins and in operon structure to an orthologous region from Enterobacter aerogenes, an enteric bacterium. How would it be possible for the phytopathogenic Xylellas to acquire DNA from enteric organisms, which occupy a different ecological niche? One possibility arises from the fact that Xylella also lives in an insect host, the sharpshooter leafhoppers, within whose foregut the pathogens reside before being injected into the plant xylem. Alternatively, this transfer might have occurred in the soil.
Concluding Remarks.
Overall, the genomewide comparison of two phytopathogenic Xf strains, from almond and oleander plants, to the complete genome sequence of the Xf citrus strain has provided us with unique information in terms of the relatedness of the three strains, the conserved gene pool of the Xf species in general, and the genomic signature of each of the strains. Eighty-two percent of the Xylella ORFs found in all three strains are distributed in common protein families, which is comparable to the pattern seen in the case of the three E. coli or Neisseria strains. These common families comprise 76% of the XFA ORFs, 83% of the XFY ORFs, and 87% of the XFX ORFs.
This study has generated useful biological insights into the plasticity, pathogenicity, and bacterial-host interactions of these close phylogenetic neighbors. We have noted extensive HGT from soil-inhabiting bacteria, including plasmid-encoded virulence (type IV secretion) components as well as conjugal DNA transfer genes. We have also detected additional virulence genes (e.g., VirB3, VirB7) compared with those reported (12). The potential lipid modification enzymes and outer membrane proteins that could serve as adhesion components will be crucial for analyzing bacterial–plant interactions. Additionally, other aspects of our functional reconstruction common to all Xf strains have revealed a potential biological control point in aerobic respiration (32). Analysis of the ORFs present in the Xf citrus strain but absent in almond and oleander revealed a 65-kb phage insertion region bearing predicted carbon-utilization gene clusters that are likely to confer host-specific functions (32). We anticipate that the studies presented here will facilitate the development of microbiological control strategies.
Supplementary Material
Acknowledgments
We thank members of the bioinformatics and genome analysis group at Integrated Genomics, including Gordon Pusch, Lynn Jablonski, Olga Vassieva, Allen Bartman, and Warren Gardner. We also acknowledge Steven Lindow (University of California, Berkeley) and Rob Edwards (University of Tennessee, Memphis) for valuable discussions. This work was supported by the Integrated Genomics research and development program and the U.S. Department of Energy, Office of Biological and Environmental Research, together with the University of California, Lawrence Livermore National Laboratory under Contract W-7405-Eng-48, Lawrence Berkeley National Laboratory under Contract DE-AC03 76SF00098, and Los Alamos National Laboratory under Contract W-7405-ENG-36.
Abbreviations
Xf, Xylella fastidiosa
XFA, Xf pv citrus
XFX, Xf pv almond
XFY, Xf pv oleander
HGT, horizontal gene transfer
RT, reverse transcriptase
References
- 1.Dow J. M. & Daniels, M. J. (2000) Yeast 17, 263-271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Purcell A. H. (1997) J. Plant Pathol. 79, 99-105. [Google Scholar]
- 3.Mircetich S. M., Lowe, S. K., Moller, J. W. & Nyland, G. (1976) Phytopathology 66, 17-24. [Google Scholar]
- 4.Davis M. J. & Thompson, S. V. (1980) Phytopathology 70, 472-475. [Google Scholar]
- 5.Purcell A. H., Saunders, S. R., Hendson, M., Grebus, M. E. & Henry, M. J. (1999) Phytopathology 89, 53-58. [DOI] [PubMed] [Google Scholar]
- 6.Hopkins D. L. (1989) Annu. Rev. Phytopathol. 27, 271-290. [Google Scholar]
- 7.Chen J., Lamikanra, O., Chang, C. J. & Hopkins, D. L. (1995) Appl. Environ. Microbiol. 61, 1688-1690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pooler M. R. & Hartung, J. S. (1995) Curr. Microbiol. 31, 134-137. [DOI] [PubMed] [Google Scholar]
- 9.Banks D., Albibi, R., Chen, J., Lamikanra, O., Jarret, R. L. & Smith, B. J. (1999) Curr. Microbiol. 39, 85-88. [DOI] [PubMed] [Google Scholar]
- 10.da Costa P. I., Franco, C. F., Miranda, V. S., Teixeira, D. C. & Hartung, J. S. (2000) Curr. Microbiol. 40, 279-282. [DOI] [PubMed] [Google Scholar]
- 11.Hendson M., Purcell, A. H., Chen, D., Smart, C., Guilhabert, M. & Kirkpatrick, B. (2001) Appl. Environ. Microbiol. 67, 895-903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Simpson A. J., Reinach, F. C., Arruda, P., Abreu, F. A., Acencio, M., Alvarenga, R., Alves, L. M., Araya, J. E., Baia, G. S., Baptista, C. S., et al. (2000) Nature (London) 406, 151-157. [DOI] [PubMed] [Google Scholar]
- 13.Selkov E., Overbeek, R., Kogan, Y., Chu, L., Vonstein, V., Holmes, D., Silver, S., Haselkorn, R. & Fonstein, M. (2000) Proc. Natl. Acad. Sci. USA 97, 3509-3514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kyrpides N. C., Ouzounis, C. A., Iliopoulos, I., Vonstein, V. & Overbeek, R. (2000) Nucleic Acids Res. 28, 4573-4576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.DelVecchio V. G., Kapatral, V., Redkar, R. J., Patra, G., Mujer, C., Los, T., Ivanova, N., Anderson, I., Bhattacharyya, A., Lykidis, A., et al. (2002) Proc. Natl. Acad. Sci. USA 99, 443-448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kapatral V., Anderson, I., Ivanova, N., Reznik, G., Los, T., Lykidis, A., Bhattacharyya, A., Bartman, A., Gardner, W., Grechkin, G., et al. (2002) J. Bacteriol. 184, 2005-2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kitajima E. W., M., Bakarcic, M. & Fernandez-Valiela, M. V. (1975) Phytopathology 65, 476-479. [Google Scholar]
- 18.Purcell A. H., Finlay, A. H. & McClean, D. L. (1979) Science 206, 839-841. [DOI] [PubMed] [Google Scholar]
- 19.Davis M. J., French, W. J. & Schaad, N. W. (1981) Curr. Microbiol. 6, 309-314. [Google Scholar]
- 20.Backus E. A. (1985) in The Leafhoppers and Planthoppers, eds. Nault, L. R. & Rodriguez, J. G. (Wiley, New York), pp. 163–194.
- 21.Purcell A. H. & Suslow, K. G. (1988) Phytopathology 78, 1541-1545. [Google Scholar]
- 22.Ojanen-Reuhs T., Kalkkinen, N., Westerlund-Wikstrom, B., van Doorn, J., Haahtela, K., Nurmiaho-Lassila, E. L., Wengelnik, K., Bonas, U. & Korhonen, T. K. (1997) J. Bacteriol. 179, 1280-1290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Barber C. E., Tang, J. L., Feng, J. X., Pan, M. Q., Wilson, T. J., Slater, H., Dow, J. M., Williams, P. & Daniels, M. J. (1997) Mol. Microbiol. 24, 555-566. [DOI] [PubMed] [Google Scholar]
- 24.Wilson T. J., Bertrand, N., Tang, J. L., Feng, J. X., Pan, M. Q., Barber, C. E., Dow, J. M. & Daniels, M. J. (1998) Mol. Microbiol. 28, 961-970. [DOI] [PubMed] [Google Scholar]
- 25.Guo L., Lim, K. B., Poduje, C. M., Daniel, M., Gunn, J. S., Hackett, M. & Miller, S. I. (1998) Cell 95, 189-198. [DOI] [PubMed] [Google Scholar]
- 26.Gibbons H. S., Lin, S., Cotter, R. J. & Raetz, C. R. (2000) J. Biol. Chem. 275, 32940-32950. [DOI] [PubMed] [Google Scholar]
- 27.Hinton J. C., Gill, D. R., Lalo, D., Plastow, G. S. & Salmond, G. P. (1990) Mol. Microbiol. 4, 1029-1036. [DOI] [PubMed] [Google Scholar]
- 28.Huang J. H. & Schell, M. A. (1990) J. Bacteriol. 172, 3879-3887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bernal A., Ear, U. & Kyrpides, N. (2001) Nucleic Acids Res. 29, 126-127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Meidanis J., Braga, M. D. & Verjovski-Almeida, S. (2002) Microbiol. Mol. Biol. Rev. 66, 272-299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.da Silva A. C., Ferro, J. A., Reinach, F. C., Farah, C. S., Furlan, L. R., Quaggio, R. B., Monteiro-Vitorello, C. B., Van Sluys, M. A., Almeida, N. F., Alves, L. M., et al. (2002) Nature (London) 417, 459-463. [DOI] [PubMed] [Google Scholar]
- 32.Bhattacharyya, A., Stilwagen, S., Reznik, G., Feil, H., Feil, W., Anderson, I., Bernal, A., D'Souza, M., Ivanova, N., Kapatral, V., et al. (2002) Genome Res., in press. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.