Abstract
The ability to manipulate the vaccinia virus (VAC) genome, as a plasmid in bacteria, would greatly facilitate genetic studies and provide a powerful alternative method of making recombinant viruses. VAC, like other poxviruses, has a linear, double-stranded DNA genome with covalently closed hairpin ends that are resolved from transient head-to-head and tail-to-tail concatemers during replication in the cytoplasm of infected cells. Our strategy to construct a nearly 200,000-bp VAC-bacterial artificial chromosome (BAC) was based on circularization of head-to-tail concatemers of VAC DNA. Cells were infected with a recombinant VAC containing inserted sequences for plasmid replication and maintenance in Escherichia coli; DNA concatemer resolution was inhibited leading to formation and accumulation of head-to-tail concatemers, in addition to the usual head-to-head and tail-to-tail forms; the concatemers were circularized by homologous or Cre–loxP-mediated recombination; and E. coli were transformed with DNA from the infected cell lysates. Stable plasmids containing the entire VAC genome, with an intact concatemer junction sequence, were identified. Rescue of infectious VAC was consistently achieved by transfecting the VAC–BAC plasmids into mammalian cells that were infected with a helper nonreplicating fowlpox virus.
Poxviruses, which comprise a large family of double-stranded DNA viruses that infect vertebrate and invertebrate hosts, are distinguished by their large size, complex morphology, cytoplasmic site of replication, and encoding of proteins for viral transcription, replication, and immune defense (1). Vaccinia virus (VAC), the best-studied member of the poxvirus family, was used for immunization against smallpox (2), and subsequently as an expression vector for laboratory investigations and recombinant vaccines (3). The introduction of new genes into the VAC genome is usually carried out by homologous recombination in infected cells. Typically, a foreign gene is cloned into a plasmid transfer vector downstream of a VAC promoter and flanked by VAC DNA from a nonessential site (4). The plasmid is then transfected into cells that are infected by VAC. Because recombination is inefficient, a variety of selection and screening methods have been devised (5). The process usually requires three to five plaque purifications over a period of several weeks. Alternative methods, involving the purification and cleavage of the VAC genome and either ligation in vitro or three-way recombination in vivo, have been described (6–10), but are cumbersome, depend on the presence of a unique restriction endonuclease site within the nearly 200,000-bp genome, and still require plaque purification. The mutagenesis of VAC genes is even more difficult, and is usually carried out by a laborious transient dominant selection recombination scheme (11).
Recently, the large DNA genomes of a baculovirus (12) and several herpesviruses (13–18) have been cloned as bacterial artificial chromosomes (BACs). These circular miniF BAC plasmids allow viral genomes to be stably maintained at low copy number and manipulated in Escherichia coli and then reconstituted as infectious virus by transfection of eukaryotic cells. The construction of baculovirus and herpesvirus BACs was relatively straightforward: plasmid sequences were inserted by recombination into the circular mature or replicating viral genomes and the latter were propagated in E. coli. Poxvirus genomes, however, are composed of linear double-stranded DNA molecules with covalently closed hairpin ends (19) that are resolved from transient head-to-head or tail-to-tail concatemers during replication (20–22). Therefore, the method used for cloning baculovirus and herpesvirus genomes seemed inapplicable.
We knew, however, that circular bacterial plasmids that contain the poxvirus concatemer junction sequence are resolved into linear minichromosomes when transfected into poxvirus-infected cells (23, 24). It seemed likely to us that a much larger plasmid containing the entire poxvirus genome and concatemer junction would also be resolved. Although it may be possible to assemble such a 200,000-bp plasmid by in vitro methods, we chose an alternative approach that depended on an observation made several years ago. It was found that poxvirus DNA concatemers accumulated under conditions in which viral late protein synthesis was specifically inhibited and that recombination occurred so that about half of the concatemers were in the head-to-tail configuration (25, 26). The latter configuration was crucial because a circle formed from a head-to-tail concatemer would contain the entire genome, whereas a circle formed from head-to-head or tail-to-tail concatemers would not. In principal, head-to-tail concatemers might undergo further recombination to form circles, though the latter have not been described. Alternatively, the highly efficient bacteriophage Cre–loxP recombinase system (27) could be used to specifically enhance circularization. A loxP site is composed of a 34-bp sequence consisting of a core spacer of 8 bp and two 13-bp palindromic flanking sequences. When two cis loxP sites are in the same orientation, the DNA between them can be excised and circularized by Cre. One strategy was to clone the loxP sites so that they would be in the same orientation in head-to-tail concatemers but opposite for head-to-head or tail-to-tail concatemers.
Here, we describe the formation and isolation of circular plasmids containing the full-length VAC genome with an intact concatemer junction sequence. These VAC–BACs were stably propagated in E. coli and converted into infectious virus in mammalian cells. VAC–BACs should allow the generation of mutant or recombinant viral genomes in bacteria, without need for recombination or plaque purification in mammalian cells.
Materials and Methods
Cells Lines and Viruses.
Monolayers of BS-C-1 and CV-1 African green monkey cells, and human A543 cells were grown in Dulbecco's modified Eagle medium or Eagle minimal medium supplemented with 5% or 10% FCS (GIBCO). The VAC strain WR ts21 mutant (28) was prepared by infecting confluent BS-C-1 monolayers at 31°C with 0.01 plaque-forming units (PFU) of virus per cell. The attenuated fowlpox virus HP1.441 (29) was provided by G. Sutter, and was grown in chicken embryo fibroblasts.
Plasmids.
The miniF BAC plasmid pMBO1374 (30), a derivative of pMBO131 (31), was provided by G. A. Smith. A DNA fragment containing NotI–loxP–BglII–HaeIII–SacII sites was formed by annealing synthetic oligonucleotides 5′ataggcatGCGGCCGCATAACTTCGTATAATGTATGCTATACGAAGTTATAGATCTGGCCCCGCGGgga and 5′tccCCGCGGGGCCAGATCTATAACTTCGTATAGCATACATTATACGAAGTTATGCGGCCGCatgcctat (sequences in italics and lowercase letters are restriction endonuclease sites and additional bases, respectively), digested with SacII and NotI, and ligated to SacII- and NotI-cleaved pMBO1374 resulting in pMBO1374–loxP.
Left (531 bp) and right (263 bp) segments of the VAC thymidine kinase (TK) gene were amplified by PCR using the primer pairs 5′-acatGCATGCATGAACGGCGGACATATTCAGTTGATAATCGGCCCC (L1–SphI) and 5′-cgcGGATCCCAACAATGTCTGGAAAGAACTGTCCTTCATCGATACCTATC (L2-BamHI) and the primer pairs 5′-agcatGGATCCAATTCTGTGAGCGTATGGCAAACGAAGGAAAAATAG (R1–BamHI) and 5′-gggGCATGCTGAGTCGATGTAACACTTTCTACACACCGAT (R2–SphI), respectively. The two fragments were digested with SphI and ligated together to form a 543-bp DNA in which the orientation of the left and right TK segments was reversed. This TKRL fragment was gel purified, digested with BamHI and then ligated to BamHI-digested and shrimp alkaline phosphatase-treated pMBO1374–loxP to form pMBO1374–loxP–TKRL (Fig. 1).
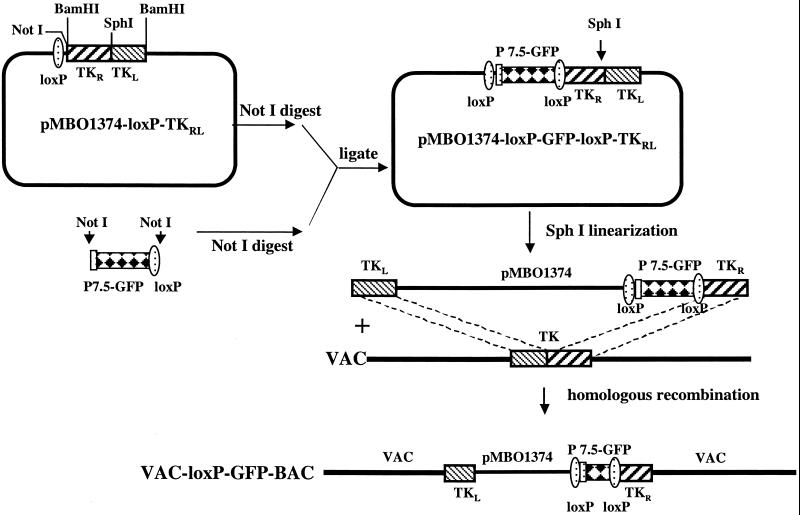
Fig 1.
Scheme for construction of VAC–loxP–GFP–BAC. The plasmid pMBO1374–loxP was constructed by addition of a synthetic oligonucleotide containing a loxP site between the NotI and SacII sites of pMBO1374. pMBO1374–loxP–TKRL was derived from pMBO1374–loxP by insertion of a BamHI fragment containing the inverted halves of the TK gene, separated by a SphI site. pMBO1374–loxP–TKRL was then cleaved with NotI and ligated to a NotI fragment containing the VAC P7.5 promoter regulating GFP to form pMBO1374–loxP–GFP–loxP–TKRL. The latter plasmid was cleaved with SphI to form a linear DNA flanked by the left and right halves of the TK gene, which was transfected into cells that were infected with either VAC strain WR or ts21 at 37°C or 31°C, respectively, to allow homologous recombination. VAC–loxP–GFP–BAC was isolated by TK-negative selection, and plaques exhibiting green fluorescence were picked several times in succession. The recombinant virus was called WR–loxP–GFP–BAC or ts21–loxP–GFP–BAC, depending on the parental virus strain.
The ORF encoding enhanced green fluoresent protein (GFP) was amplified by PCR from pEGFP-N1 (CLONTECH) using 5′-tgaGTCGACATGGTGAGCAAGGGCGAGGAGCTGTTCA and 5′-ataggcatGCGGCCGCCCGCGGTTACTTGTACAGCTCGTCCATGCCGAGAGTGATCC primers, digested with SalI and NotI, and gel purified. A three-way ligation was carried out with the latter fragment, the SacII- and NotI-digested NotI–loxP–BglII–HaeIII–SacII DNA, and SalI- and NotI-digested pSC11 S-B-A-K-N (5) to form pSC11-GFP–loxP in which the VAC P7.5 promoter regulates GFP. The P7.5–GFP–loxP fragment was amplified by PCR from the latter plasmid by using the primers 5′-ataggcatGCGGCCGCCACTAATTCCAAACCCACCCGCTTTTTATAGTAAGT and 5′-ataggcatGCGGCCGCATAACTTCGTATAATGTATG. The PCR product was digested with NotI and inserted in the unique NotI site of pMBO1374–loxP–TKRL to form pMBO1374–loxP–GFP–loxP–TKRL (Fig. 1), which was used to produce a recombinant VAC.
The pOG231 expression plasmid, containing Cre-fused to a nuclear localization signal (31), was provided by G. A. Smith. The Cre-expression cassette, excluding the nuclear localization signal, was subcloned by PCR using the following primers: 5′-ccgCTCGAGGCCACCATGTCCAATTTACTGACCGTACACCAAAATTTGCCTGCATTACCGGTC and 5′-tccCCCGGGCTAATCGCCATCTTCCAGCAGGCGCACCATTGCCCC. The Cre cassette was inserted in XhoI- and SmaI-digested pCIneo (Promega) to form pCI–Cre in which the Cre gene was under the control of a cytomegalovirus promoter.
All constructs were analyzed by restriction enzyme digestions, and the inserted fragments and junctions were sequenced.
Construction of VAC WR- or ts21–loxP–GFP–BAC.
BS-C-1 cells in a 6-well plate were infected with 0.01 PFU of VAC WR or ts21 per cell. At 2 h after infection, the cells were transfected with 10 μl of Lipofectamine (Life Technologies) and 2–3 μg of pMBO1374-loxP–GFP-loxP–TKRL that had been cut once with SphI. After 48 h at 37°C for WR or the permissive temperature of 31°C for the ts mutant, the cells were harvested and the virus was released by three freeze–thaw cycles. TK− recombinant VAC was selected in TK− A543 cells in the presence of 25 μg of bromodeoxyuridine per ml of medium. Plaques that exhibited green fluorescence were picked at 48 h after infection and purified by 5 cycles of plaque isolation. The DNA of the recombinant VAC was analyzed by PCR and by digestion with the HindIII restriction enzyme.
Analysis of Concatemers by Pulsed-Field Gel Electrophoresis.
BS-C-1 cells in a 6-well plate were infected at a multiplicity of 3 PFU of ts21 or ts21–loxP–GFP–BAC and incubated for 24 h at 31°C or 40°C. The cells were harvested and recovered by low-speed centrifugation (1,000 × g for 5 min at 4°C), and 2 million cells were embedded per agarose plug. The DNA was extracted by using CHEF Genomic DNA Plug kit (Bio-Rad). Slices from the agarose plugs were sealed in 1% agarose and analyzed by pulsed-field electrophoresis in 1% agarose (pulsed-field certified agarose, Bio-Rad) in 0.5× TBE (Bio-Rad) at 4–5 V/cm for 20–22 h, with a switch time of 50 and 90 s.
In some experiments, the plugs were incubated with 50 units of HindIII in 0.5 ml of digestion buffer for 12 h at 37°C. The buffer was replaced with 0.5 ml of fresh digestion buffer and 50 units of HindIII for 8 h more at 37°C. After electrophoresis, the gel was stained with ethidium bromide, photographed, exposed to UV light for 5 min, transferred to a nylon membrane, and hybridized to 32P-labeled probes containing part of the VAC HindIII B, C, or A fragment.
Isolation of Viral DNA.
Infected cells from an individual well of a 6-well plate were harvested, collected by low-speed centrifugation, rinsed 2–3 times with PBS, suspended in 50 μl of 0.15 M NaCl/0.02 M Tris⋅HCl, pH 8.0/0.01 M EDTA buffer, followed by addition of 250 μl of 0.02 M Tris⋅HCl (pH 8.0)/0.01 M EDTA/0.75% SDS containing 0.65 mg of proteinase K per ml. After incubation for 10–12 h at 37°C, the samples were extracted twice with 1 volume of phenol, once with phenol-chloroform, and once with chloroform, and precipitated with ethanol, and the DNA was dissolved in 20 μl of Tris⋅EDTA.
Detection of Concatemer Junction Fragments.
DNA was digested with BstEII, subjected to electrophoresis through a 1% agarose gel, and analyzed by Southern blotting using a 32P-labeled probe containing the terminal 70-bp repeat sequence.
Cre-loxP Recombination and Transformation of E. coli.
BS-C-1 cells in individual wells of a 6-well plate were transfected with 2 μg of pCI–Cre and 10 μl of Lipofectamine and incubated at 37°C. At 24 h after transfection, the cells were infected with 5 PFU per cell of ts21–loxP–GFP–BAC and incubated for 24 h at 40°C. Alternatively, cells were infected with WR–loxP–GFP–BAC and incubated at 37°C in the presence of 45 μM of isatin-β-thiosemicarbazone per ml. The cells were harvested by scraping, and DNA was extracted as described above, dissolved in 20 μl of Tris⋅EDTA, and used to transform E. coli DH10B electrocompetent cells (GIBCO/BRL) in 0.1-cm cuvettes with the Gene Pulser electroporation system (Bio-Rad) at 1.8 kV, 200 ohms and 25 μF. Bacteria were recovered in 1 ml of SOC medium (GIBCO) at 37°C for 1 h, plated on LB-agar containing 50 μg of chloramphenicol and incubated overnight at 37°C. DNA was isolated by the alkaline lysis method, and in some cases was purified by polyethylene glycol precipitation.
Rescue of Infectious Virus.
CV-1 cells in a 12-well plate were infected with fowlpox virus (0.005 PFU per cell), transfected with 2 μg of VAC–BAC DNA, and incubated for 4–8 days at 31°C.
Results and Discussion
General Strategy for Constructing VAC–BACs.
The process consisted of (i) inserting miniF plasmid, loxP, and GFP sequences into the VAC genome by homologous recombination; (ii) infecting cells with the recombinant VAC and promoting the accumulation of head-to-tail concatemers by inhibiting viral late protein synthesis (25, 26) by using ts21, a temperature-sensitive VAC with a mutation in the RPO18 subunit of RNA polymerase (28, 32), or by adding isatin-β-thiosemicarbazone (33, 34); (iii) allowing genome circularization to occur by homologous or Cre-mediated recombination; and (iv) transforming E. coli. The TK site was chosen for insertion of DNA sequences because it is nonessential and has been used extensively for this purpose (4).
Cloning BAC Sequences into the VAC Genome.
A multistep procedure was used for construction of the plasmid transfer vector pMBO1374–loxP–GFP–loxP–TKRL, which contains a split VAC TK ORF with the two halves in reverse orientation and a GFP expression cassette flanked by loxP sites (Fig. 1). By digesting the plasmid with SphI, a linear DNA with the TK left (TKL) and TK right (TKR) sequences at the two ends was produced. This DNA was transfected into cells infected with wild-type VAC strain WR or ts21 at the permissive temperature of 37°C or 31°C, respectively, to allow homologous recombination (4). Recombinant virus was repeatedly purified from plaques that formed under TK− selection conditions and exhibited green fluorescence. Restriction enzyme analysis showed that the HindIII J fragment of the VAC genome, which contains the TK ORF, increased in size from about 5 kbp to 14 kbp (data not shown). The insertion of DNA into the TK locus was also demonstrated by PCR. The recombinant VAC–loxP–GFP–BACs were designated WR- or ts21- depending on the parental VAC strain.
Circularization of the VAC Genome.
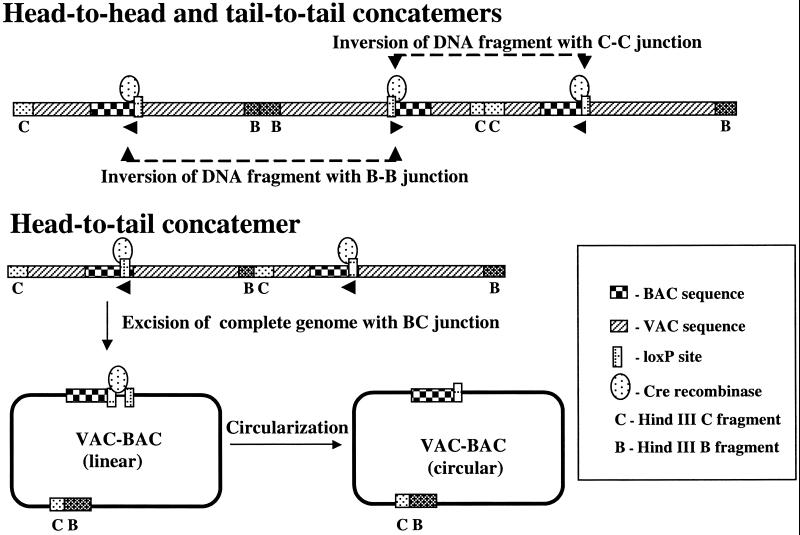
Initial efforts to make a VAC–BAC were carried out by using ts21–loxP–GFP–BAC. The accumulation of concatemeric DNA in cells infected with ts21–loxP–GFP–BAC at the nonpermissive temperature of 40°C was demonstrated by pulsed-field gel electrophoresis and by detection of concatemer junctions by restriction enzyme analysis and DNA hybridization (data not shown). To enhance circularization of the VAC genome, a Cre-expression plasmid was transfected into cells before infecting them with ts21–loxP–GFP–BAC. The GFP cassette, which is flanked by similarly oriented loxP sites, should be efficiently excised leaving a single loxP site in each VAC unit genome. A unit-length genome flanked by loxP sites in the same orientation would also be excised from the concatemer, whereas DNA inversion would occur between oppositely oriented loxP sites. As seen in Fig. 2, loxP sites in the same orientation occur only in head-to-tail (BC) concatemers. After excision, the DNA would be circularized by Cre to form a plasmid containing a complete VAC genome. Note that even if excision occurred with the oppositely oriented loxP sites flanking BB or CC junctions, the products would not include an entire VAC genome and would either lack BAC sequences or contain two unstable copies.
Fig 2.
Representation of the action of Cre on head-to-head, tail-to-tail, and head-to-tail concatemers. B and C represent the terminal HindIII fragments of the VAC genome. CC, BB, and CB represent head-to-head, tail-to-tail, and head-to-tail concatemer junctions, respectively. Because of the orientation of the loxP sites, excision, and circularization of a complete viral genome only occurs with head-to-tail junctions.
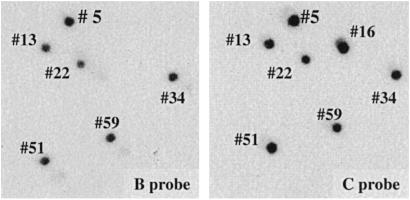
E. coli were electroporated with DNA from the lysates of cells that had been infected with ts21–loxP–GFP–BAC and then plated on chloramphenicol selective medium. Bacterial colonies were picked, transferred to duplicate membranes, and hybridized with 32P-labeled probes containing sequences from the terminal HindIII C or B fragments of the VAC genome. Of 117 colonies analyzed, DNA from six (nos. 5, 13, 22, 34, 51, and 59) reacted with both probes, one (no. 16) reacted with the C probe alone, and none reacted with the B probe alone (Fig. 3). Colonies that did not react with either probe were not extensively examined, but most had relatively small fragments of VAC DNA.
Fig 3.
Screening of bacterial colonies for VAC–BACs. After transformation of E. coli with DNA from cells infected with ts21–loxP–GFP–BAC in the presence of Cre, individual colonies were screened by hybridization to 32P-labeled probes containing DNA from the HindIII C and B terminal fragments of the VAC genome. Colonies number 5, 13, 16, 22, 34, 51, and 59 reacted with the C probe and, except for number 16, reacted with the B probe.
Analysis of VAC–BAC DNA.
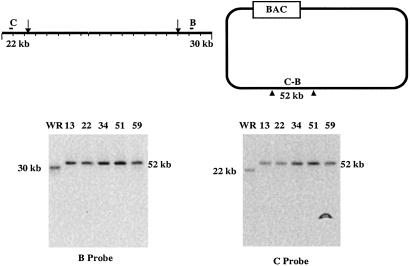
As diagrammed in Fig. 4, digestion of the linear VAC genome with HindIII should release terminal B and C fragments of 30 and 22 kbp, respectively (35). By contrast, the HindIII C and B sequences of the circular VAC–BAC genome should be fused through the concatemer junction resulting in a combined HindIII CB fragment of 52 kbp. DNA from five VAC–BAC clones and that of authentic VAC genomic DNA were analyzed. The C and B sequences of the VAC–BAC clones were released as a single fragment of the expected size (Fig. 4).
Fig 4.
The terminal HindIII C and B sequences of VAC DNA are fused in the VAC–BAC. Representations of the linear VAC genome and circular VAC–BAC DNA are shown in the upper left and right, respectively. DNA from five VAC–BAC clones and that of VAC genomic DNA were digested with HindIII, resolved by agarose gel electrophoresis, transferred to a membrane and hybridized to 32P-labeled probes containing sequences from the HindIII B or C fragment of the VAC genome. The B and C probes reacted with fragments of 30 and 22 kbp of VAC WR genomic DNA but with a 52-kbp fragment of the VAC–BACs.
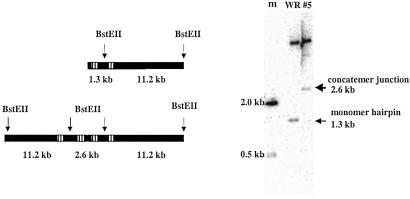
Evidence that the concatemer junction links the C and B sequences was obtained by analyzing the digestion products of BstEII, a restriction enzyme that cuts near the hairpin ends of the mature genome. As depicted in Fig. 5 Left, BstEII cleaves 1.3 kbp from the hairpin end of the mature genome or 1.3 kbp on either side of the concatemer junction. As expected, a 1.3-kbp terminal fragment was detected when WR genomic DNA was analyzed, whereas a double-size 2.6-kbp junction fragment was detected from VAC–BAC DNA (Fig. 5 Right).
Fig 5.
Detection of the concatemer junction fragment in VAC–BAC DNA. The hairpin terminus and concatemer junctions are represented in the left upper and lower portions of the figure. The sites of BstEII cleavage and the resulting size fragments are indicated. The boxes with vertical lines represent tandem repeat sequences. DNA from VAC (WR) and VAC–BAC number 5 were digested with BstEII, and the products were resolved by agarose gel electrophoresis, transferred to a membrane, and probed with 32P-labeled DNA containing tandem repeat sequences located close to the two ends of the genome. An autoradioraph is shown on the right. m, is DNA size markers. The 1.3-kbp hairpin terminus was detected in WR DNA, and the 2.6-kbp concatemer junction in the VAC–BAC. The upper band in both the WR and #5 lanes was generated by cleavage at a distal BstEII site not shown in the diagram.
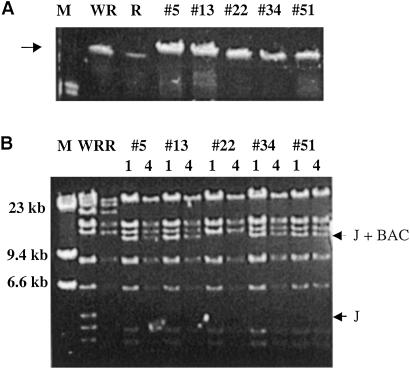
To determine whether the VAC–BACs contained full-length viral genomes, five were digested with ApaI restriction enzyme, which only cuts within the BAC sequence, and analyzed by pulsed-field gel electrophoresis. As seen in Fig. 6A, the mobilities of the cloned DNAs were similar to each other and to VAC genomic DNA.
Fig 6.
Size and stability of VAC–BAC DNA. (A) DNA of VAC (WR), ts21–loxP–GFP–BAC (R) and numbered VAC–BACs were analyzed by pulsed-field gel electrophoresis. Similar bands (arrow) of approximately 200,000 bp were detected by ethidium bromide staining. Only a segment of the gel is shown. (B) The same DNA samples were digested with HindIII and analyzed by agarose gel electrophoresis. Note that the BC fragments of the VAC–BACs migrate with the A fragment. In addition, the J-size fragment of WR (lower arrow) is missing in ts21–loxP–GFP–BAC and all of the VAC–BACS because of inserted DNA. In VAC–BACs 5, 13, 34, and 51, the positions of the more slowly migrating J fragment containing BAC sequences is shown by the upper arrow. In ts21–loxP–GFP–BAC and VAC–BAC 22, the J fragment also contains the GFP gene, and therefore migrates more slowly and coincides with the next higher band.
DNA stability was determined for five clones during four successive overnight cultures. The HindIII restriction patterns of DNA isolated from the first and the fourth culture were identical in each case (Fig. 6B). As expected, the 53-kbp HindIII A fragment and the 52-kbp HindIII CB junction fragment comigrated at the top of the gel. The 12.8-kbp fragment, indicated by the upper arrow, was derived from the 5-kbp HindIII J fragment of VAC and contained BAC sequences. In clone 22 (and ts21–loxP–GFP–BAC), the corresponding fragment also contained the GFP cassette, which in this case had not been excised by Cre and therefore migrated with the next higher band of about 14 kbp. The latter result suggested that the level of Cre may have been limiting and that the efficiency of VAC–BAC formation might be improved by higher expression.
Rescue of Infectious Virus.
The next step was reconstitution of infectious virus from the VAC–BACs. Poxviruses replicate and transcribe their genomes in the cytoplasm, rather than the nucleus of infected cells. To accomplish this, poxviruses encode a unique transcription system that includes a multisubunit DNA-dependent RNA polymerase, stage-specific promoters, and cognate transcription factors (1). The viral RNA polymerase and early transcription factors are packaged within infectious particles to begin a new infection. Therefore, an infection cannot be initiated with viral DNA alone, and consequently, a helper virus that provides early functions must be provided. For example, rabbitpox virus was recovered from rabbitpox DNA when the transfected cells were infected with a temperature-sensitive mutant of ectromelia virus (36). Similarly, wild-type VAC was recovered when VAC DNA was transfected into cells that were infected with a temperature-sensitive mutant of VAC (6) or the genetically distant fowlpox virus, which cannot replicate in mammalian cells or undergo recombination with VAC (7).
The previous rescue experiments were done with linear viral genomes with hairpin ends. Based on studies with cloned concatemer junctions (23, 24), however, we thought that the VAC–BACs would be replicated and resolved into linear genomes. CV-1 cells were infected with a low multiplicity of fowlpox virus, transfected with VAC–BAC DNA, and incubated for 4–8 days at 31°C. The recovery of infectious virus was determined by plaque assay on BS-C-1 cells, which like CV-1 cells, are nonpermissive for fowlpox virus. Comparable amounts of infectious virus were recovered from each of the VAC–BAC clones, whereas no virus was detected when either fowlpox virus or VAC–BAC was omitted (Table 1). As expected from the analysis of the parental plasmid, virus derived from clone 22 expressed GFP (data not shown). The HindIII restriction enzyme digestion patterns of the virus clones were similar to each other, and no fowlpox virus DNA was detected by Southern blotting (data not shown). Thus, a poxvirus genome was successfully cloned and replicated as a bacterial plasmid and rescued as infectious virus.
Table 1.
Rescue of infectious VAC
| DNA (2 μg) | Fowlpox virus | PFU/well |
|---|---|---|
| VAC–BAC #5 | + | 675 |
| VAC–BAC #13 | + | 900 |
| VAC–BAC #22 | + | 1,125 |
| VAC–BAC #51 | + | 338 |
| VAC–BAC #59 | + | 495 |
| VAC–BAC #5 | − | 0 |
| None | + | 0 |
Alternative Methods of Producing VAC–BACs.
In principal, circular genomes could also arise by homologous recombination of head-tail-concatemers either in the infected cells or in the bacteria after transfection. To evaluate this possibility, cells were transfected with ts21–loxP–GFP–BAC without the Cre-expressing plasmid. Under these conditions, one of 100 plasmids analyzed contained a complete VAC genome capable of producing infectious virus.
The VAC–BACs described thus far contain the ts21 mutation. Although this mutation could be eliminated by homologous recombination, we succeeded in producing VAC–BACs starting from the wild-type WR genome. A similar strategy was used except that isatin-β-thiosemicarbazone was used to accumulate head-to-tail concatemers.
We also attempted to make VAC–BACs without preventing concatemer resolution, with the thought that low amounts of head-to-tail concatemers might occur naturally. Several hundred plasmid clones were analyzed and some with nearly complete genomes were identified, leading us to believe that complete genomes might be recovered after a more extensive search. Thus, although Cre-mediated recombination and inhibition of concatemer resolution increase the efficiency of VAC genome circularization, they are not essential for producing VAC–BACs.
Summary and Conclusions.
The ability to propagate the entire VAC genome as a BAC makes it possible to modify or delete VAC genes or add foreign DNA by using a variety of methods developed for bacterial systems (37). Moreover, by eliminating the need for plaque purification, production of recombinant viruses for library screening or other high throughput purposes should be facilitated.
Acknowledgments
We thank Mark Challberg for critical reading of the manuscript and helpful suggestions, and G. A. Smith and L. W. Enquist for plasmids.
Abbreviations
VAC, vaccina virus
BAC, bacterial artificial chromosome
TK, thymidine kinase
PFU, plaque-forming units
References
- 1.Moss B. (2001) in Fields Virology, eds. Knipe, D. M. & Howley, P. M. (Lippincott, Philadelphia), Vol. 2, pp. 2849–2883. [Google Scholar]
- 2.Fenner F., Henderson, D. A., Arita, I., Jezek, Z. & Ladnyi, I. D., (1988) Smallpox and its Eradication (World Health Organization, Geneva).
- 3.Moss B. (1996) Proc. Natl. Acad. Sci. USA 93, 11341-11348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mackett M., Smith, G. L. & Moss, B. (1984) J. Virol. 49, 857-864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Earl P. L., Moss, B., Wyatt, L. S. & Carroll, M. W. (1998) in Current Protocols in Molecular Biology, eds. Ausubel, F. M., Brent, R., Kingston, R. E., Moore, D. D., Seidman, J. G., Smith, J. A. & Struhl, K. (Greene & Wiley, New York), Vol. 2, pp. 16.17.1–16.17.19. [Google Scholar]
- 6.Merchlinsky M. & Moss, B. (1992) Virology 190, 522-526. [DOI] [PubMed] [Google Scholar]
- 7.Scheiflinger F., Dorner, F. & Falkner, F. G. (1992) Proc. Nat. Acad. Sci. USA 89, 9977-9981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Merchlinsky M., Eckert, D., Smith, E. & Zauderer, M. (1997) Virology 238, 444-451. [DOI] [PubMed] [Google Scholar]
- 9.Smith E. S., Mandokhot, A., Evans, E. E., Mueller, L., Borrello, M. A., Sahasrabudhe, D. M. & Zauderer, M. (2001) Nat. Med. 7, 967-972. [DOI] [PubMed] [Google Scholar]
- 10.Timiryasova T. M., Chen, B., Fodor, N. & Fodor, I. (2001) BioTechniques 31, 534-540. [DOI] [PubMed] [Google Scholar]
- 11.Falkner F. G. & Moss, B. (1990) J. Virol. 64, 3108-3111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Luckow V. A., Lee, S. C., Barry, G. F. & Olins, P. O. (1993) J. Virol. 67, 4566-4579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Messerle M., Crnkovic, I., Hammerschmidt, W., Ziegler, H. & Koszinowski, U. H. (1997) Proc. Natl. Acad. Sci. USA 94, 14759-14763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Borst E. M., Hahn, G., Koszinowski, U. H. & Messerle, M. (1999) J. Virol. 73, 8320-8329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Delecluse H. J., Hilsendegen, T., Pich, D., Zeidler, R. & Hammerschmidt, W. (1998) Proc. Natl. Acad. Sci. USA 95, 8245-8250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Horsburgh B. C., Hubinette, M. M., Qiang, D., MacDonald, M. L. & Tufaro, F. (1999) Gene Ther. 6, 922-930. [DOI] [PubMed] [Google Scholar]
- 17.Saeki Y., Ichikawa, T., Saeki, A., Chiocca, E. A., Tobler, K., Ackermann, M., Breakefield, X. O. & Fraefel, C. (1998) Hum. Gene Ther. 9, 2787-2794. [DOI] [PubMed] [Google Scholar]
- 18.Smith G. A. & Enquist, L. W. (1999) J. Virol. 73, 6405-6414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Baroudy B. M., Venkatesan, S. & Moss, B. (1982) Cell 28, 315-324. [DOI] [PubMed] [Google Scholar]
- 20.Moyer R. W. & Graves, R. L. (1981) Cell 27, 391-401. [DOI] [PubMed] [Google Scholar]
- 21.Baroudy B. M., Venkatesan, S. & Moss, B. (1982) Cold Spring Harbor Symp. Quant. Biol. 47, 723-729. [DOI] [PubMed] [Google Scholar]
- 22.Merchlinsky M., Garon, C. & Moss, B. (1988) J. Mol. Biol. 199, 399-413. [DOI] [PubMed] [Google Scholar]
- 23.Merchlinsky M. & Moss, B. (1986) Cell 45, 879-884. [DOI] [PubMed] [Google Scholar]
- 24.Delange A. M., Reddy, M., Scraba, D., Upton, C. & McFadden, G. (1986) J. Virol. 59, 249-259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Merchlinsky M. & Moss, B. (1989) J. Virol. 63, 1595-1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.DeLange A. M. (1989) J. Virol. 63, 2437-2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hoess R., Abremski, K. & Sternberg, N. (1984) Cold Spring Harb. Symp. Quant. Biol. 49, 761-768. [DOI] [PubMed] [Google Scholar]
- 28.Seto J., Celenza, L. M., Condit, R. C. & Niles, E. G. (1987) Virology 160, 110-119. [DOI] [PubMed] [Google Scholar]
- 29.Mayr A. & Malicki, K. (1966) Zentralbl. Veterinarmed. Reihe B 13, 1-13. [PubMed] [Google Scholar]
- 30.O'Connor M., Peifer, M. & Bender, W. (1989) Science 244, 1307-1312. [DOI] [PubMed] [Google Scholar]
- 31.O'Gorman S., Dagenais, N. A., Qian, M. & Marchuk, Y. (1997) Proc. Natl. Acad. Sci. USA 94, 14602-14607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ahn B.-Y., Jones, E. V. & Moss, B. (1990) J. Virol. 64, 3019-3024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Woodson B. & Joklik, W. K. (1965) Proc. Natl. Acad. Sci. USA 54, 946-953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pennington T. H. (1977) J. Gen. Virol. 35, 567-571. [DOI] [PubMed] [Google Scholar]
- 35.Wittek R., Menna, A., Schumperli, D., Stoffel, S., Muller, H. K. & Wyler, R. (1977) J. Virol. 23, 669-678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sam C. K. & Dumbell, K. R. (1981) Ann. Virol. (Inst. Pasteur) 135 E, 135-150. [Google Scholar]
- 37.Lee E. C., Yu, D., Martinez de Velasco, J., Tessarollo, L., Swing, D. A., Court, D. L., Jenkins, N. A. & Copeland, N. G. (2001) Genomics 73, 56-65. [DOI] [PubMed] [Google Scholar]