Abstract
GTP cyclohydrolase I (GCHI) mediates the first and committing step of the pterin branch of the folate-synthesis pathway. In microorganisms and mammals, GCHI is a homodecamer of ≈26-kDa subunits. Genomic approaches identified tomato and Arabidopsis cDNAs specifying ≈50-kDa proteins containing two GCHI-like domains in tandem and indicated that such bimodular proteins occur in other plants. Neither domain of these proteins has a full set of the residues involved in substrate binding and catalysis in other GCHIs. The tomato and Arabidopsis cDNAs nevertheless encode functional enzymes, as shown by complementation of a yeast fol2 mutant and by assaying GCHI activity in extracts of complemented yeast cells. Neither domain expressed separately had GCHI activity. Recombinant tomato GCHI formed dihydroneopterin triphosphate as reaction product, as do other GCHIs, but unlike these enzymes it did not show cooperative behavior and was inhibited by its substrate. Denaturing gel electrophoresis verified that the bimodular GCHI polypeptide is not cleaved in vivo into its component domains, and size-exclusion chromatography indicated that the active enzyme is a dimer. The deduced tomato and Arabidopsis GCHI polypeptides lack overt targeting sequences and thus are presumably cytosolic, in contrast to other plant folate-synthesis enzymes, which are mitochondrial proteins with typical signal peptides. GCHI mRNA and protein are strongly in expressed unripe tomato fruits, implying that fruit folate is made in situ rather than imported. As ripening advances, GCHI expression declines sharply, and folate content drops, suggesting that folate synthesis fails to keep pace with turnover.
Tetrahydrofolate and its derivatives (folates) are essential cofactors for one-carbon transfer reactions in all organisms. Similar to bacteria and yeasts, plants make folates de novo from pterin, p-aminobenzoate (PABA), and glutamate moieties (1, 2). In contrast, humans and other mammals lack a complete folate-synthesis pathway and thus need dietary folate. Because plant foods are major folate sources, and folate deficiency is a global health problem, enhancing plant folate content is a prime target for metabolic engineering (1, 3). This engineering demands knowledge of the biosynthetic pathway.
The plant folate-synthesis pathway is not understood fully, but is most probably similar to that in bacteria (1, 2). The pterin hydroxymethyldihydropteroate is formed from GTP, and PABA from chorismate. The pterin and PABA units are condensed, glutamylated, and reduced to give tetrahydrofolate, and a polyglutamyl tail is added (4). Plant genes and enzymes for the final five steps have been characterized (5, 6), and the enzymes have been shown to be mitochondrial (7, 8). Far less is known for plants about the early steps that produce the pterin and PABA moieties.
The first step of pterin synthesis is of special interest, because it commits GTP to pterin production and is considered to control flux into the pathway (9, 10). This step, mediated by GTP cyclohydrolase I (GCHI, EC 3.5.4.16), is a complex ring expansion that converts GTP to dihydroneopterin triphosphate and formate (Fig. 1). GCHI has been cloned and characterized from Escherichia coli, yeast, and mammals (in mammals it participates in tetrahydrobiopterin synthesis), and the crystal structures of the E. coli and human enzymes have been solved (11–13). All the cloned GCHI polypeptides have 235 ± 15 residues and a molecular mass of 26 ± 2 kDa. E. coli and human GCHIs are homodecamers comprising five tightly associated dimers. There are 10 active sites per decamer, each located at the interface of three subunits and each with a catalytically essential zinc ion bound by histidine and cysteine side chains (11, 12). Plant GCHIs have not been cloned, but the enzyme has been isolated from spinach leaves and partially characterized (14).
Fig 1.
The reaction catalyzed by GCHI. This reaction entails opening of the five-membered imidazole ring, release of C-8 of GTP (•) as formate, and use of C-1′ and C-2′ of the ribose moiety of GTP to rebuild a six-membered dihydropyrazine ring. DHNTP, 7,8-dihydro-d-neopterin-3′-triphosphate.
Fruits are promising targets for folate engineering to improve human nutrition (1, 2). Fruit folate levels are so low to begin with that large gains seem possible. Fruit folate is generally highly bioavailable (15), and much fruit is consumed fresh, eliminating the large folate losses that occur in cooking (1). We have accordingly selected the model fruit, tomato, as well as Arabidopsis to investigate folate synthesis. In this study, we identified tomato and Arabidopsis GCHI cDNAs, and showed that they encode unique bimodular enzymes. Because nothing was known about folate synthesis in fruits, we analyzed GCHI expression and folate levels during fruit development.
Materials and Methods
Plants and Growth Conditions.
Tomato (Lycopersicon esculentum Mill. cv. MicroTom) plants were grown in a greenhouse (maximum temperature, 27°C) or a growth chamber (16-h days, irradiance 200 μmol of photons/m−2⋅s−1, 23°C/8-h nights, 20°C) in potting mix with appropriate fertilizer and pesticide regimes. Fruits and leaves were frozen in liquid N2 and held at −80°C until use.
Yeast Strains and Culture Conditions.
Strains 971/6c (Mata ade2-1 his3-11,15 leu2-3,112 ura3-1 can1) and 971/6a (Matα ade2-1 his3-11,15 leu2-3,112 ura3-1 can1 fol2:HIS3) (13) were obtained from M. L. Agostoni Carbone (Università di Milano, Milan). Yeast was cultured at 30°C in synthetic minimal medium (0.7% Difco yeast nitrogen base without amino acids, 2% glucose) plus appropriate supplements. When needed, (6R,6S)-5-formyltetrahydrofolate (Ca2+ salt, Schircks Laboratories, Jona, Switzerland) was added at a final concentration of 50 μg⋅ml−1. For enzyme extraction, 1-liter cultures were grown to an A600 value of ≈1.5. Cells were washed twice in water and once in 50 mM Tris⋅HCl, pH 8.0, frozen in liquid N2, and stored at −80°C.
cDNA Cloning and Expression.
PCR was made with Pfu Turbo (Stratagene) or DyNAzyme EXT (Finnzymes, Helsinki). Constructs were verified by sequencing. Expressed sequence tag AW738516 [encoding Lycopersicon esculentum (Le)GCHI] was obtained from CUGI (Clemson, SC). An Arabidopsis cDNA encoding Arabidopsis thaliana (At)GCHI was obtained from leaf mRNA by using SuperScriptII reverse transcriptase (Invitrogen) and amplified with the primers 5′-ATAATAGAATTCATTAAAGAGGAGAAATTAACTATGGGCGCATTAGATGAGGGATGTTTG-3′ and 5′-TATTATTATAGATCTTCAAAATGGAGAGCTTGACTCTGTCTTC-3′. The amplificate was digested with EcoRI and BglII and cloned into pNCO113 (16). The vector for yeast expression was pVT103-U (17). The LeGCHI coding sequence was amplified by using primers 5′-ACGCGGATCCATGGGCGCATTAGATGAA-3′ and 5′-CTGTCATCTTCCTGCAGAACC-3′; the separate domains were amplified by using these plus the internal primers 5′-CTGTCATTTCAGTAGACTCCAGAA-3′ or 5′-ACGCGGATCCATGTTCAGAGGTATTAGCATAGA-3′ for N- or C-terminal domains, respectively. The BamHI-digested amplificates were ligated between the BamHI and PvuII sites of pVT103-U and introduced into E. coli DH10B. For AtGCHI, pVT103-U was digested with BamHI and SacI and ligated with a synthetic double-stranded oligonucleotide (5′-GATCCATGGGCGCATTAGATGAGGGATGTTTGAATCTGGAGCT- 3′/5′-CCAGATTCAAACATCCCTCATCTAATGCGCCCATG-3′) to give pVT103-U-N, which was transformed into E. coli XL-1. The 1,380-bp SacI–PstI fragment of the pNCO113 construct then was cloned into pVT103-U-N, and the resulting plasmid was transformed into XL-1 cells. Yeast was transformed by using the YEASTMAKER system (CLONTECH). For retransformation tests, plasmids were isolated from yeast cells by a phenol-chloroform procedure and amplified in E. coli DH10B cells before reintroduction into yeast. To produce histidine-tagged LeGCHI in E. coli, the coding sequence was amplified by using the primers 5′-ACGTGGATCCAATGGGCGCATTAGATGAA-3′ and 5′-GCATCTCGAGTCTTCCTGCAGAACCAGA-3′, digested with BamHI and XhoI, cloned into pET-28b (Novagen), and transformed into DH10B cells. This construct then was expressed in BL21 (DE3) cells.
Protein Extraction and Analysis.
Operations were at 0–4°C. Pelleted yeast cells from 0.5 liters of culture were suspended in 2 ml of 0.1 M Tris⋅HCl, pH 8.0, and shaken with 0.5-mm zirconia-silica beads in a MiniBeadBeater (Biospec Products, Bartlesville, OK) at top speed for 6 × 30 s. The extracts were cleared by centrifugation (25,000 × g, 30 min) and desalted on PD-10 columns (Amersham Pharmacia) equilibrated in 50 mM Tris⋅HCl, pH 8.0, containing 0.1 M KCl and 10% glycerol. Extracts were routinely frozen in liquid N2 and stored at −80°C, which preserves GCHI activity. Tomato fruits were ground in liquid N2, extracted with 0.1 M Tris⋅HCl, pH 8.0, containing 5 mM EDTA and 3% polyvinyl-polypyrrolidone) and centrifuged (10,000 × g, 15 min) to clear. Native molecular mass was estimated by using a Waters 626 HPLC system equipped with a Superdex 200 HR 10/30 column (Amersham Pharmacia). The elution buffer was 50 mM Tris⋅HCl, pH 8.0, containing 0.1 M KCl. Protein was estimated by Bradford's method (18) using BSA as standard. Histidine-tagged LeGCHI from isopropyl β-D-thiogalactoside-induced E. coli cultures was isolated under denaturing conditions by using Ni2+-nitrilotriacetic acid resin (Qiagen); antibodies against the purified protein were raised in rabbits. SDS/PAGE and immunoblotting were as described (19); antiserum was diluted 1:1,000.
GCHI Assays.
GCHI activity was assayed by a procedure in which the reaction product, dihydroneopterin triphosphate, is oxidized and dephosphorylated to yield neopterin for HPLC analysis (20). Standard assays (40–200 μl) contained 50 mM Tris⋅HCl, pH 8.0, 0.1 M KCl, 1 mM GTP, and enzyme and were run at 37°C for 1 h. Reactions were stopped on ice with oxidizing solution (0.5% I2/1% KI in 1 M HCl, 10 μl per 100-μl reaction) and incubated for 1 h at 23°C before alkaline phosphatase treatment and deproteinization. [Tests showed that the high pH of the phosphatase step converts dihydroneopterin triphosphate to the phosphatase-resistant cyclic monophosphate (21, 22). Unless the triphosphate is hydrolyzed during the assay, as occurs by phosphatase action in yeast extracts, the cyclic form is therefore the major product.] Chromatography was a modification of the method of Lee et al. (23). Briefly, samples were injected onto an Ultramex C18 (ion pair) column (5 μm, 250 × 4.6 mm, Phenomenex, Belmont, CA) and eluted isocratically with 10 mM Na2HPO4, pH 6.0, at 1.2 ml⋅min−1. Peaks were detected by a Jasco FP 920 fluorescence detector (350-nm excitation, 450-nm emission). The neopterin peak was quantified relative to D-neopterin standards. Data were corrected for the recovery of 7,8-dihydro-D-neopterin-3′-monophosphate spikes added to assays before incubation; recoveries were ≈50%. Pterin standards were from Schircks Laboratories. Dihydroneopterin triphosphate was prepared by using recombinant E. coli FolE (12), oxidized, and converted to the cyclic monophosphate at pH ≈ 10 (21).
Folate Analysis.
Pilot studies using deconjugated samples (ref. 24, with modifications) showed that >90% of fruit folate was present as 5-methyltetrahydrofolate. However, because deconjugation can be inhibited by organic ions, the assay was adapted to measure 5-methyltetrahydrofolate polyglutamates. Folate was purified from 1.0–1.5 g of fruit (24) and analyzed by using reverse-phase fluorometric HPLC (25). Authentic 5-methyltetrahydrofolate monoglutamate (Eprova, Schaffhausen, Switzerland) was used as a quantitative standard, and folate extracts from human erythrocytes were used to identify the retention times of the 5-methyltetrahydrofolate polyglutamates.
Northern Analysis.
Total RNA was isolated from fruit and leaf samples (0.5–1.0 g) as described (26), separated on formaldehyde gels containing 1% agarose, and blotted to Nitropure membrane (Osmonics, Minnetonka, MN). Blots were hybridized overnight at 65°C in 5× SSPE/5× Denhardt's solution/0.5% SDS/100 μg⋅ml−1 fragmented salmon sperm DNA and washed at 37°C in 0.1× SSPE containing 1% SDS (standard saline phosphate/EDTA: 0.75 M NaCl/0.05 M phosphate, pH 7.4/5 mM EDTA. Blots were probed with the PCR-amplified coding sequence of LeGCHI and then stripped and reprobed with an 18S rRNA sequence (27). Probes were labeled with 32P by random priming. Radioactive LeGCHI mRNA bands were detected by autoradiography for 10 days.
Results
Higher Plant Homologs of GCHI Are Bimodular.
BLAST searches of GenBank and TIGR databases using the protein sequences of E. coli or mammalian GCHIs detected GCHI-like proteins encoded by an Arabidopsis gene (At3g07270) and by expressed sequence tag contigs from tomato (TC91226) and Medicago truncatula (TC36937). Remarkably, all three predicted proteins have two tandem GCHI domains that share more identity with various eukaryotic GCHIs than with each other (Fig. 2A). To confirm that such bimodular GCHI homologs actually exist in mRNA populations, we sequenced full-length Arabidopsis and tomato cDNAs. These cDNAs indeed coded for bimodular proteins (Fig. 2B). Alignments of the domains of the Arabidopsis and tomato proteins (henceforth, AtGCHI and LeGCHI) with the well characterized E. coli and human GCHIs revealed a second exceptional feature: neither domain has a complete set of the residues deemed essential for substrate binding and catalysis (11, 12), although all are present in the protein as a whole (Fig. 2B). The deduced M. truncatula protein shares these heterodox features (not shown). Furthermore, both domains of the plant enzymes lack an EF-hand-like calcium-binding motif that is conserved in all other eukaryotic GCHIs (28) and in fact have small insertions in the motif region (Fig. 2B).
Fig 2.
Primary structures of deduced plant GCHI polypeptides. (A) Scheme showing that the tandem plant GCHI-like domains share 34–42% identity with various eukaryotic GCHIs but only 26–28% identity with each other (arrows and % values). Red bars denote conserved sets of active site residues, and white bars show where these sets are not conserved in plant GCHIs. (B) Amino acid sequence alignment of E. coli GCHI (Ec, GenBank accession no. P27511), human GCHI (Hs, CAB77392), and the N- and C-terminal domains of tomato (Le, AY069920) and Arabidopsis (At, AF489530) GCHIs. Numbering is that of E. coli GCHI. Residues 1–30 of human GCHI are omitted. Identical residues are shaded in black, and similar ones are shaded in gray. Dashes are gaps introduced to maximize alignment. Triangles mark the conserved active site residues of the E. coli and human enzymes; red color denotes participation in zinc binding (11). Blue circles mark the Ca2+-coordinating residues of the EF-hand-like motif conserved in animal and yeast GCHIs (25).
AtGCHI and LeGCHI Are Functional Enzymes but Their Separate Domains Are Not.
To find whether the plant proteins have GCHI activity, their entire coding sequences were cloned into the yeast expression vector pVT103-U and introduced into yeast strain 971/6a. This strain is a fol2 deletant that lacks GCHI activity and is auxotrophic for folate (supplied as 5-formyltetrahydrofolate; ref. 13). Expression of the full-length plant proteins restored the ability to grow without folate (Fig. 3A). No complementation was seen with vector alone (Fig. 3A), and retransformation of 971/6a with rescued plasmid restored folate prototrophy, confirming that the complementation is caused by the encoded plant protein (not shown).
Fig 3.
Evidence that LeGCHI and AtGCHI encode functional enzymes. (A) Complementation of a yeast fol2 mutant. Similar numbers of cells of the wild-type 971/6c (WT), the fol2 deletant 971/6a (M), and 971/6a transformed with pVT103-U alone (V) or containing LeGCHI (Le) or AtGCHI (At) were plated on minimal medium plus or minus 50 μg⋅ml−1 (6R,6S)-5-formyltetrahydrofolate. The minimal medium contained adenine, histidine, leucine, uracil, and tryptophan. Plates were incubated for 3 days at 30°C. (B) GCHI activities in desalted extracts of wild-type yeast strain 971/6a (WT), and the fol2 deletant harboring pVT103-U (V) alone or containing LeGCHI (Le), AtGCHI (At), or the N-terminal (Le-N) or C-terminal (Le-C) domain of LeGCHI. The data are means of three replicates ± SE. (C) Response of recombinant LeGCHI activity to GTP concentration. Assays contained 28 μg of yeast-extract protein and were incubated at 37°C for 1 h; product formation was linear during this time. Data points are means of 3–6 replicates ± SE. (D) Comigration in reverse-phase HPLC of the GCHI assay product with authentic neopterin. N, neopterin; WT, product of the wild-type yeast enzyme; Le, product of LeGCHI. (E) Comigration of authentic neopterin triphosphate (T) and of the oxidized but not phosphatase-treated product of partially purified LeGCHI (Le).
GCHI activity in extracts of complemented 971/6a cells was assayed by a standard HPLC-fluorescence method in which the dihydroneopterin triphosphate reaction product is measured as its oxidized and dephosphorylated derivative, neopterin (20). Activity was detected readily in complemented cells at levels higher than in wild-type yeast but was not found in empty-vector controls (Fig. 3B). The assay product cochromatographed with authentic neopterin (Fig. 3D). To confirm that the direct product of plant GCHI is a triphosphate, the recombinant tomato enzyme was subjected to HPLC to remove endogenous phosphatases (see below), and phosphatase treatment was omitted from the sample work-up; the oxidation step was retained. As expected, this procedure yielded a reaction product that cochromatographed with neopterin triphosphate (Fig. 3E). Further confirmation that this product is the triphosphate was obtained by exposing it to alkaline conditions (pH ≈ 10), which gave a product that cochromatographed with cyclic neopterin monophosphate (not shown). This cyclization reaction is diagnostic for the triphosphate (21).
To test whether either of the domains can function on its own, those of LeGCHI were expressed separately in yeast strain 971/6a. The two domains were as shown in Fig. 2B except for the addition of a methionine at the start of the C-terminal domain. Neither domain conferred folate-independent growth (not shown) or produced detectable GCHI activity in cell extracts (Fig. 3B).
LeGCHI Does Not Show Cooperative Behavior and Is Inhibited by GTP.
Because mammalian and bacterial GCHIs show positive and negative substrate cooperativity, respectively (29, 30), we investigated the kinetic properties of recombinant LeGCHI. The GTP substrate used was chromatographically homogeneous by HPLC. Plots of initial velocity vs. GTP concentration showed no sign of cooperative behavior (which other GCHIs manifest within the range of GTP concentrations we used) but exhibited marked substrate inhibition (Fig. 3C). The apparent Km and Ki values for GTP were calculated as described by Cleland (31) to be 46 and 173 μM, respectively. The Km value is close to the GTP concentration that gives half-maximal velocity (K0.5) for the rat enzyme (29) and 50-fold higher than the K0.5 for E. coli GCHI (30). Substrate inhibition has not been reported for other GCHIs.
The LeGCHI Polypeptide Remains Intact in Vivo and Forms Active Dimers.
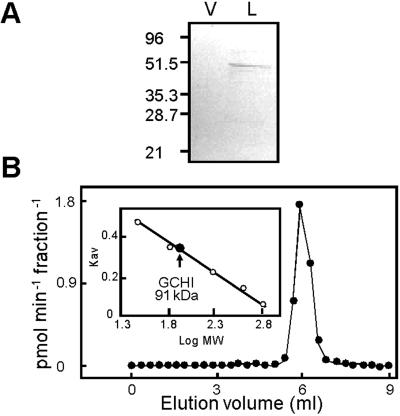
To check that plant GCHIs are not posttranslationally cleaved into their constituent domains, for which there is precedent in other proteins (32), antibodies raised against LeGCHI were used to probe immunoblots of recombinant LeCGHI from yeast extracts (Fig. 4A). The only band detected had a molecular mass of 51 kDa, which is very close to the predicted value of 50 kDa. This result confirms that the bimodular polypeptide exists as such in vivo.
Fig 4.
Molecular masses of denatured and native LeGCHI. (A) Immunoblot of proteins extracted from yeast strain 971/6a transformed with pVT103-U alone (V) or containing LeGCHI (L). Lanes contained 25 μg of protein; the positions of molecular mass markers (kDa) are shown. (B) Elution profile of recombinant LeGCHI activity from a Superdex 200 HR 10/30 column loaded with 0.25 mg of protein extracted from strain 971/6a expressing LeGCHI. The column calibration is shown (Inset). The molecular mass of GCHI was determined to be 91 ± 5 kDa (mean ± SE of four experiments). Standards were carbonic anhydrase (29 kDa), BSA (66 kDa), β-amylase (200 kDa), apoferritin (443 kDa), and thyroglobulin (669 kDa).
Because other GCHIs are decamers (11, 12), we investigated the native molecular mass of the plant enzyme. Size-exclusion HPLC showed that recombinant LeGCHI behaved as a single species with an apparent mass of 91 ± 5 kDa (Fig. 4B). The same value was obtained when the substrate GTP (0.1 mM) was added to the column buffer (not shown). These data indicate that the native enzyme is a dimer of 50-kDa subunits.
LeGCHI Is Expressed Strongly in Unripe but Not Ripe Fruit.
Because nothing is known about expression of the folate-synthesis pathway in fruits, GCHI transcript and protein levels were analyzed in tomatoes harvested at various developmental stages. Leaves were included as a benchmark, because they are known to have active folate synthesis (7, 14). We also measured the fruit levels of 5-methyltetrahydrofolate, which was shown to be >90% of total fruit folate. GCHI mRNA level was highest in unripe (mature green and breaker stage) fruit, when it was near that in leaves (Fig. 5A). As ripening advanced, the mRNA level fell sharply and was barely detectable when the fruit reached full color and was softening (red-ripe stage). GCHI protein likewise was abundant only in unripe fruit and disappeared even faster than the transcript (Fig. 5B). Levels of 5-methyltetrahydrofolate showed a significant downward trend (P = 0.06) during ripening (Fig. 5C).
Fig 5.
Developmental changes in levels of LeGCHI mRNA, LeGCHI protein, and folate in tomato fruits. (A) Northern analysis of LeGCHI mRNA. Lanes contained 10 μg of total RNA from fruits at mature green (MG), breaker (Br), ripe (R), and red-ripe (RR) stages or from young leaves. Blots were probed with the LeGCHI coding sequence (Upper) and then with an 18S ribosomal DNA sequence (Lower) as a loading control. The sizes (kb) of the hybridizing RNAs are indicated. (B) Western analysis of LeGCHI protein in fruits. Lanes contained 20 μg of protein; the positions of molecular mass markers (kDa) are shown. The Upper blot was probed with anti-GCHI serum and the Lower blot with control antiserum depleted in anti-GCHI antibodies by preincubation with purified recombinant LeGCHI. (C) 5-Methyltetrahydrofolate (methyl-THF) levels in fruits. The data are means of three replicates ± SE. FW, fresh weight.
Discussion
We have identified tomato and Arabidopsis cDNAs specifying GCHI, the first and committing enzyme in the pterin branch of the folate-synthesis pathway. This brings the total number of cloned plant folate-synthesis enzymes to six of a probable total of nine (2). The primary structure of plant GCHIs is without precedent in other organisms, comprising dual GCHI-like domains that are more diverged from one another than from other eukaryotic GCHIs. Such divergence implies that bimodular GCHIs are evolutionarily ancient, as do the phylogenetic distances between the species shown to have them—i.e., tomato, Arabidopsis, and M. truncatula (33).
Because biochemical and crystallographic evidence indicates that the E. coli and mammalian GCHI decamers can be viewed as pentamers made up of tightly associated dimers (11, 12, 34), it might seem that the dimer has merely become a covalently bonded unit in plants; but this is not so, because neither of the domains of the plant protein has all the residues identified in other GCHIs as essential for substrate binding and catalysis, and neither domain alone is enzymatically competent. Also, the bimodular plant protein dimerizes to form the active enzyme, and this quaternary structure precludes formation of the active site in the same way as in E. coli and mammalian GCHIs. Table 1 summarizes how the active site residues in these nonplant GCHIs come from three different subunits and shows for the plant enzymes how residues missing from one domain are present in the other such that the protein as a whole has a full set. The two cysteine and one histidine residues that coordinate the catalytically essential zinc ion (Table 1, bold italics) are a special case. Whereas all three are in canonical positions in the plant GCHI C-terminal domain, Cys-181 and His-113 have switched places in the N-terminal domain. Both domains of plant GCHIs thus may be able to bind zinc. Because plant GCHIs seem to violate principles upon which other GCHIs are constructed, determining their molecular structure will be of much interest from the standpoint of reaction mechanism.
Table 1.
Conservation of active site residues in the domains of LeGCHI and AtGCHI
| Residue |
Monomer |
Conservation of residues | |
|---|---|---|---|
| N-terminal domain | C-terminal domain | ||
| Arg-65 | A | + | + |
| Lys-68 | A | + | + |
| Cys-110 | B | + | + |
| His-112 | B | − | + |
| His-113 | B | (Cys) | + |
| Ile-132 | C | + | + |
| Ser-135 | C | + | +/− |
| Lys-136 | C | + | − |
| Arg-139 | C | + | − |
| Gln-151 | B | − | + |
| Glu-152 | B | − | + |
| His-179 | B | + | + |
| Cys-181 | B | (His) | + |
| Arg-185 | B | − | + |
Numbered by reference to E. coli GCHI. Residues involved in zinc binding are in bold italics.
A, B, and C denote the three different GCHI monomers that contribute the active site residues in the E. coli enzyme.
Plus (+) indicates conserved or conservatively replaced residues, and minus (−) indicates nonconserved residues; +/− indicates conservation in Arabidopsis but not tomato. Amino acids that replace His-113 and Cys-181 in the N-terminal domain are in parentheses.
In mammals, feedback inhibition of GCHI by the end product tetrahydrobiopterin is mediated by a pentameric GCHI feedback-regulatory protein (GFRP) that binds specifically to each of the two faces of the toroid-shaped GCHI decamer (35, 36). That plant GCHI has primary and quaternary structures so unlike those of mammals makes it improbable that the plant enzyme has the same type of feedback-inhibitory mechanism. Consistent with this, there are no obvious GFRP homologs in plant genome or expressed sequence tag databases. Moreover, plants do not produce tetrahydrobiopterin. Were mammalian GCHI expressed in plants, it therefore might be deregulated through lack of feedback control, which could provide a way to engineer increased flux to pterins and folate.
The last five steps in folate synthesis, from pterin activation onward, are all mitochondrial in plants, four of them exclusively so, and the corresponding enzymes have obvious transit peptides (5, 6). The lack of a transit sequence in GCHI thus makes it an exception and indicates that it is most probably a cytosolic enzyme. If GCHI indeed is cytosolic, it follows that mitochondria must import the pterin moiety of folate from the cytosol. Because genomic data suggest that the PABA moiety of folate is made in chloroplasts (2), coordinated reactions in three subcellular compartments and three membrane transport steps may be needed to produce folates in plants. This arrangement is not found in other eukaryotes, the folate pathway enzymes of which are predominantly cytosolic.
The strong expression of GCHI in unripe fruits implies that fruits synthesize their own supply of pterin—and, by extension, folate—rather than rely on import from leaves. This inference is corroborated by the existence of tomato fruit expressed sequence tags specifying three other folate-synthesis enzymes (e.g., GenBank AW223881, BF050827, and BE433834). The steep decline in GCHI expression after the mature green stage, when folate levels begin to fall, implies a collapse of the fruit's capacity to maintain its folate-synthesis machinery, and hence to sustain its folate levels in the face of ongoing turnover. The collapse in GCHI expression presumably is programmed, because ripening-related genes are still being induced when it occurs (37). Fruit folate content therefore might be enhanced by prolonging the expression of GCHI and perhaps other folate-synthesis enzymes.
Acknowledgments
We thank M. L. Agostoni Carbone for yeast strains and D. M. Tieman and M. G. Taylor for advice and help. This work was supported in part by the Florida Agricultural Experiment Station, by an endowment from the C. V. Griffin, Sr., Foundation, and by grants from the National Science Foundation (to A.D.H. and J.F.G.) and National Institutes of Health (to J.F.G.). Journal series no. R-08850.
Abbreviations
PABA, p-aminobenzoate
GCHI, GTP cyclohydrolase I
Le, Lycopersicon esculentum
At, Arabidopsis thaliana
References
- 1.Scott J. M., Rébeillé, F. & Fletcher, J. (2000) J. Sci. Food Agric. 80 795-824. [Google Scholar]
- 2.Hanson A. D. & Gregory, J. F., III (2002) Curr. Opin. Plant Biol. 5 244-249. [DOI] [PubMed] [Google Scholar]
- 3.DellaPenna D. (1999) Science 285 375-379. [DOI] [PubMed] [Google Scholar]
- 4.Green J. M., Nichols, B. P. & Matthews, R. G. (1996) in Escherichia coli and Salmonella—Cellular and Molecular Biology, ed. Neidhardt, F. C. (Am. Soc. Microbiol., Washington, DC), pp. 665–673.
- 5.Ravanel S., Cherest, H., Jabrin, S., Grunwald, D., Surdin-Kerjan, Y., Douce, R. & Rébeillé, F. (2001) Proc. Natl. Acad. Sci. USA 98 15360-15365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rébeillé F. & Douce, R. (1999) in Regulation of Primary Metabolic Pathways in Plants, eds. Kruger, N. T., Hill, S. A. & Ratcliffe, R. G. (Kluwer, Dordrecht, The Netherlands), pp. 53–99.
- 7.Neuburger M., Rébeillé, F., Jourdain, A., Nakamura, S. & Douce, R. (1996) J. Biol. Chem. 271 9466-9472. [DOI] [PubMed] [Google Scholar]
- 8.Rébeillé F., Macherel, D., Mouillon, J.-M., Garin, J. & Douce, R. (1997) EMBO J. 16 947-957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schoedon G., Redweik, U., Frank, G., Cotton, R. G. H. & Blau, N. (1992) Eur. J. Biochem. 210 561-568. [DOI] [PubMed] [Google Scholar]
- 10.Yoneyama T., Wilson, L. M. & Hatakeyama, K. (2001) Arch. Biochem. Biophys. 388 67-73. [DOI] [PubMed] [Google Scholar]
- 11.Auerbach G., Herrmann, A., Bracher, A., Bader, G., Gütlich, M., Fischer, M., Neukamm, M., Garrido-Franco, M., Richardson, J., Nar, H., et al. (2000) Proc. Natl. Acad. Sci. USA 97 13567-13572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nar H., Huber, R., Auerbach, G., Fischer, M., Hösl, C., Ritz, H., Bracher, A., Meining, W., Eberhardt, S. & Bacher, A. (1995) Proc. Natl. Acad. Sci. USA 92 12120-12125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nardese V., Gütlich, M., Brambilla, A. & Carbone, M. L. (1996) Biochem. Biophys. Res. Commun. 218 273-279. [DOI] [PubMed] [Google Scholar]
- 14.Sohta Y., Ohta, Y. & Masada, M. (1997) Biosci. Biotechnol. Biochem. 61 1081-1085. [Google Scholar]
- 15.Wei M.-M., Bailey, L. B., Toth, J. P. & Gregory, J. F., III (1996) J. Nutr. 126 3100-3108. [DOI] [PubMed] [Google Scholar]
- 16.Stüber D., Matile, H. & Garotta, G. (1990) in Immunological Methods, eds. Lefkovits, I. & Pernis, P. (Academic, New York), Vol. IV, pp. 121–152. [Google Scholar]
- 17.Vernet T., Dignard, D. & Thomas, D. Y. (1987) Gene 52 225-233. [DOI] [PubMed] [Google Scholar]
- 18.Bradford M. M. (1976) Anal. Biochem. 72 248-254. [DOI] [PubMed] [Google Scholar]
- 19.Nuccio M. L. & Thomas, T. L. (1999) Plant Mol. Biol. 39 1153-1163. [DOI] [PubMed] [Google Scholar]
- 20.Hatakeyama K. & Yoneyama, T. (1998) Methods Mol. Biol. 100 265-272. [DOI] [PubMed] [Google Scholar]
- 21.Plowman J., Cone, J. E. & Guroff, G. (1974) J. Biol. Chem. 249 5559-5564. [PubMed] [Google Scholar]
- 22.Cone J. & Guroff, G. (1971) J. Biol. Chem. 246 979-985. [PubMed] [Google Scholar]
- 23.Lee S. W., Lee, H. W., Chung, H. J., Kim, Y. A., Kim, Y. J., Hahn, Y., Chung, J. H. & Park, Y. S. (1999) FEMS Microbiol. Lett. 176 169-176. [DOI] [PubMed] [Google Scholar]
- 24.Gregory J. F., Manley, D. B. & Day, B. P. F. (1984) J. Nutr. 114 341-353. [DOI] [PubMed] [Google Scholar]
- 25.Pfeiffer C. M. & Gregory, J. F. (1996) Clin. Chem. 42 1847-1854. [PubMed] [Google Scholar]
- 26.Hamilton A. J., Lycett, G. W. & Grierson, D. (1990) Nature (London) 346 284-287. [Google Scholar]
- 27.Nuccio M. L., Ziemak, M. J., Henry, S. A., Weretilnyk, E. A. & Hanson, A. D. (2000) J. Biol. Chem. 275 14095-14101. [DOI] [PubMed] [Google Scholar]
- 28.Steinmetz M. O., Plüss, C., Christen, U., Wolpensinger, B., Lustig, A., Werner, E. R., Wachter, H., Engel, A., Aebi, U., Pfeilschifter, J. & Kammerer, R. A. (1998) J. Mol. Biol. 279 189-199. [DOI] [PubMed] [Google Scholar]
- 29.Hatakeyama K., Harada, T., Suzuki, S., Watanabe, Y. & Kagamiyama, H. (1989) J. Biol. Chem. 264 21660-21664. [PubMed] [Google Scholar]
- 30.Bracher A., Fischer, M., Eisenreich, W., Ritz, H., Schramek, N., Boyle, P., Gentili, P., Huber, R., Nar, H., Auerbach, G. & Bacher, A. (1999) J. Biol. Chem. 274 16727-16735. [DOI] [PubMed] [Google Scholar]
- 31.Cleland W. W. (1971) in The Enzymes, ed. Boyer, P. D. (Academic, New York), Vol. 2, pp. 35–43. [Google Scholar]
- 32.Sabourin L. A., Tamai, K., Seale, P., Wagner, J. & Rudnicki, M. A. (2000) Mol. Cell. Biol. 20 684-696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Takhtajan A., (1997) Diversity and Classification of Flowering Plants (Columbia Univ. Press, New York).
- 34.Yim J. J. & Brown, G. M. (1976) J. Biol. Chem. 251 5087-5094. [PubMed] [Google Scholar]
- 35.Yoneyama T. & Hatakeyama, K. (1998) J. Biol. Chem. 273 20102-20108. [DOI] [PubMed] [Google Scholar]
- 36.Bader G., Schiffmann, S., Herrmann, A., Fischer, M., Gütlich, M., Auerbach, G., Ploom, T., Bacher, A., Huber, R. & Lemm, T. (2001) J. Mol. Biol. 312 1051-1057. [DOI] [PubMed] [Google Scholar]
- 37.Gray J., Picton, S., Shabbeer, J., Schuch, W. & Grierson, D. (1992) Plant Mol. Biol. 19 69-87. [DOI] [PubMed] [Google Scholar]