Abstract
Tocopherol (vitamin E) is a plant chloroplast lipid presumed to be involved in the response to oxidative stress. A tocopherol-deficient mutant (vte1) was isolated from Arabidopsis thaliana by using a TLC-based screening approach. Mutant plants lacked all four tocopherol forms and were deficient in tocopherol cyclase activity. Genetic mapping of vte1 and a genomics-based approach led to the identification of the ORF At4g32770 as a candidate gene for tocopherol cyclase. In vte1, At4g32770 contains a splicing site mutation and the corresponding mRNA expression is reduced. Expression of VTE1 in Escherichia coli resulted in the production of a protein with high tocopherol cyclase and tocotrienol cyclase activity. The VTE1 sequence shows no similarities to genes with known function, but is similar to that of SXD1, a gene that was recently isolated based on the availability of the sucrose export defective1 maize mutant (sxd1). Growth of the vte1 mutant, chlorophyll content, and photosynthetic quantum yield were similar to wild type under optimal growth conditions. Therefore, absence of tocopherol has no large impact on photosynthesis or plant viability, suggesting that other antioxidants can compensate for the loss of tocopherol. During photo-oxidative stress, chlorophyll content and photosynthetic quantum yield were slightly reduced in vte1 as compared with wild type indicating a potential role for tocopherol in maintaining an optimal photosynthesis rate under high-light stress.
During their life cycle, higher plants are exposed to a variety of abiotic and biotic stress factors, including high temperature, drought, high light, and senescence. Under these stress conditions, reactive oxygen species derived from molecular oxygen (peroxide, hyperoxide, hydroxyl radicals, singlet oxygen) can accumulate in the leaves resulting in the oxidation of many important cellular components, including proteins, chlorophyll, and lipids, and may finally result in cell death (e.g., refs. 1–4). Different biochemical redox systems participate in the plant's response to oxidative stress—e.g., ascorbate, glutathione, salicylate, and tocopherol (5–8). Furthermore, transcription of stress-related genes is induced, resulting in the expression of enzymes involved in scavenging reactive oxygen species (9, 10). The role of the stress-induced accumulation of specific lipids is only poorly understood (11–13). In contrast to galactolipids and phospholipids, which are abundant in plant membranes, different lipids (triacylglycerol, free fatty acids, and tocopherol) accumulate in the leaves during stress.
Tocopherol represents an amphiphilic lipid that was proposed to be crucial for the oxidative stress response in plant membranes (5). The hydrophilic chromanol ring that is localized close to the lipid/water interface of membranes encompasses a hydroquinone ring system capable of undergoing redox reactions with reactive oxygen species (5, 14). Based on the number and position of methyl groups on the hydroquinone ring, four forms of tocopherol can be distinguished (α-, β-, γ-, δ-tocopherol). In addition to tocopherols, tocotrienols are found in small amounts in plant tissues. Tocotrienols carry an unsaturated geranylgeranyl side chain bound to the chromanol ring instead of the saturated phytyl chain. Their exact function in plants is unknown. In in vitro experiments, tocopherol was shown to be responsible for scavenging reactive oxygen species, thereby preventing the oxidative degradation of fatty acids in membranes (15–17). Because tocopherol is particularly enriched in chloroplast membranes (18, 19), it was proposed to be involved in the protection of chloroplast lipids and of chlorophyll against oxidative damage.
The eight different forms of tocopherol and tocotrienol (commonly referred to as vitamin E) represent an essential component of the human diet, with α-tocopherol showing the highest vitamin E activity (20, 21). Much effort was devoted to unraveling the role of vitamin E during oxidative stress in animals. Similar to plants, it is assumed that the main vitamin E function in mammals includes the scavenging of reactive oxygen species. Vitamin E deficiency in rats leads to infertility and fetal death (22). However, similar to the plant system, no clear evidence was obtained for an in vivo role of tocopherol during oxidative stress.
High-throughput biochemical screening methods have successfully been used to isolate plant mutants affected in lipid biosynthesis (e.g., refs. 23 and 24). The availability of such mutants was the basis for the isolation of the corresponding genes by chromosome walking (e.g., refs. 25 and 26). To elucidate the function of tocopherol during the oxidative stress response pathway, we initiated a screening approach for Arabidopsis plants carrying mutations in tocopherol biosynthesis. Employing a biochemical, TLC-based screening procedure, we were able to isolate a mutant totally devoid of tocopherol.
Materials and Methods
Isolation and Molecular Characterization of a Tocopherol-Deficient Mutant.
Arabidopsis thaliana (ecotype Col-2) M2 plants derived from ethyl methanesulfonate mutagenesis were grown on soil under standard conditions (120 μmol⋅m−2⋅s−1, 20°C, 60% air humidity, 16 h light/8 h dark). To induce the stress-related synthesis of lipids, leaves detached from individual plants were incubated overnight on a wet filter paper at 37°C. For screening, lipids were isolated from stressed leaves with 1 M KCl/0.2 M H3PO4 and diethylether, separated by TLC on silica plates with hexane/diethylether/acetic acid (90:10:1, vol/vol), and stained with iodine.
The vte1 mutant was four times back-crossed to WT Col-2 to reduce the number of background mutations. Genomic DNA was isolated from F2 plants derived from a cross to WT Landsberg erecta and used for PCR marker mapping (27, 28). The gene At4g32770 on chromosome 4 (BAC F4D11, GenBank accession no. AL022537) was amplified by PCR from vte1 with the oligonucleotides PD202 (AAA GGA AGG ATT TGT TTT GTT TGC GAC TG) and PD203 (GGA CCG AAC GAA CTA AAA TCA TAC ATA CA) and used for sequencing.
Northern Analysis, Heterologous Expression in Escherichia coli.
For Northern analysis, total RNA was isolated from leaves of WT and vte1 by using standard protocols (29, 30). The blot was hybridized to a 32P-labeled At4g32770 probe obtained by PCR amplification from genomic Arabidopsis WT DNA with the primers PD202 and PD203.
The mature part of the protein without the apparent signal peptide (31) was amplified by PCR from first-strand cDNA, using the oligonucleotide primers PD224 (TAA TGG ATC CAC TCC GGA CTC CTC ACA GTG G) and PD225 (GCT CAA GCT TTT ACA GAC CCG GTG GCT TGA A). The PCR fragment was ligated into the BamHI and HindIII sites of pQE31 and transferred into E. coli M15(pREP4) cells for protein expression (Qiagen, Hilden, Germany).
Enzyme Assay.
Tocopherol cyclase activity was measured with 2,3-dimethyl-5-phytyl-1,4-hydroquinone (DMPQ) or 2,3-dimethyl-5-geranylgeranyl-1,4-hydroquinone (DMGQ) (32). The tocopherol cyclase reaction with these substrates resulted in the synthesis of γ-tocopherol and γ-tocotrienol, respectively (32). Reaction products were separated by HPLC (Waters 2690) on a RP30 column (Bischoff Chromatography, Leonberg, Germany; 230 × 4.6 mm, 3.0 μm) with isocratic elution in 100% methanol and detected by fluorescence (excitation at 295 nm; emission at 332 nm).
Tocopherol Analysis.
For GC/MS analysis, Arabidopsis leaves were homogenized in liquid nitrogen and lipid extracted with 100 μl of 1 M KCl/0.2 M H3PO4 and 400 μl of diethylether. The organic phase was removed and the solvent evaporated in a nitrogen gas stream. After chemical modification with N-methyl-N-trimethylsilyltrifluoroacetamide, the trimethylsilyl derivatives of lipids were analyzed by GC/MS as described (33). Tocopherol was quantified by fluorescence HPLC with standards (34).
Measurement of Chlorophyll and Chlorophyll Fluorescence.
Chlorophyll was extracted from leaves with 80% (vol/vol) acetone and quantified by measuring the absorbances at 646 and 663 nm (35). In vivo chlorophyll fluorescence was determined with a pulse-amplitude modulation fluorimeter (PAM-2000, Heinz Walz, Effeltrich, Germany). The quantum yield was calculated from (Fm′ − Ft)/Fm′, where Ft and Fm′ are the fluorescence emissions of a light-adapted leaf under measuring light or after application of a saturating light pulse, respectively (36).
Results
Isolation of a Mutant Deficient in Tocopherol Synthesis.
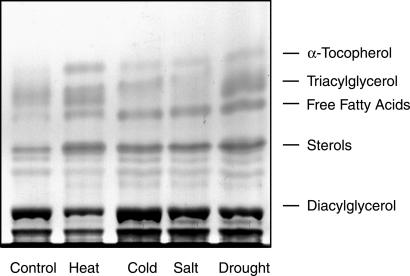
A genetic approach was chosen to further dissect the pathway of tocopherol synthesis. Different neutral lipids (free fatty acids, triacylglycerols, tocopherols) were found to accumulate during oxidative stress in leaves of higher plants (refs. 11–13; Fig. 1). The increase in tocopherol was particularly prominent, because its amount in leaves exposed to stress was up to 5 times higher as compared with control conditions. Based on these findings, a screening procedure for Arabidopsis mutants affected in tocopherol synthesis was developed. Detached leaves from single Arabidopsis plants were incubated overnight at 37°C resulting in a combination of different stress factors (i.e., heat, wounding). After screening 2,000 individual Arabidopsis M2 mutant plants for changes in lipid composition by TLC, a tocopherol-deficient line was identified. This line was tentatively designated vte1 (for vitamin E deficient).
Fig 1.
Accumulation of neutral lipids during stress conditions in leaves. Arabidopsis WT plants were exposed to different stress conditions (heat, 3 days at 37°C; cold, 5 days at 4°C; drought, 3 days without watering; salt, watering with 0.5 M NaCl for 3 days). Lipids were extracted from leaves, separated by TLC, and stained with iodine.
The vte1 Mutant Is Deficient in All Four Forms of Tocopherol and Contains Reduced Activity of Tocopherol Cyclase.
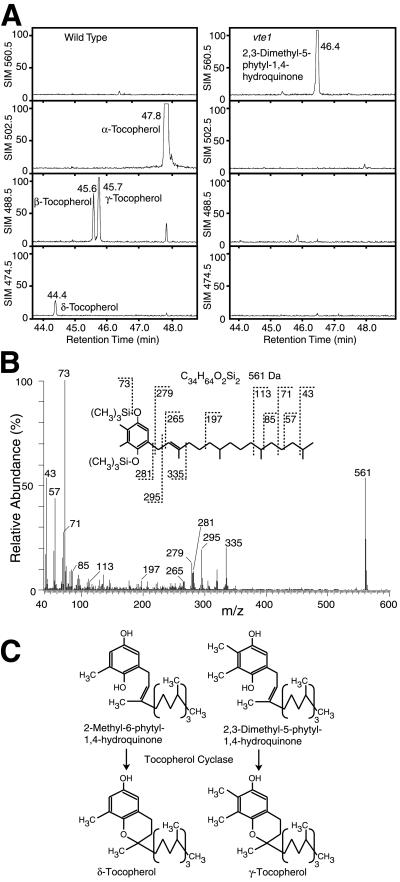
To unravel the biosynthetic block in the vte1 mutant, lipids were isolated from WT and mutant leaves and analyzed by GC/MS after trimethylsilylation (Fig. 2A). In contrast to WT, which contained mostly α-tocopherol and small amounts of the other three forms of tocopherol (37), the vte1 mutant was completely devoid of all four forms of tocopherol. However, in vte1, a compound with a molecular mass of 560.5 Da accumulated that was not detectable in WT (Fig. 2A). The mass spectrum of this compound is in agreement with the fragmentation pattern of the di(trimethylsilyl) derivative of DMPQ (Fig. 2B). This finding suggests that the vte1 mutation affects the activity of DMPQ cyclase (tocopherol cyclase), because this enzyme catalyzes the conversion of 2-methyl-6-phytyl-1,4-hydroquinone and DMPQ to δ- and γ-tocopherol, respectively (Fig. 2C). Because these two forms of tocopherol are the precursors for β- and α-tocopherol, a reduction in tocopherol cyclase activity is expected to result in the loss of all four forms of tocopherol.
Fig 2.
All four forms of tocopherol are absent from vte1 leaves. (A) GC/MS chromatograms of trimethylsilylated lipids from Arabidopsis WT and vte1 leaves. For each sample (WT or vte1 mutant), four single ion monitoring traces (SIM; m/z = 560.0, 502.5, 488.5, and 474.5) derived from one GC/MS chromatogram are presented. The signals are drawn to the same relative scale (100%). (B) The compound with the retention time t = 46.4 min accumulating in vte1 leaves was identified as the di(trimethylsilyl) derivative of DMPQ based on its fragmentation pattern. (C) Conversion of 2-methyl-6-phytyl-1,4-hydroquinone or DMPQ by tocopherol cyclase yields δ- and γ-tocopherol, respectively.
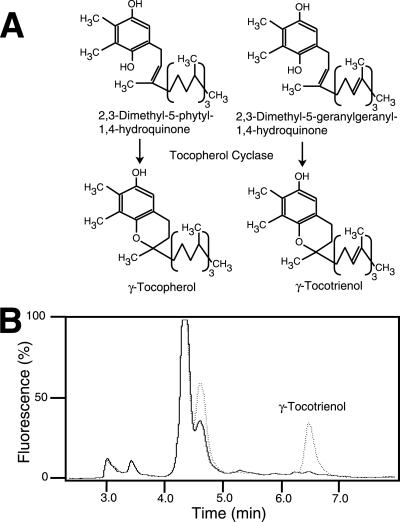
Enzyme activity for tocopherol cyclase was measured in leaf extracts from WT and vte1 (Fig. 3 A and B). Because enzyme activities in plant extracts using DMPQ or DMGQ were shown to be very similar (ref. 32 and data not shown), only one of these substrates, DMGQ, was used. Whereas in WT, a large amount of DMGQ was converted to γ-tocotrienol, the tocopherol cyclase activity was very low in vte1 (407 ± 20 and 6.6 ± 0.7 ng of γ-tocotrienol per mg of protein per h for WT and vte1, respectively). The activity for 2-methyl-6-phytyl-1,4-hydroquinone methyltransferase was very similar in vte1 (data not shown). Therefore, the vte1 mutation specifically affects the activity of tocopherol cyclase and does not cause a general down-regulation of the enzymes of tocopherol biosynthesis.
Fig 3.
The vte1 mutant is deficient in tocopherol cyclase activity. (A) Tocopherol cyclase catalyzes the conversion of DMPQ and 2,3-dimethyl-5-geranylgeranyl-1,4-hydroquinone (DMGQ) to γ-tocopherol and γ-tocotrienol, respectively. (B) Tocopherol cyclase assays of WT and vte1. Protein extracts were incubated with DMGQ, and the reaction products separated by HPLC and detected by fluorescence emission. The retention time of γ-tocotrienol was confirmed with a standard. Dotted line, WT; solid line, vte1.
Isolation of the VTE1 Gene.
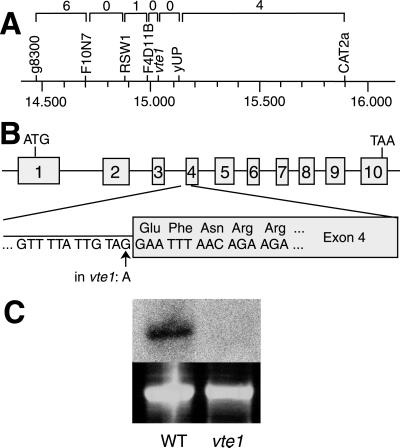
F2 plants derived from a cross of vte1 with WT Landsberg erecta were used for mapping the mutation to the Arabidopsis genome. The VTE1 locus was localized to the lower arm of chromosome 4, between the markers RSW1 and CAT2a (Fig. 4A). These two markers span a region of 106 bp on the Arabidopsis genome. In this region, no genes encoding enzymes of tocopherol biosynthesis were found (37–41). Therefore, the vte1 locus represents a novel gene involved in tocopherol biosynthesis. The gene At4g32770 on BAC F4D11 (GenBank accession no. AL022537) represented a candidate gene for VTE1, because it encodes a protein similar to the ORF slr1737 of Synechocystis (52.2% and 41.1% identity on DNA and amino acid level, respectively). In Synechocystis, slr1737 is localized in the same operon as slr1736, which encodes the homogentisate phytyltransferase, another enzyme of tocopherol synthesis (38, 39). Because in prokaryotes, genes encoding enzymes of the same biosynthetic pathway are oftentimes combined in operons, the ORF slr1737 was a likely candidate for another gene of tocopherol synthesis. A similar genomics-based approach has already successfully been used to isolate the structural gene for γ-tocopherol methyltransferase (37). For this reason, the gene At4g32770 from Arabidopsis was amplified by PCR from genomic DNA of vte1 and completely sequenced. The sequence of the mutant gene showed only one mismatch as compared with WT at the splicing site of intron 3/exon 4. The last base at the 3′ end of intron 3 of At4g32770 was changed from G to A (Fig. 4B). By comparing 97 intron sequences from dicots, the G at this position was found to be 100% conserved (42). Therefore, it is very likely that in vte1, splicing of intron 3 is abolished, which results in the synthesis of a truncated, inactive protein.
Fig 4.
Isolation of the VTE1 gene. (A) Mapping of the vte1 mutation to the lower arm of chromosome 4. The values on top represent the numbers of recombinations between two adjacent markers in a mapping population of 384 chromosomes. The numbers at the bottom indicate the position on the physical map of chromosome 4 in 106 bp according to the MIPS database (http://mips.gsf.de/proj/thal/). (B) Exon/intron structure of the gene At4g32770. In vte1, this gene carries a G to A point mutation at the 3′ boarder of intron 3. (C) Northern analysis of total RNA from WT and vte1. (Upper) The hybridization signal with At4g32770. (Lower) The ethidium bromide-stained 25S rRNA bands.
To test whether expression of At4g32770 was altered in vte1, Northern analysis was done with RNA isolated from leaves. A band with a size of about 1.6 kb hybridized to the At4g32770 probe in WT, but in vte1 no band was detectable at this position (Fig. 4C). Therefore, transcription of At4g32770 or mRNA stability was strongly reduced in vte1.
VTE1 Encodes an Enzyme with Tocopherol Cyclase and Tocotrienol Cyclase Activity.
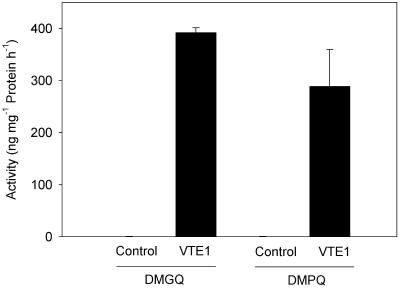
To provide further evidence that the VTE1 protein of Arabidopsis codes for a functional enzyme with tocopherol cyclase activity, the corresponding cDNA was heterologously expressed in E. coli and used for enzyme assays. The protein contains an apparent N-terminal signal sequence for the chloroplast (ref. 31; see below). Therefore, only the mature part of the protein lacking the N-terminal 74 aa was used for E. coli expression. As depicted in Fig. 5, the VTE1 protein showed high tocopherol cyclase activity with the substrates DMPQ and DMGQ, resulting in the synthesis of γ-tocopherol and γ-tocotrienol, respectively. These findings provide clear evidence that VTE1 encodes tocopherol cyclase and that this enzyme is involved in the synthesis of both tocopherols and tocotrienols.
Fig 5.
Tocopherol cyclase activity of VTE1 after heterologous expression in E. coli. The mature part of the VTE1 cDNA was expressed in E. coli and used for tocopherol cyclase activity assays with DMGQ or DMPQ. The products, γ-tocotrienol and γ-tocopherol, respectively, were quantified by fluorescence HPLC. The values represent the means of two independent measurements and standard errors. Control, E. coli transformed with empty vector (pQE31); VTE1, E. coli transformed with VTE1 in pQE31. The tocopherol cyclase activity of control cells was below 1 ng per mg of protein per h for the two substrates.
The vte1 Mutant Phenotype During Oxidative Stress.
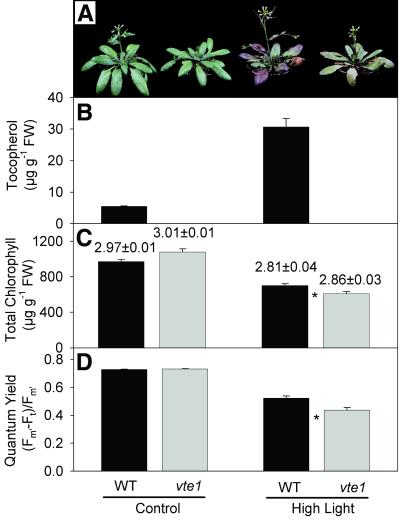
When raised under optimal growth conditions, the overall stature, chlorophyll content, and photosynthetic yield of WT and vte1 plants were very similar (Fig. 6). The fresh weight of 50-day-old vte1 plants was reduced by only 10–15%. The flowering time as determined from the number of rosette leaves at the time of the opening of the first flower (43) was similar in WT (16.9 ± 0.9; mean ± SE, n = 10) and vte1 (18.1 ± 1.1). Therefore, under optimal growth conditions, absence of all forms of tocopherol had no large impact on photosynthesis, growth, and development of the mutant plant. We considered the possibility that tocopherol deficiency might result in a decrease of photosynthetic efficiency only during oxidative stress. Plants were exposed to high-light conditions (850 μmol⋅m−2⋅s−1) and used for measurements of tocopherol, chlorophyll content, and chlorophyll fluorescence. Exposure to high light resulted in the occurrence of necrotic areas on the leaves of the vte1 mutant and WT (Fig. 6A). In contrast to WT, the stress-induced accumulation of anthocyanin in the mutant was much less pronounced (Fig. 6A). Photo-oxidative stress resulted in an increase in α-tocopherol in WT, but no tocopherol was detectable in vte1 under these conditions (Fig. 6B). Total chlorophyll was slightly reduced in vte1 leaves during oxidative stress (Fig. 6C), but the chlorophyll a to chlorophyll b ratio was not changed. The photosynthetic quantum yield determined by measurement of chlorophyll fluorescence was lower for WT and vte1 under high-light stress as compared with control conditions, and it was even further decreased in vte1 (Fig. 6D). Therefore, under conditions of photo-oxidative stress, chlorophyll content and photosynthesis in vte1 mutant plants were more severely affected as compared with WT. The differences in chlorophyll content and quantum yield for vte1 under high light were also observed with a set of independently raised plants exposed to 1,050 μmol⋅m−2⋅s−1, and the difference in chlorophyll content was found to be more pronounced in old leaves as compared with young leaves (data not shown).
Fig 6.
Stress-induced changes in plant morphology, tocopherol, chlorophyll, and chlorophyll fluorescence. Thirty-five-day-old plants were transferred to high light (850 μmol⋅m−2⋅s−1) for 5 days. (A) Visible phenotype of plants. (B) Total tocopherol content measured by fluorescence HPLC. (C) Total chlorophyll content. The numbers indicate the chlorophyll a to chlorophyll b ratio. (D) Photosynthetic quantum yield. Data represent the means ± SE of three (B), 10 (C), or 30 (D) measurements each. Values for chlorophyll content and quantum yield that were different between WT and mutant as judged by Student's t test (P < 0.02) are indicated with an asterisk.
In addition to high light, vte1 mutant plants were also analyzed during different stress conditions—e.g., paraquat treatment, high temperature (5 days at 37°C), and cold treatment (5 days at 4°C) (data not shown). Growth, chlorophyll content, and chlorophyll fluorescence of vte1 plants were always very similar to that of WT.
Discussion
The vte1 mutant of Arabidopsis represents the first higher plant mutant carrying a block that specifically affects tocopherol synthesis. The reduction in tocopherol cyclase activity accompanied with the accumulation of its substrate, DMPQ, and a concomitant decrease in γ-tocopherol strongly suggest that the vte1 mutant carries a mutation in the structural gene encoding tocopherol cyclase.
Several lines of evidence indicate that the gene At4g32770 on chromosome 4 of Arabidopsis encodes the enzyme with tocopherol cyclase activity: (i) The vte1 mutation was mapped to an interval of 106 bp on chromosome 4 encompassing the At4g32770 locus; (ii) the orthologous gene from Synechocystis, slr1737, is localized in the same operon as homogentisate phytyltransferase, another gene of tocopherol synthesis (38, 39); (iii) At4g32770 from vte1 carries a splicing site mutation at a position absolutely conserved in dicots; (iv) At4g32770 mRNA abundance and tocopherol cyclase activity were strongly reduced in vte1; (v) heterologous expression of At4g32770 in E. coli resulted in the synthesis of a protein with high tocopherol cyclase activity. Because the vte1 mutant was four times back-crossed to WT, it is extremely unlikely that this plant carries another mutation in a gene involved in tocopherol synthesis. The point mutation found at the highly conserved splicing site of At4g32770 strongly suggests that vte1 represents a null allele. This was confirmed by the finding that the amounts of all forms of tocopherol were reduced below detection limit.
Growth, chlorophyll content, and photosynthetic quantum yield of vte1 under optimal conditions were very similar to WT. Similarly, the homogentisate phytyltransferase mutant of Synechocystis deficient in all forms of tocopherol was not reduced in growth or chlorophyll content (38). However, during photo-oxidative stress, chlorophyll content and quantum yield were decreased in vte1 as compared with WT. This result is in agreement with the presumed role of tocopherol in protecting photosynthetic complexes in thylakoids from oxidative stress. However, the stress-induced changes observed for vte1 were rather small. Therefore, other redox systems (e.g., ascorbate, glutathione, carotenoids) might be involved in scavenging reactive oxygen species under high-light conditions.
The reduction in geranylgeranyl reductase activity by antisense expression in tobacco plants also resulted in a partial reduction of tocopherol synthesis (44, 45). Similar to the Arabidopsis vte1 mutant, only under conditions of oxidative stress, photosynthesis in the transgenic tobacco lines was affected. However, in contrast to vte1, which is exclusively affected in tocopherol synthesis, chlorophyll biosynthesis in the geranylgeranyl reductase antisense plants was also reduced. Therefore, it was not clear whether the phenotypical changes observed for the tobacco antisense lines were derived from deficiency in tocopherol or chlorophyll. Similarly to vte1, an Arabidopsis mutant deficient in ascorbate synthesis (soz1/vtc1) showed an increased sensitivity to environmental stress (46). The two mutants, vte1 and vtc1, open the way to study the in vivo role of both vitamin E and vitamin C during oxidative stress in higher plants.
Interestingly, VTE1 from Arabidopsis shows high sequence similarity (62% identity on amino acid level) to the maize SXD1 gene (31, 47). The findings that only one VTE1-like locus is found in the Arabidopsis genome and that in maize all SXD1-like expressed sequence tags present in the public database have a sequence identical to SXD1 suggest that VTE1 and SXD1 represent (orthologous) single copy genes, both encoding enzymes with the same function—i.e., tocopherol cyclase activity. The sucrose export defective mutant1 of maize (sxd1) shows alterations in the ultrastructure of plasmodesmata, which was implied to be the cause for the reduced photoassimilate export from mutant leaves. The VTE1/SXD1 cDNA was predicted to encode a hydrophilic protein of 488 aa (54.7 kDa) with an N-terminal plastid signal sequence (31). blast searches in GenBank revealed the presence of expressed sequence tags in many different plant species, but no similarities to animal or yeast sequences. The fact that the VTE1/SXD1 protein was imported into pea chloroplasts (47) is in agreement with the presumed localization of tocopherol cyclase to the inner chloroplast envelope (19). During import experiments, the pea protein was processed indicating the presence of a cleavable N-terminal signal peptide. It was proposed that VTE1/SXD1 represents a protein factor involved in chloroplast-to-nucleus signaling of the metabolic status of chloroplasts (31). The finding that VTE1/SXD1 encodes an enzyme of tocopherol synthesis raises the question of the exact nature of the factor(s) responsible for the morphological and metabolic changes observed for the maize sxd1 mutant. Therefore, comparative experiments with the two mutants, sxd1 from maize and vte1 from Arabidopsis, are required to clarify the role of lipid antioxidants during oxidative stress, as well as for primary reactions of photosynthesis and photoassimilate transport.
Acknowledgments
We would like to thank Conny Wagner and Joachim Kopka for their help with GC/MS. This work was in part supported by a Research Fellowship from the Deutsche Forschungsgemeinschaft (Grant Po757/1 to S.P.).
Abbreviations
DMGQ, 2,3-dimethyl-5-geranylgeranyl-1,4-hydroquinone
DMPQ, 2,3-dimethyl-5-phytyl-1,4-hydroquinone
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Alscher R. G. & Hess, J. L., (1993) Antioxidants in Higher Plants (CRC, Boca Raton, FL).
- 2.Foyer C. H., Descourvieres, P. & Kunert, K. J. (1994) Plant Cell Environ. 17, 507-523. [Google Scholar]
- 3.Chool Boo Y. & Jung, J. (1999) J. Plant Physiol. 155, 255-261. [Google Scholar]
- 4.Knight H. & Knight, M. R. (2001) Trends Plant Sci. 6, 262-267. [DOI] [PubMed] [Google Scholar]
- 5.Fryer M. J. (1992) Plant Cell Environ. 15, 381-392. [Google Scholar]
- 6.Noctor G. & Foyer, C. H. (1998) Annu. Rev. Plant Physiol. Plant Mol. Biol. 49, 249-279. [DOI] [PubMed] [Google Scholar]
- 7.Mallick N. & Mohn, F. H. (2000) J. Plant Physiol. 157, 183-193. [Google Scholar]
- 8.Borsani O., Valpuesta, V. & Botella, M. A. (2001) Plant Physiol. 126, 1024-1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Willekens H., van Camp, W., van Montagu, M., Inzé, D., Lagerbartels, C. & Sandermann, H. (1994) Plant Physiol. 106, 1007-1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Desikan R., Mackerness, S. A.-H., Hancock, J. T. & Neill, S. J. (2001) Plant Physiol. 127, 159-172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wilson R. F., Burke, J. J. & Quisenberry, J. E. (1987) Plant Physiol. 84, 251-254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rise M., Cojocaru, M., Gottlieb, H. E. & Goldschmidt, E. E. (1989) Plant Physiol. 89, 1028-1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sakaki T., Kondo, N. & Yamada, M. (1990) Plant Physiol. 94, 781-787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Grau A. & Ortiz, A. (1998) Chem. Phys. Lipids 91, 109-111. [DOI] [PubMed] [Google Scholar]
- 15.Fukuzawa K., Tokumura, A., Ouchi, S. & Tsukatani, H. (1982) Lipids 17, 511-513. [DOI] [PubMed] [Google Scholar]
- 16.Niki E., Saito, T., Kawakami, A. & Kamiya, Y. (1984) J. Biol. Chem. 259, 4177-4182. [PubMed] [Google Scholar]
- 17.Liebler D. C., Kling, D. S. & Reed, D. J. (1986) J. Biol. Chem. 261, 12114-12119. [PubMed] [Google Scholar]
- 18.Lichtenthaler H., Prenzel, U., Douce, R. & Joyard, J. (1981) Biochim. Biophys. Acta 641, 99-105. [DOI] [PubMed] [Google Scholar]
- 19.Soll J., Schultz, G., Joyard, J., Douce, R. & Block, M. A. (1985) Arch. Biochem. Biophys. 238, 290-299. [DOI] [PubMed] [Google Scholar]
- 20.DellaPenna D. (1999) Science 285, 375-379. [DOI] [PubMed] [Google Scholar]
- 21.Grusak M. A. & DellaPenna, D. (1999) Annu. Rev. Plant Physiol. Plant Mol. Biol. 50, 133-161. [DOI] [PubMed] [Google Scholar]
- 22.Evans H. M. & Bishop, K. S. (1922) J. Metabol. Res. 1, 319-355. [Google Scholar]
- 23.Hugly S., Kunst, L. & Somerville, C. (1991) J. Hered. 82, 484-488. [Google Scholar]
- 24.Dörmann P., Hoffmann-Benning, S., Balbo, I. & Benning, C. (1995) Plant Cell 7, 1801-1810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Arondel V., Lemieux, B., Hwang, I., Gibson, S., Goodman, H. M. & Somerville, C. R. (1992) Science 258, 1353-1355. [DOI] [PubMed] [Google Scholar]
- 26.Dörmann P., Balbo, I. & Benning, C. (1999) Science 284, 2181-2184. [DOI] [PubMed] [Google Scholar]
- 27.Konieczny A. & Ausubel, F. M. (1993) Plant J. 4, 403-410. [DOI] [PubMed] [Google Scholar]
- 28.Bell C. J. & Ecker, J. R. (1994) Genomics 19, 137-144. [DOI] [PubMed] [Google Scholar]
- 29.Logemann J., Schell, J. & Willmitzer, L. (1987) Anal. Biochem. 163, 16-20. [DOI] [PubMed] [Google Scholar]
- 30.Sambrook J., Fritsch, E. F. & Maniatis, T., (1989) Molecular Cloning: A Laboratory Manual (Cold Spring Harbor Lab. Press, Plainview, NY).
- 31.Provencher L. M., Miao, L., Sinha, N. & Lucas, W. J. (2001) Plant Cell 13, 1127-1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stocker A., Rüttimann, A. & Woggon, W.-D. (1993) Helv. Chim. Acta 76, 1729-1738. [Google Scholar]
- 33.Fiehn O., Kopka, J., Trethewey, R. N. & Willmitzer, L. (2000) Anal. Chem. 72, 3573-3580. [DOI] [PubMed] [Google Scholar]
- 34.Thompson J. N. & Hatina, G. (1979) J. Liquid Chromatogr. 2, 327-344. [Google Scholar]
- 35.Lichtenthaler H. K. (1987) Methods Enzymol. 148, 350-382. [Google Scholar]
- 36.Schreiber U., Schliwa, U. & Bilger, W. (1986) Photosynth. Res. 10, 51-62. [DOI] [PubMed] [Google Scholar]
- 37.Shintani D. & DellaPenna, D. (1998) Science 282, 2098-2100. [DOI] [PubMed] [Google Scholar]
- 38.Collakova E. & DellaPenna, D. (2001) Plant Physiol. 127, 1113-1124. [PMC free article] [PubMed] [Google Scholar]
- 39.Schledz M., Seidler, A., Beyer, P. & Neuhaus, G. (2001) FEBS Lett. 499, 15-20. [DOI] [PubMed] [Google Scholar]
- 40.Shintani D. K., Cheng, Z. & DellaPenna, D. (2002) FEBS Lett. 511, 1-5. [DOI] [PubMed] [Google Scholar]
- 41.Norris S. R., Barrette, T. R. & DellaPenna, D. (1995) Plant Cell 7, 2139-2149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hanley B. A. & Schuler, M. A. (1988) Nucleic Acids Res. 16, 7159-7176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Reeves P. H. & Coupland, G. (2001) Plant Physiol. 126, 1085-1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tanaka R., Oster, U., Kruse, E., Rüdiger, W. & Grimm, B. (1999) Plant Physiol. 120, 695-704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Graßes T., Grimm, B., Koroleva, O. & Jahns, P. (2001) Planta 213, 620-628. [DOI] [PubMed] [Google Scholar]
- 46.Conklin P. L., Williams, E. H. & Last, R. L. (1996) Proc. Natl. Acad. Sci. USA 93, 9970-9974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Russin W. A., Evert, R. F., Vanderveer, P. J., Sharkey, T. D. & Briggs, S. P. (1996) Plant Cell 8, 645-658. [DOI] [PMC free article] [PubMed] [Google Scholar]