Abstract
Objective: To create “extensional definitions” of laboratory codes from derived characteristics of coded values in a clinical database and then use these definitions in the automated mapping of codes between disparate facilities.
Design: Repository data for two laboratory facilities in the Intermountain Health Care system were analyzed to create extensional definitions for the local codes of each facility. These definitions were then matched using automated matching software to create mappings between the shared local codes. The results were compared with the mappings of the vocabulary developers.
Measurements: The number of correct matches and the size of the match group were recorded. A match was considered correct if the corresponding codes from each facility were included in the group. The group size was defined as the total number of codes in the match group (e.g., a one-to-one mapping is a group size of two).
Results: Of the matches generated by the automated matching software, 81 percent were correct. The average group size was 2.4. There were a total of 328 possible matches in the data set, and 75 percent of these were correctly identified.
Conclusions: Extensional definitions for local codes created from repository data can be utilized to automatically map codes from disparate systems. This approach, if generalized to other systems, can reduce the effort required to map one system to another while increasing mapping consistency.
Electronic data exchange between different computer systems in the health care industry is hampered by the variety of local codes used for representing the data in different systems. Communication between systems requires that shared codes be identified and mapped to each other. This mapping process is a tedious and labor-intensive effort requiring many hours of expert review. The task is complicated by the lack of widely available tools for mapping coded data. Methodologies have been developed for matching terms to controlled vocabularies using various lexical and morphologic text matching techniques.1
In this article, we explore a novel approach to the mapping of structured data by using extensional definitions created from the data available for each code.2 We hypothesize that similar codes can be identified by similar extensional definitions and that the comparison can be automated using commercially available matching software.
Background
The proliferation of computerized medical record systems throughout the health care industry has created a need for combining laboratory results from many different sources. Vast amounts of structured data, which can be utilized for patient care and pooled for research purposes, are now available. Unfortunately, combining similar data from different systems can be difficult and time consuming. As the number of systems increases, the difficulty increases geometrically.
One solution is to create a standard set of codes for data exchange. Once mapped to the standard, a system would be able to communicate with any other similarly mapped system. A suitable code set must be universally available, consistently mapped, and capable of expressing all the shared concepts contained in the data collections. Recognizing this need, the Logical Observation Identifiers, Names, and Codes (LOINC) coding database was created as a pre-coordinated vocabulary for observations.3,4 As a pre-coordinated vocabulary, it explicitly defines what combinations of concepts are allowed. New combinations may not be created by the user. In contrast, a post-coordinated vocabulary allows the user to combine concepts to create more expressive concepts. For example, the modifier “left” can be combined with the concept “arm” to create the concept “left arm.”
In one study, Baorto et al.5 attempted to use LOINC for the exchange of laboratory tests between three academic hospitals.5 Their results showed that the mapping of local tests to LOINC was complex and inconsistent between institutions. When the three different pairings of institutions were examined only, 3, 12, and 62 percent of the true matches were identified by matching LOINC codes. They concluded that the potential for the unambiguous pooling of data without human inspection can be achieved “only if a careful, standard LOINC coding procedure is used at all sites, performed by individuals with significant domain-specific expertise and well-educated in the LOINC system.” Others have suggested using a post-coordinated vocabulary, in which terms can be combined to create more complex terms, to allow greater flexibility of expression.6 This approach would still require compositional guidelines to achieve consistent coding.
Another solution for achieving consistent coding procedures is automating the process of mapping to standardized codes. Theoretically, variability in the coding process will be reduced as human intervention is reduced. The difficulty is creating an automated tool with sufficient accuracy to provide consistent mapping between codes.
A number of automated tools have been developed to map concepts to standardized vocabularies.1,7–9 All of these utilize the name of the concept, with different lexical, morphologic, and semantic matching techniques, to map the concepts to controlled terminologies. These techniques require large knowledge bases, sometimes unique to the system being mapped, to provide the information for normalization, synonymy, and semantic structure. Although these approaches are promising, they limit the information for matching to the name of the concept. Laboratory test names generally do not include all the information utilized by LOINC, such as units of measure, specimen type, and source. Thus, without participation of expert laboratory personnel, this approach is not likely to be highly accurate for mappings to LOINC.
We propose a different approach. The laboratory tests represented by the institutional codes are defined in two ways. First, each has an “intentional definition,” which is the definition given by the vocabulary designers. For example, the local system code 1443 might be defined as “the concentration of sodium in a sample of serum.” Notice that this definition contains fewer details and is much less specific than the LOINC definition for this measurement, but it is typical of the kind of description that is usually available in clinical systems.
Second, for each code a number of entries (instances of data) exist in the clinical database. This collection of entries can be thought of as an “extensional definition” of what the code means in the system. For example, the mean, standard deviation, and units of measure for the entries characterize the collection of entries for code 1443. By analyzing the database entries associated with the local codes, we create an extensional definition for the code that can be compared from institution to institution. We hypothesize that codes with similar extensional definitions are likely to have the same intentional definitions. If the elements used for comparison are present in both collections and a method for comparing definitions exists, then we propose that this technique can be used to map different data collections.
Methods
Data Sources
Intermountain Health Care (IHC) is a large health care organization in northern Utah that comprises three laboratory hubs utilizing the 3M Healthcare Data Dictionary. The local names and codes for each hub have been independently mapped to numeric concept identifiers (NCIDs) and corresponding LOINC codes.10 Health care data collected by IHC facilities is stored in the IHC longitudinal data repository (LDR). While aspects of the data storage are standardized throughout IHC, each hub's data can be considered to represent a single institution. For this study, the two largest hubs were used. Hub 1 represents data from a large tertiary-care hospital, a children's hospital, two community hospitals, and two clinics. Hub 2 represents data from three community hospitals.
All LDR entries for a one-month period (March 1999) were obtained and stored, after removal of patient identifiers, in a Microsoft Access database. Entries were grouped by hub and NCID for preprocessing, with each entry representing a single observation. The fields utilized for each entry are shown in Table 1▶.
Table 1 .
Fields Used from the LDR Data Set to Create the Table of Extensional Definitions
| Field | Description |
|---|---|
| Observation ID | Identifies a single observation |
| Observation NCID | Numeric coded observation identifier for the test. |
| Event ID | Identifies a single event. A number of observations can be part of the same event. |
| Numeric Value | The value of the result if it is numeric. |
| Coded Value | The coded representation of the result if it is not numeric. |
| Units Code | The coded representation for the unit of measure for the result value. |
| Facility | Specifies the facility for the observation. Used to determine the hub. |
Automatch Software
Automated matching was performed using Automatch 4.0 by Vality Technologies, Inc. (Boston, Massachusetts). Automatch is a record linkage program designed to compare identifying and geographic data for individuals, households, or events, matching those that are the same.11,12 To match two collections of records, the Automatch program uses a series of passes, each of which employs blocking (or grouping) and matching parameters. Blocking parameters limit the number of comparisons that are required for each pass by specifying the subset of records that will be compared. Records that are identical for all the blocking fields specified for that particular pass are compared. For example, if the unit of measure is a blocking field, then only records that have identical units of measure are compared (e.g., a record that has mg/dl as its unit of measure would be compared only with other records that have mg/dl as their unit of measure). The comparison tests are specified by the matching parameters for each record field involved in the match.
All records in the block set are compared pair-wise. As each pair of records is compared, each field in the records is compared, and a probabilistic weight is assigned on the basis of how closely the values of the given field from the different records match. The weights for all field-wise comparisons are summed, and the sum represents the total weight for that match pair. A match is defined as a total weight greater than the user specified cutoff for each pass.
Automatch considers similar records contained in only one collection to be duplicates of the same record. This allows many-to-many mappings, when “duplicates” from one collection are mapped to “duplicates” in the other. Once a record is matched in a given pass, the record is removed from consideration in further passes.
Preprocessing
Entries in the LDR were preprocessed to create the extensional definition tables for each hub used for the match. Definitions were created for the laboratory tests of each hub. Table 2▶ shows the fields contained in the resulting table.
Table 2 .
Fields Used for the Extensional Definitions of Each Numeric Concept Identifier (NCID)
| Field | Description |
|---|---|
| NCID | Numeric concept identifier used for evaluating match results. |
| Name | The local laboratory name for the observation. |
| Frequency | Percentile for the frequency of this NCID in the data set. |
| Mean | The mean of the numeric result values, if numeric. |
| Standard Deviation | The standard deviation of the numeric result values, if numeric. |
| Codes | An array of the 10 most frequent coded values. |
| Co-occurrences | An array of the 14 most frequent co- occurring NCIDs. |
| Units | Unit of measure for the result value as a coded entry. |
| Mean Group | Group labels created to allow blocking for the mean. |
| SD Group | Group labels created to allow blocking for the standard deviation. |
| Frequency Group | Group labels created to allow blocking for the frequency. |
Each laboratory test is identified by a unique NCID. Laboratory tests can have numeric result entries, coded result entries, or both. The NCID was considered to represent a numeric laboratory test if the number of numeric result entries for that NCID was greater than the number of coded result entries. The mean and standard deviation were calculated for each numeric laboratory test. The frequency percentile of each NCID represents the percentage of NCIDs from that hub with an equal or smaller number of entries.
For each NCID representing a coded item, the ten most commonly occurring result codes, entered as coded result values, were obtained. The units field for numeric items was defined as the most commonly occurring unit of measure for that NCID. Co-occurrences were defined as NCIDs that shared the same event-ID (i.e., laboratory tests performed at the same time as part of the same battery). The 14 most commonly co-occurring NCIDs were obtained for each NCID. Finally, the names for each NCID (up to six names) were obtained from the regional laboratory systems.
Fields can serve as blocking fields if a number of NCIDs share identical values for the field entries. To allow blocking using fields that contain few identical values, values had to be grouped. Groups were created for the mean, standard deviation, and frequency fields by sorting the larger data set (Hub1) in increasing order and selecting group cutoff values, which created groups of approximately 40 entries. For example, NCIDs with a mean of 0 to 1.1 would be in mean group “1.” The same cutoff points were used in the smaller data set to create the same groupings. The group labels were recorded in three new fields—the mean group, SD group, and frequency group fields, respectively—which were used as blocking fields.
Matching
The extensional definitions for Hub 1 were matched to the definitions for Hub 2 using Automatch. For our data, identical blocking parameters where created by grouping fields as described in the preprocessing section. The comparison tests we used were selected from those available in the Automatch program.
Numeric comparisons were accomplished using an algebraic numeric comparison or a prorated comparison, which allows disagreement by a specified absolute amount. Character comparisons used an information-theoretic character comparison that tolerates phonetic errors, transpositions, random insertion, deletion, and replacement of characters.
The algorithm is proprietary to the software, but descriptions are available in the literature.13 First, the strings are tested to see whether they are identical, except for any trailing blanks. If they are, then the full weight is given. If the strings are different, the length of each is determined and the number of unassigned characters is determined. Each letter in one string is compared with the letters of the other that are located in the same position plus or minus the acceptable match distance. An acceptable match distance is defined as half the longer string minus one.
If the same character is found, it is considered matched (assigned) to the other string. All unassigned letters are flagged and counted. Transpositions are then counted by comparing the first assigned character in one string with the first assigned character in the other. If these characters disagree, this is considered one half of a transposition. The comparison continues for the remainder of the matched characters.
The number of transpositions is then determined by dividing the number of disagreements by two. The weight is given by the following formula:
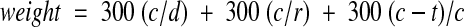 |
where c is the number of matched letters; d, the length of the data file string; r, the length of the reference string; and t, the number of transpositions. Exact matches were given the full weight, whereas partial matches were given partial weight. Weights falling below a specified cutoff (we used 500) were considered non-matches. No attempt was made to correct for word order or synonymy. This was mainly done for reasons of expediency, but we also wanted to see how the technique performed independent of the knowledge base required for concept normalization.
The matching algorithm used a total of five passes with progressively less restrictive blocking. The cutoffs were established to optimize the overall results. A concise description of the methodology used by Automatch appears in a paper by Jaro.12
Outcome Measures
For each pass, the total number of matches made and the NCIDs correctly matched were recorded. An NCID was correctly matched when identical NCIDs from each hub were included in a match grouping. The size of the match group was the total number of NCIDs returned for each match from both hubs. For example, a one-to-one match would have a size of two, with one NCID returned for each hub.
Results
The data set for March 1999 contained 992,910 entries from Hub 1 and 287,694 entries from Hub 2, representing 767 and 372 unique NCIDs, respectively. Of these NCIDs, 328 are shared, representing potential matches.
The results for the matching algorithm are shown in Table 3▶. Overall, 245 (75 percent) of the 328 possible matches were correctly identified by this automated mapping technique. When a match was made, it contained an average of 2.4 NCIDs and was correct 81 percent of the time. Of the 328 possible matches contained in the two data collections, 221 were numeric tests, with 170 of these matched correctly. There was no significant difference in success of matching between the numeric tests and the coded tests (χ2P > 0.18).
Table 3 .
Results of the Matching Algorithm for Each Pass and Overall
| Pass | Average Size | Correct Matches (%) | Total Matches | |
|---|---|---|---|---|
| 1 | 2.0 | 17 | (94) | 18 |
| 2 | 2.1 | 58 | (84) | 69 |
| 3 | 2.3 | 39 | (89) | 44 |
| 4 | 2.4 | 84 | (84) | 100 |
| 5 | 2.9 | 47 | (66) | 71 |
| All | 2.4 | 245 | (81) | 302 |
Figure 1▶ shows the success of matching for each frequency decile. Matching was significantly less accurate for NCIDs with a frequency decile of 30 or less (χ2P < 0.00001). A frequency decile of 30 represented fewer than 18 entries for Hub 1 and fewer than 26 entries for Hub 2.
Figure 1.
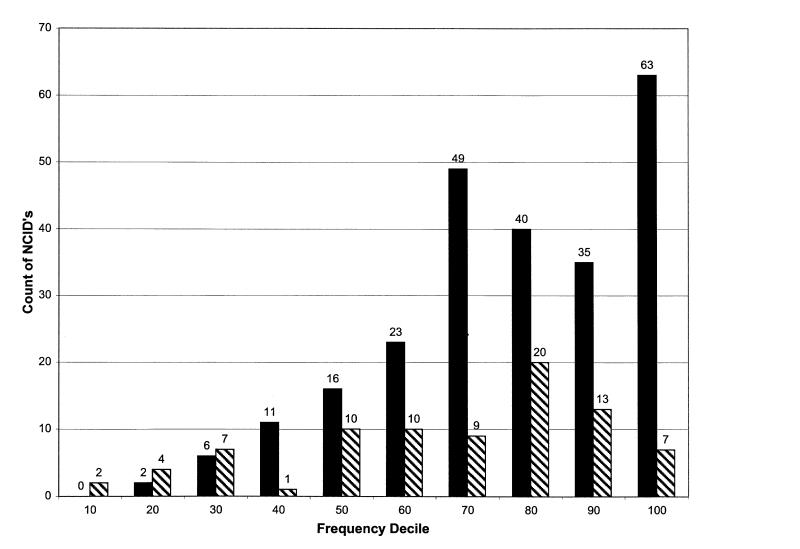
Number of matched (solid) and unmatched (hatched) numeric concept identifiers (NCIDs), by frequency rank.
Discussion
The mapping technique used in this study demonstrated a recall (correct matches made/correct matches possible) of 75 percent and a precision (correct matches/total matches made) of 81 percent, which is comparable with other mapping algorithms employing lexical and morphologic text-matching techniques.1 This is achieved with little use of any domain-specific knowledge, demonstrating the usefulness of extensional definitions. One benefit to our approach is that it can utilize commercially available matching software. Automating the process significantly reduces the effort, expense, and time required for mapping.
Conservatively estimated, mapping the codes in Hub 1 and Hub 2 manually would require two months of work by an expert. With this automated process, once data from each system is made available, all preprocessing of the data and generation of the matching results can be accomplished with little human intervention. Domain experts can then review the matches for correctness. Using this process, we estimate that 75 percent of the concepts in our test sets could be mapped in a few days. The remaining 25 percent would still require manual mapping. This would reduce the time required by approximately 65 percent, from eight weeks to three weeks.
The data sets represent realistic examples of the real-world uses for such a tool. The variations in number of local codes, amounts of data, and institutional size suggest that the technique can be generally used. Since the extensional definitions depend, to some extent, on the nature of the institution providing the data, it is possible that the entries in a database for a tertiary-care center may differ from entries for the same local code at a smaller institution. For example, the average serum sodium level at a tertiary-care center, which has patients with severe physiologic problems, may be different from that at a smaller center, which does not care for severely ill patients. Pediatric hospitals may also have values that differ from those in which the majority of patients are adults. In addition, differences in laboratory techniques (i.e., normal ranges) may also restrict the usefulness of this technique. In this study, the institutional size difference between the two hubs does not appear to have been a problem. Laboratory differences may have benefited from standardization of practices in IHC.
To a certain extent, the data collections used for this study are idealized. Result comparability across IHC has been a corporate goal for more than 20 years. Although the data elements we used are readily available from the Health Level 7 (HL7) messages for any laboratory system, the comparisons benefited from the standardization of the codes being compared. Non-numeric result values and units of measure were coded using the same code sets in the two hubs. This allowed a direct comparison of the field entries. These would need to be mapped, prior to performing the concept matching, in systems in which units of measure and result value codes had not previously been normalized. However, effective exchange of information would require that these codes be mapped anyway. For this reason, some have suggested standardizing these elements by using snomed codes.6
The co-occurrence code comparison also benefited from the standardization of codes between the two hubs. If test A1 in Hub 1 is often run with test B1 and test A2 in Hub 2 is often run with B2, this information is useful only when test B1 is mapped to test B2. Thus, the use of co-occurrence codes would require a different approach, perhaps an iterative approach that allows the system to take advantage of mappings from earlier iterations to interpret the co-occurrence information.
Cutoffs were established that optimized the results for this data set. The same cutoffs, applied to a validation set, would not be expected to perform as well. However, our main goal was to demonstrate that these techniques can be useful and that the effort required to further improve, validate, and generalize the process is worthwhile.
Refinements can certainly be made to our matching algorithms. In particular, we made little attempt to exploit the information available in the names beyond character comparisons. The match results would probably be improved by combining our technique with mapping strategies that employ more sophisticated normalized text comparisons for the names. The use of mean and standard deviation for comparing the distribution of numeric laboratory results could be improved by adding skewness and kurtosis. Codes with a small number of entries were not mapped as well as codes with a larger number of entries. Additional sampling could be employed for values that have a limited number of entries.
If either system is mapped to LOINC, the method described in this paper could serve to map the remaining system to LOINC by cross-referencing the matching codes. In a sense, the extensional definition of a code correctly mapped to a LOINC code is the extensional definition of the LOINC code itself. These definitions, along with the mapping process, can serve as a standard definition of LOINC, providing mapping consistency for existing data repositories.
In conclusion, this study demonstrates the feasibility of using extensional definitions for mapping HL7 structured repository data using commercially available automated matching software. The technique can be easily implemented when structured data are available for the codes being mapped. Further studies will be needed to determine whether it can be generalized to mappings between other systems and to standard coding sets themselves.
References
- 1.Barrows RC Jr, Cimino JJ, Clayton PD. Mapping clinically useful terminology to a controlled medical vocabulary. Proc Annu Symp Comput Appl Med Care. 1994:211–5. [PMC free article] [PubMed]
- 2.Sowa JF. Knowledge Representation: Logical, Philosophical, and Computational Foundations. Pacific Grove, Calif: Brooks/Cole, 2000.
- 3.Forrey AW, McDonald CJ, DeMoor G, et al. Logical observation identifiers, names, and codes (LOINC) database: a public use set of codes and names for electronic reporting of clinical laboratory test results. Clin Chem. 1996;42(1):81–90. [PubMed] [Google Scholar]
- 4.Huff SM, Rocha RA, McDonald CJ, et al. Development of the Logical Observation Identifiers, Names, and Codes (LOINC) vocabulary. J Am Med Inform Assoc. 1998;5(3):279–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baorto DM, Cimino JJ, Parvin CA, Kahn MG. Using Logical Observation Identifiers, Names, and Codes (LOINC) to exchange laboratory data among three academic hospitals. Proc AMIA Annu Fall Symp. 1997:96–100. [PMC free article] [PubMed]
- 6.White MD, Kolar LM, Steindel SJ. Evaluation of vocabularies for electronic laboratory reporting to public health agencies. J Am Med Inform Assoc. 1999;6(3):185–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yang Y, Chute CG. An application of least squares fit mapping to clinical classification. Proc Annu Symp Comput Appl Med Care. 1992:460–4. [PMC free article] [PubMed]
- 8.Sherertz DD, Tuttle MS, Olson NE, Erlbaum MS, Nelson SJ. Lexical mapping in the UMLS Metathesaurus. Proc Annu Symp Comput Appl Med Care. 1989:494–9.
- 9.Bodenreider O, Nelson SJ, Hole WT, Chang HF. Beyond synonymy: exploiting the UMLS semantics in mapping vocabularies. Proc AMIA Annu Symp. 1998:815–9. [PMC free article] [PubMed]
- 10.Rocha RA, Huff SM. Coupling vocabularies and data structures: lessons from LOINC. Proc AMIA Annu Fall Symp. 1996:90–4. [PMC free article] [PubMed]
- 11.Fellegi IP, Sunter AB. A theory for record linkage. J Am Stat Assoc. 1969;64(328):1183–210. [Google Scholar]
- 12.Jaro MA. Advances in record-linkage methodology as applied to matching the 1985 census of Tampa, Florida. J Am Stat Assoc. 1989;84(406):414–20. [Google Scholar]
- 13.Jaro MA. Unimatch: A Record Linkage System—User's Manual. Washington, DC: U.S. Bureau of the Census, 1978.


