Abstract
Objective: To evaluate the performance of a natural language processing system in extracting pneumonia-related concepts from chest x-ray reports.
Methods: Design: Four physicians, three lay persons, a natural language processing system, and two keyword searches (designated AAKS and KS) detected the presence or absence of three pneumonia-related concepts and inferred the presence or absence of acute bacterial pneumonia from 292 chest x-ray reports. Gold standard: Majority vote of three independent physicians. Reliability of the gold standard was measured. Outcome measures: Recall, precision, specificity, and agreement (using Finn's R statistic) with respect to the gold standard. Differences between the physicians and the other subjects were tested using the McNemar test for each pneumonia concept and for the disease inference of acute bacterial pneumonia.
Results: Reliability of the reference standard ranged from 0.86 to 0.96. Recall, precision, specificity, and agreement (Finn R) for the inference on acute bacterial pneumonia were, respectively, 0.94, 0.87, 0.91, and 0.84 for physicians; 0.95, 0.78, 0.85, and 0.75 for natural language processing system; 0.46, 0.89, 0.95, and 0.54 for lay persons; 0.79, 0.63, 0.71, and 0.49 for AAKS; and 0.87, 0.70, 0.77, and 0.62 for KS. The McNemar pairwise comparisons showed differences between one physician and the natural language processing system for the infiltrate concept and between another physician and the natural language processing system for the inference on acute bacterial pneumonia. The comparisons also showed that most physicians were significantly different from the other subjects in all pneumonia concepts and the disease inference.
Conclusion: In extracting pneumonia related concepts from chest x-ray reports, the performance of the natural language processing system was similar to that of physicians and better than that of lay persons and keyword searches. The encoded pneumonia information has the potential to support several pneumonia-related applications used in our institution. The applications include a decision support system called the antibiotic assistant, a computerized clinical protocol for pneumonia, and a quality assurance application in the radiology department.
Pneumonia is an infectious disease of the lung and a major cause of morbidity and mortality that affects approximately four million persons in North America each year.1 About one third to one half of pneumonia cases are caused by bacteria. Despite technologic advances in medicine, the diagnosis and treatment of pneumonia remain challenging for clinicians. Empiric antibiotic treatment is usually initiated before a definitive microbiologic diagnosis is available.
Automatic methods, such as computerized clinical guidelines and decision support systems, have been developed to assist the physicians in the diagnosis of pneumonia and the management of patients with the disease.2–4 A recent evaluation showed that one of these systems, as measured by significant reductions in several outcome measures (such as length of stay, adverse drug events, allergies, and costs), improves the quality of patient care.4
To determine whether a patient has acute bacterial pneumonia, automatic systems search the hospital information system for relevant clinical data, such as laboratory results, microbiology data, and chest x-ray reports. The majority of laboratory and microbiology data, such as hemograms, white blood cell counts, and results of sputum and blood cultures, are stored in a coded or numeric format. However, chest x-ray reports are usually stored as free-text reports. Information in free-text reports is not in a computable format. To perform any type of logic, computerized applications require coded data from a defined clinical vocabulary in which the concepts are represented in an unambiguous format.
Several methods exist to encode the information in free-text reports. Manual coding of the reports by trained personnel is rarely done, because it is expensive and time intensive. In addition, a time delay for the availability of the encoded data prevents its use by real-time applications. Radiologists could encode the findings from a patient's film in real time. Encoding findings in real time requires an appropriate user interface. However, developing of interfaces that map findings to a clinical vocabulary is not trivial, and even if such interfaces exist, radiologists prefer the simplicity of dictating a report.
None of these methods addresses past reports already stored in hospital information system as free-text reports. Keyword searches can be implemented to extract information from the free-text reports. Keyword searches are susceptible to all the problems that result from the complexities of natural language, such as grammatical ambiguities, synonymy, negation of concepts, and distribution of concepts.5
Natural language processing (NLP) is a methodology for automatically encoding clinical data from narrative reports. Such systems receive free-text reports as input, process the reports using syntactic and semantic information, and output coded data that can be mapped to a controlled terminology. Several NLP systems have been developed to encode clinical information from chest x-ray reports and other narrative reports.6–10 However, few formal evaluation studies have been reported.11
In this study, we evaluated the accuracy of an NLP system in the extraction of pneumonia-related concepts from chest x-ray reports. The concepts were selected on the basis of the requirements of a decision support system called the antibiotic assistant, which has long been used in our institution.3,4
Methods
We evaluated the accuracy of an NLP system called SymText9,10 in extracting pneumonia-related concepts from chest x-ray reports. We compared SymText against four physicians, two different keyword searches, and three lay persons. The performance of the study subjects (SymText, physicians, keyword searches, and lay persons) was established with respect to an independent gold standard consisting of three physicians. Our evaluation study was based on a methodology proposed by Hripcsak et al.11
We first describe the characteristics of the study subjects and the gold standard. Second, we define the pneumonia concepts and describe how the concepts were extracted from the radiology reports. Third, we describe report selection, calculation of the gold standard reliability, and the outcome measures.
Subjects and Gold Standard
Seven physicians (two radiologists and five internists) independently read the chest x-ray reports to establish the presence or absence of the pneumonia concepts. None of the reading physicians was from LDS Hospital, where the reports were originally produced. Three of the seven physicians (one radiologist and two internists) were randomly selected to provide the gold standard. The gold standard interpretation for the pneumonia concepts was established by the majority vote of the three physicians. The readings of the remaining four physicians (one radiologist and three internists) were tested against the gold standard.
Two different keyword searches were tested. The first keyword search is part of the antibiotic assistant, a computerized decision support systems implemented at LDS Hospital. The antibiotic assistant helps physicians select appropriate antibiotics for infectious diseases. One of the diseases is acute bacterial pneumonia. To decide on the presence or absence of acute bacterial pneumonia, the antibiotic assistant searches the HELP hospital information system12 for all the available pertinent clinical information. To extract the required pneumonia information from the chest x-ray reports, the developers of the antibiotic assistant applied a keyword search (AAKS, for antibiotic assistant keyword search). The second keyword search (KS) was developed by the authors who have background in NLP. Later, we describe the differences between the keyword searches.
Three lay persons served as baseline subjects. The lay persons did not have any background in biomedical sciences.
SymText is the NLP system developed at LDS hospital.9,10 SymText was developed to encode information in chest x-ray reports but has been used for admission diagnoses and ventilation/perfusion lung scan reports.13,14 The underlying structure has been previously described.9,10 SymText has a syntactic and a semantic component. The syntactic component is implemented as a set of augmented transition network grammars15 followed by the application of a transformational grammar. The semantic component consists of three different Bayesian networks.16 The first Bayesian network, shown in Figure 1▶, represents radiographic findings such as infiltrates, pleural effusions, and mediastinal widening. The second Bayesian network models the diseases that can be described in the reports, such as pneumonia, congestive heart failure, and atelectasis. The third Bayesian network models the devices that are frequently described in the chest x-ray report, such as Swanz-Ganz catheters, intravenous lines, and nasogastric tubes. SymText is able to extract 76 different radiographic findings and 89 different diseases from chest x-ray reports. In this study, SymText was evaluated only in the context of pneumonia.
Figure 1.
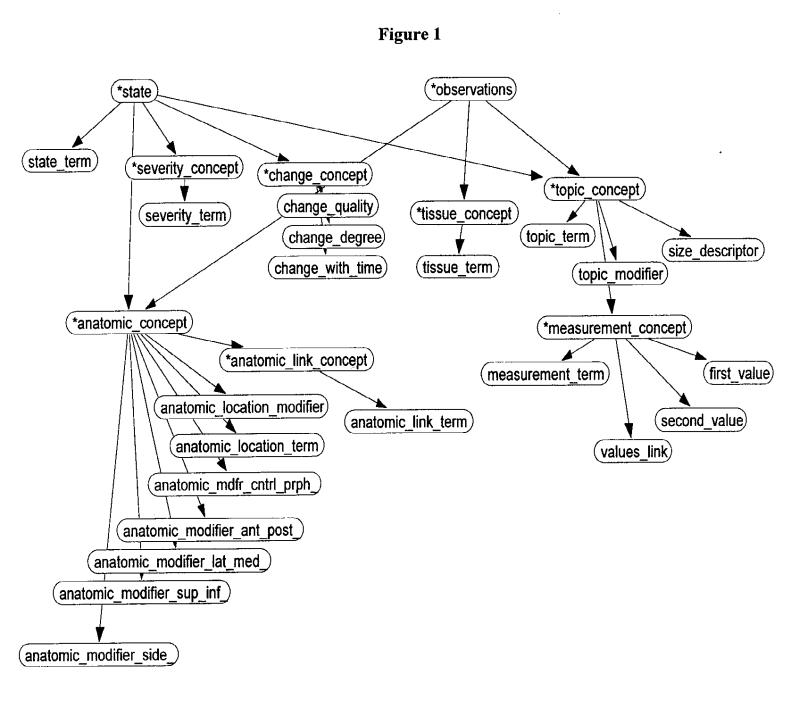
Bayesian network for radiographic findings.
For every sentence in the report, SymText makes an interpretation using the semantic model in the Bayesian networks as an attribute-value template. Values of the nodes (attributes) in the networks are either words taken directly from a sentence or broader concepts inferred from words. For example, Figure 2▶ shows the instantiation of the utterance “dense infiltrative opacity in the right upper lobe” resulting from use of the Bayesian network in Figure 1▶ (radiographic findings) as a semantic template.
Figure 2.
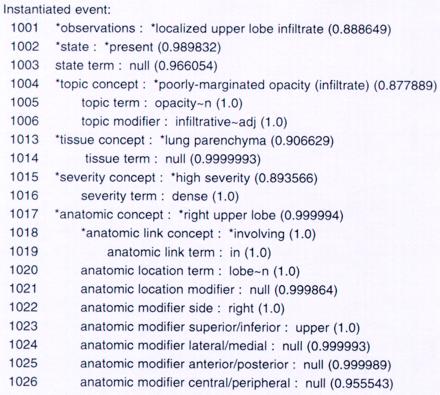
Partial view of the template instantiation for the sentence “dense infiltrative opacity in the right upper lobe.” The numbers from 1001 to 1026 are node identifiers. The numbers inside the parentheses are the probabilities given by the Bayesian network that model findings (see Figure 1▶) from the chest x-ray report. First, the individual words of the sentence are instantiated in the lower-lever nodes (indicated by a probability of 1.0). Then the conceptual nodes (indicated by asterisks) are inferred through probability propagation.
Pneumonia Concepts
All the subjects (human and automatic) were tested for their ability to identify each of the following concepts from the chest x-ray reports: pneumonia, infiltrate compatible with acute bacterial pneumonia, and aspiration. The three concepts were selected on the basis of actual requirements of the antibiotic assistant. In addition, all the subjects were asked to make an inference, on the basis of the report only, on acute bacterial pneumonia as a disease in the patient. We distinguished between pneumonia as a concept and acute bacterial pneumonia as a disease inferred from the report.
The gold standard physicians typically considered the concept pneumonia to be present if the radiologist stated the term “pneumonia” or a synonym such as “pneumonitis.” For the inference of acute bacterial pneumonia, the radiologist might not explicitly mention “pneumonia” in the report. Consider, as an example, the following two phrases from two different reports: “pneumonia in the right lower lobe” and “consolidation in the left lower lobe.” In the first sentence, the pneumonia concept is present and the disease pneumonia is easily inferred. In the second sentence, although pneumonia is not mentioned explicitly (pneumonia as a concept is absent), the entire report may allow the inference of the disease pneumonia. This distinction was made on the basis of the requirements of the antibiotic assistant. The antibiotic assistant does not try to infer whether the whole report supports acute bacterial pneumonia, but rather only searches for the three pneumonia-related concepts. The pneumonia concepts from chest x-ray reports are then combined with other sources, including laboratory and microbiology data, to determine the presence of the disease. The ability to infer the presence of acute bacterial pneumonia on the sole basis of chest x-ray reports might be important for other computer applications.
A block diagram of the flow of information for this project is shown in Figure 3▶. First, the subjects identified the three concepts required by the antibiotic assistant. Then the subjects inferred whether the whole report supports pneumonia. In the following paragraphs we explain how the automatic systems (SymText, AAKS, and KS) identify the three concepts and make the inference.
Figure 3.
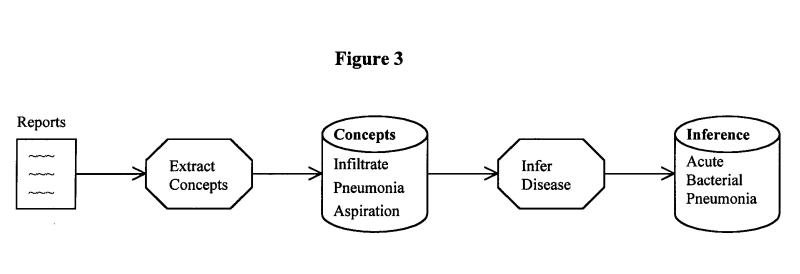
Flow of information for all subjects (human and automatic) in the study.
For each report, a rule-based algorithm was applied to SymText's output, to determine the presence or absence of the three literal pneumonia concepts. These rules are shown in Figure 4▶. The rules search the concepts in our semantic model (the Bayesian networks) and translate to the concepts needed by the antibiotic assistant. For example, the rule for infiltrate searches for the presence of such concepts as localized infiltrate, localized lower lobe infiltrate, and localized consolidation.
Figure 4.

Rules applied to SymText's output for a report to extract the three pneumonia concepts.
The two keyword searches (AAKS and KS) look for specific words or a combination of words inside the reports. For instance, both search for the strings “pneumoni” and “aspirati” in the report. If they cannot negate these strings with words such as “no,” “no evidence of,” or “not,” then pneumonia or aspiration are considered to be present.
The keyword searches differ in how they extract the infiltrate concept. AAKS simply searches for the string “infiltr.” KS is more general and uses more terms, such as “opacit,” to define the concept infiltrate. Radiologists frequently describe infiltrates by using a general term like “opacity” and modifiers such as “hazy,” “ill-defined,” and “patchy.”
To infer whether the report supports acute bacterial pneumonia as a disease in the patient, we applied a simple rule to all the automatic methods after they processed the three literal concepts. If one of the three concepts (pneumonia, aspiration, and infiltrate) was present, then the report supported pneumonia; if not, the report did not support pneumonia. Other studies have tested different methods for making inferences from automatically extracted concepts.17,18
Reports
For this study, we selected 292 reports from about 15,000 chest x-ray reports produced during a six -month period (October 1998 to March 1999) at LDS Hospital, Salt Lake City, Utah. Of the 292 reports, 217 were randomly selected from all the reports stored in the HELP System in the first three-month period (October to December). From the following three-month period (January to March), the remaining 75 reports were randomly selected from a list of patients with a primary ICD-9 hospital discharge diagnosis of bacterial pneumonia. We used this approach to increase the prevalence of pneumonia-related reports in our sample. We did not constrain the selected reports to the patient's admission chest x-ray report, because the antibiotic assistant searches for pneumonia information in all available chest x-ray reports during a patient's hospital encounter.
Gold Standard Reliability
Measuring a system's performance requires a reliable gold standard. Reliability19 is a measure of gold standard quality that quantifies the agreement among the experts who generated the standard. To assess the reliability of the gold standard, we followed a methodology based on generalizability theory that was proposed by Shavelson et al.20 The methodology was adapted to the NLP domain by Hripcsak et al.21 Following this methodology, a generalizability coefficient (ρ) was computed. The coefficient reflects the reliability of the gold standard and ranges from 0 to 1. The higher the coefficient, the greater the confidence in the reference standard.
We calculated the generalizability coefficient for each of the three concepts and the disease inference using the following variance component model:
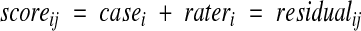 |
1 |
where score is the answer provided by the physician for a concept, i is an index on the random facet case (292 reports), and j is an index on the random facet rater (three raters).
Estimated variance components (σ2) were calculated from the output of a two-way ANOVA based on the model of equation (1) as explained in the review by Shavelson and Webb.20 From the estimated variance components we calculated the generalizability coefficient per rater for each concept using the formula:
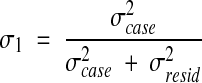 |
2 |
The numerator contains the estimated variance components of the facet of interest. The denominator sums the estimated variance components of the facet of interest with the estimated variance components of all the sources of errors.
We obtained the actual generalizability coefficient for our study by dividing the residual component by three raters as follows:
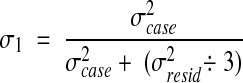 |
3 |
Outcome Measures
We calculated recall (sensitivity), precision (positive predictive value), and specificity with their respective 95 percent confidence intervals (95%CI) for each of the three concepts and the inference on acute bacterial pneumonia as follows:
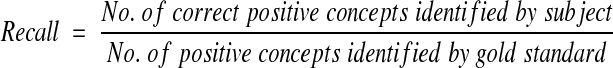 |
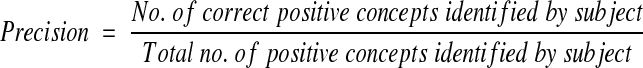 |
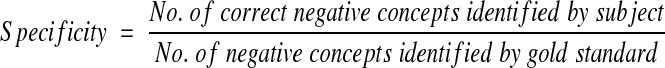 |
Recall and specificity were plotted on receiver operator characteristics axes (ROC plots). Finn's R statistic was applied to measure the agreement between each of the subjects (physicians, SymText, AAKS, KS, lay persons) and the gold standard.22
We used the McNemar test23 to determine whether the subjects were different from the physicians on any of the concepts. The gold standard was used to determine whether the answers provided by the physicians and by the other subjects were correct or incorrect (i.e., whether the subject's answer for a concept matched the gold standard). An alpha of 0.05 was used with a Bonferroni correction for 96 multiple comparisons among the four physicians and the other six subjects (SymText, AAKS, KS, and three lay persons). Since the main objective of this paper is to compare the NLP system with the physician experts, a less conservative Bonferroni correction (16 multiple comparisons) is also reported in comparisons of SymText against the four physicians.
In addition, we used the McNemar test to compare the NLP system with the other five subjects on the disease inference for pneumonia. In this analysis, an alpha of 0.05 was used with a Bonferroni correction for five multiple comparisons.
A power calculation to detect a difference (including the Bonferroni correction) was performed for the 16 comparisons between the NLP system and the physicians whenever the null hypothesis was not rejected.
Results
The reliability measures for the reference standard are shown in Table 1▶. For each concept in the study, the table presents the generalizability coefficient per rater (equation 1) and the generalizability coefficient for the three raters who constituted the gold standard (equation 2). The gold standard reliability ranged from 0.86 (for the infiltrate concept) to 0.96 (for the pneumonia concept).
Table 1.
Reliability Measures for the Reference Standard
| Generalizability Coefficient per Rater (ρ1) | Actual Generalizability Coefficient (ρ3), 3 Raters | |
|---|---|---|
| Pneumonia | 0.91 | 0.96 |
| Aspiration | 0.82 | 0.93 |
| Infiltrate | 0.68 | 0.86 |
| Support pneumonia | 0.72 | 0.89 |
Recall, precision, and specificity for every subject are presented with their respective 95 percent confidence intervals in Table 2▶. In Figure 5▶, recall (sensitivity) is plotted against specificity on ROC axes for all the concepts.
Table 2.
Performance Measures for All Subjects in the Study
| MD1 | MD2 | MD3 | MD4 | SymText | AAKS | KS | Lay1 | Lay2 | Lay3 | |
|---|---|---|---|---|---|---|---|---|---|---|
| Pneumonia (N = 47): | ||||||||||
| Recall | 0.98 (0.94–1.00) | 0.96 (0.90–1.00) | 1 (0.80–0.98) | 0.98 (0.94–1.00) | 0.94 (0.87–1.00) | 0.94 (0.87–1.00) | 0.83 (0.72–0.94) | 0.89 (0.80–0.98) | 0.72 (0.59–0.85) | 0.89 (0.80–0.98) |
| Precision | 0.96 (0.90–1.00) | 0.94 (0.87–1.00) | 0.96 (0.91–1.00) | 0.98 (0.94–1.00) | 0.96 (0.90–1.00) | 0.52 (041–0.63) | 0.83 (0.72–0.94) | 0.81 (0.70–0.92) | 0.87 (0.76–0.98) | 0.95 (0.89–1.00) |
| Specificity | 0.99 (0.98–1.00) | 0.99 (0.98–1.00) | 0.99 (0.98–1.00) | 0.99 (0.98–1.00) | 0.99 (0.98–1.00) | 0.83 (0.78–0.88) | 0.97 (0.95–0.99) | 0.96 (0.94–0.98) | 0.98 (0.96–1.00) | 0.99 (0.98–1.00) |
| Aspiration (N = 6): | ||||||||||
| Recall | 1 | 1 | 1 | 1 | 1 | 0.83 (0.53–1.00) | 1 | 0.66 (0.28–1.00) | 0.83 (0.53–1.00) | 0.83 (0.53–1.00) |
| Precision | 1 | 1 | 1 | 1 | 0.75 (0.45–1.00) | 0.62 (0.28–0.96) | 0.85 (0.59–1.00) | 0.80 (0.45–1.00) | 0.35 (0.10–0.60) | 1 |
| Specificity | 1 | 1 | 1 | 1 | 0.99 (0.98–1.00) | 0.98 (0.96–1.00) | 0.99 (0.98–1.00) | 0.99 (0.98–1.00) | 0.97 (0.95–1.00) | 1 |
| Infiltrate (N = 132): | ||||||||||
| Recall | 0.80 (0.73–0.87) | 0.93 (0.89–0.97) | 0.79 (0.72–0.86) | 0.77 (0.70–0.84) | 0.84 (0.78–0.90) | 0.56 (0.48–0.64) | 0.76 (0.69–0.83) | 0.045 (0.01–0.08) | 0.13 (0.07–0.19) | 0.14 (0.08–0.20) |
| Precision | 0.97 (0.94–1.00) | 0.92 (0.87–0.97) | 0.96 (0.92–1.00) | 0.93 (0.88–0.98) | 0.87 (0.81–0.93) | 0.92 (0.86–0.98) | 0.82 (0.75–0.89) | 0.75 (0.45–1.0) | 0.94 (0.83–1.00) | 0.63 (0.46–0.80) |
| Specificity | 0.98 (0.96–1.00) | 0.94 (0.90–0.98) | 0.97 (0.94–1.00) | 0.95 (0.92–0.98) | 0.90 (0.85–0.95) | 0.96 (0.93–0.99) | 0.86 (0.81–0.91) | 0.98 (0.96–1.00) | 0.99 (0.97–1.00) | 0.93 (0.89–0.97) |
| Support Pneumonia (N = 112): | ||||||||||
| Recall | 0.94 (0.90–0.98) | 0.93 (0.88–0.98) | 0.97 (0.94–1.00) | 0.91 (0.86–0.96) | 0.95 (0.91–0.99) | 0.79 (0.71–0.87) | 0.87 (0.81–0.93) | 0.38 (0.29–0.47) | 0.58 (0.49–0.67) | 0.41 (0.32–0.50) |
| Precision | 0.83 (0.76–0.90) | 0.92 (0.87–0.97) | 0.88 (0.82–0.94) | 0.84 (0.77–0.91) | 0.78 (0.71–0.85) | 0.63 (0.55–0.71) | 0.70 (0.62–0.78) | 0.86 (0.76–0.96) | 0.85 (0.77–0.93) | 0.97 (0.92–1.00) |
| Specificity | 0.88 (0.83–0.93) | 0.95 (0.92–0.98) | 0.92 (0.88–0.96) | 0.89 (0.84–0.94) | 0.85 (0.80–0.90) | 0.71 (0.64–0.78) | 0.77 (0.71–0.83) | 0.96 (0.93–0.99) | 0.93 (0.89–0.97) | 0.99 (0.98–1.00) |
note: The 95 percent confidence intervals (95% CI) are shown in parentheses. N is the number of reports containing the concept, as judged by the gold standard.
Figure 5.
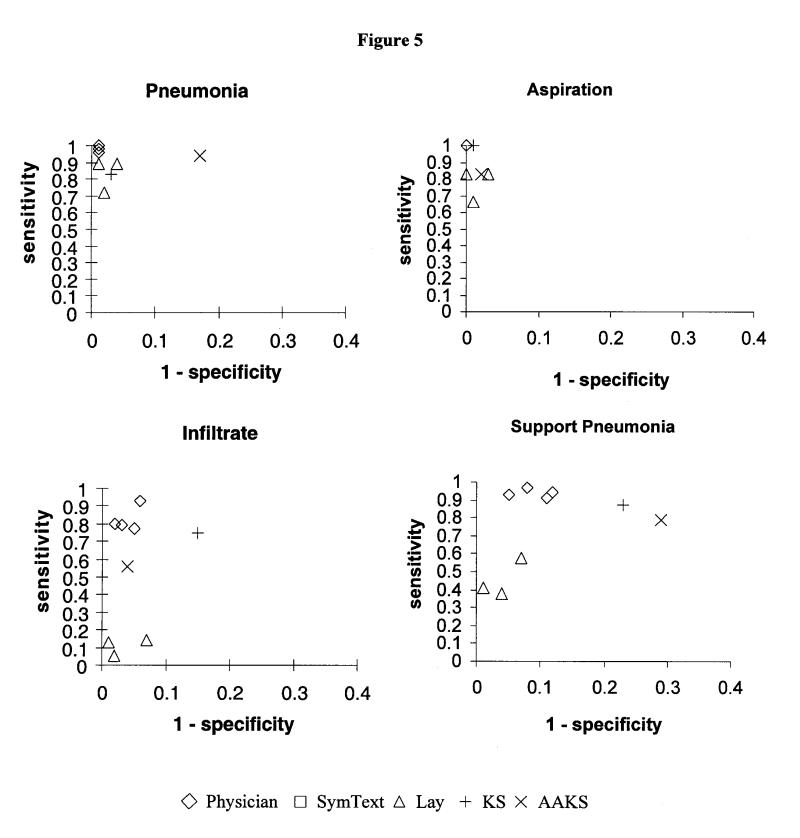
Performance as indicated by sensitivity x (1 – specificity) for every subject plotted on receiver operator characteristics (ROC) curve axes. Diamonds indicate physicians; squares, SymText; triangles, lay persons. Keyword searches are indicated by plus signs (KS) and times signs (AAKS).
For the disease inference in acute bacterial pneumonia, the physicians had an average recall of 94 percent (CI, 91–96 percent), an average precision of 87 percent (CI, 83–91 percent), and average specificity of 91 percent (CI, 88–94 percent). SymText had recall of 95 percent (CI, 91–99 percent), precision of 78 percent (CI, 71–85 percent) and specificity of 85 percent (CI, 80–90 percent). The keyword searches followed with an average recall of 83 percent, average precision of 65 percent, and average specificity of 74 percent. The KS slightly outperformed the AAKS, but they were not statistically different from each other. The lay persons had a lower performance with 45 percent recall but higher precision and specificity (89 and 96 percent, respectively).
For the infiltrate concept, the physicians had an average recall of 84 percent (CI, 77–91 percent), an average precision of 95 percent (CI, 93–97 percent), and average specificity of 96 percent (CI, 94–98 percent). SymText had recall of 84 percent (CI, 78–90 percent), precision of 87 percent (CI, 81–93 percent) and specificity of 90 percent (CI, 85–95 percent). The other methods did not perform as well. The keyword search from the antibiotic assistant (AAKS) had 56 percent recall (CI, 48–64 percent) for the infiltrate concept. The other keyword search (KS) had higher recall for infiltrate with 76 percent (CI, 69–83 percent) but lower specificity with 86 percent (CI, 81–91 percent). The average lay person achieved 10 percent recall for infiltrate, but specificity was higher with 97 percent.
In Figure 5▶, SymText is clustered with the physicians in a position of higher performance on the ROC plots for all the concepts and for the disease inference. The lay persons are clustered in a substantially lower performance position. The keyword searches were in an intermediate position, but neither of them achieved the performance exhibited by SymText.
The agreement between each subject and the gold standard, as measured by Finn's R statistic, is shown in Figure 6▶. For the pneumonia inference, Finn's R statistic for the four physicians ranged from 0.8 to 0.89, with an average of 0.84 (CI, 0.83–0.85). For the concept infiltrate, Finn's R for the four physicians ranged from 0.74 to 0.87, with an average of 0.80 (CI, 0.75–0.85). SymText had a Finn's R of 0.75 for the disease inference and 0.74 for the infiltrate concept. The other methods had lower measures of agreement with the gold standard. The three lay persons had an average Finn's R of 0.16 for the infiltrate concept and 0.54 for the pneumonia inference. The KS slightly outperformed the AAKS (0.62 compared with 0.49 for the disease inference and 0.64 compared with 0.56 for the infiltrate concept).
Figure 6.
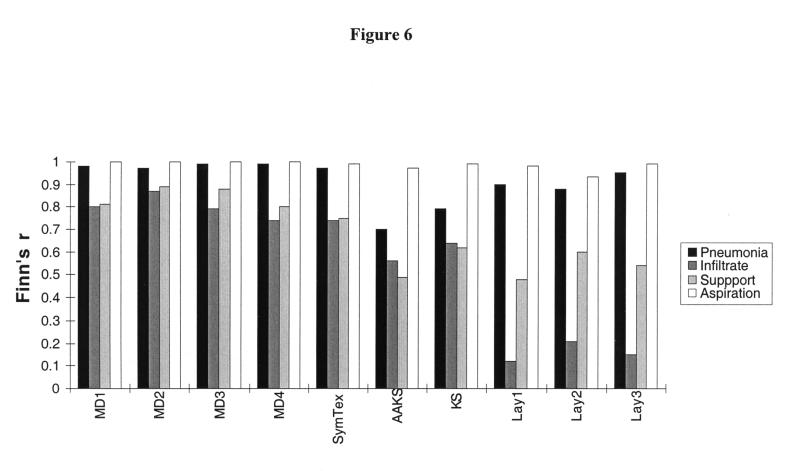
Agreement with reference standard, as measured by Finn's R statistic. Physicians are indicated by MD1, MD2, and MD3; keyword searches by AAKS and KS; and lay persons by Lay1, Lay2, and Lay3.
A matrix of P values produced by the McNemar test for the pairwise comparisons between physicians and subjects is shown in Table 3▶. SymText did not differ significantly from the physicians after Bonferroni corrections for 96 multiple comparisons. However, the less conservative approach of 16 multiple comparison showed differences between one physician and SymText for the infiltrate concept and between another physician and SymText on the disease inference for pneumonia (represented by an asterisk [*] in Table 3▶). The comparisons also showed that most physicians were significantly different from the other subjects (keyword searches and lay persons) in the disease inference and in all concepts except aspiration. SymText outperformed the other subjects on the disease inference for pneumonia (P < 0.01 for five comparisons between SymText and the other subjects).
Table 3.
Matrix of P Values for Pairwise Comparison Using the McNemar Test.
| SymText | AAKS | KS | Lay1 | Lay2 | Lay3 | ||
|---|---|---|---|---|---|---|---|
| MD1 | Pneumonia | 0.68750 | 0.00000 | 0.00235 | 0.34375 | 0.00149 | 0.00753 |
| Aspiration | 0.50000 | 0.12500 | 1.00000 | 1.00000 | 0.00149 | 0.25000 | |
| Infiltrate | 0.17417 | 0.00000 | 0.00150 | 0.00000 | 0.00000 | 0.00000 | |
| Support pneumonia | 0.26819 | 0.00000 | 0.00014 | 0.00002 | 0.00053 | 0.00000 | |
| MD2 | Pneumonia | 1.00000 | 0.00000 | 0.00000 | 0.77412 | 0.00719 | 0.02127 |
| Aspiration | 0.50000 | 0.12500 | 1.00000 | 1.00000 | 0.00149 | 0.25000 | |
| Infiltrate | 0.00222* | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | |
| Support pneumonia | 0.00366 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | |
| MD3 | Pneumonia | 0.37500 | 0.00000 | 0.00012 | 0.17968 | 0.00040 | 0.00097 |
| Aspiration | 0.50000 | 0.12500 | 1.00000 | 1.00000 | 0.00149 | 0.25000 | |
| Infiltrate | 0.32224 | 0.00000 | 0.00382 | 0.00000 | 0.00000 | 0.00000 | |
| Support pneumonia | 0.00294* | 0.00000 | 0.00000 | 0.00000 | 0.00000 | 0.00000 | |
| MD4 | Pneumonia | 0.25000 | 0.00000 | 0.00050 | 0.17960 | 0.00040 | 0.00234 |
| Aspiration | 0.50000 | 0.12500 | 1.00000 | 1.00000 | 0.00149 | 0.25000 | |
| Infiltrate | 1.00000 | 0.00120 | 0.06200 | 0.00000 | 0.00000 | 0.00000 | |
| Support pneumonia | 0.39160 | 0.00000 | 0.00068 | 0.00002 | 0.00051 | 0.00000 |
note: Keyword searches are indicated by AAKS and KS, laypersons by Lay1, Lay2, and Lay3. Bold font indicates statistical significance (p < 0.000521) after Bonferroni correction for 96 overall comparisons. Asterisks (*) indicate statistical significance for P < 0.003125 after a less conservative Bonferroni correction for 16 overall comparisons between the NLP system and the physicians.
In 14 of the 16 multiple comparisons between the NLP system and the physicians, the null hypothesis was not rejected. The power to detect a difference was greater than 80 percent in all 14 comparisons and was most often greater than 90 percent. Therefore, the study had enough power and sample size to detect real differences between the physicians and the NLP system.
Discussion
We studied the ability of an NLP system post-processed by a rule-based algorithm to identify pneumonia-related concepts from chest x-ray reports. The performance of the NLP system was closer to the performance of the physicians than were any other subjects in the study. Pairwise comparisons showed minor differences between some physicians and the NLP system, but the other subjects were clearly inferior to the physician experts. The NLP system outperformed the other subjects when inferring whether the report supported pneumonia, and it demonstrated a physician-like agreement profile with the gold standard for all the pneumonia concepts. The only exception was the aspiration concept, for which all subjects had similar performance. Given the low prevalence of aspiration (6 of 292 reports), we probably did not have enough cases to make any conclusion on this concept.
The surprising result was the low performance of the already implemented AAKS. Recall is probably the most important performance measure for the antibiotic assistant application, because the program collects information from other sources (laboratory and microbiology data) before it decides whether a patient has acute bacterial pneumonia. Although AAKS had good recall for the pneumonia and aspiration concepts, it demonstrated low performance for infiltrates, with 54 percent recall. A recall of 54 percent means that AAKS missed a subset of the infiltrates compatible with acute bacterial pneumonia.
AAKS also had low precision (52 percent) for the pneumonia concept. It was not able to detect negations of the concept pneumonia when negations were distributed across a complex phrase or sentence. For example, in the sentence “I see no evidence of atelectasis, scarring, or pneumonia,” the phrase “no evidence” must be distributed among the three concepts (atelectasis, scarring, pneumonia) to detect the negation of pneumonia. The other keyword search (KS) had a higher performance but did not achieve the physician-like performance of SymText.
Describing radiographic support for pneumonia is complex, and keyword searches may not perform on a level sufficient for the successful extraction of pneumonia concepts. If we were targeting a different disease, like pneumothorax, for which description of radiographic findings is explicit, then the keyword approaches might have been sufficient. However, designing keyword searches for every clinical concept is less desirable than using a general-purpose encoding mechanism.
The three lay persons in our study had good levels of precision and specificity but lower recall, particularly for the infiltrate concept. Although the lay persons did not have a medical background, they were able to recognize the presence and absence of the pneumonia concepts if the concepts were explicitly mentioned in the report. For example, in the sentences “infiltrates in the left lower lobe” and “no evidence of pneumonia,” the lay persons recognized the presence of infiltrates and the absence of pneumonia. However, the lay persons failed to recognize concepts when medical vocabulary and induction was required. For example, the lay persons did not identify an infiltrate in the sentence “ill-defined patchy opacity with air bronchograms in the left upper lobe.”
A generalizability coefficient of 0.7 or higher is considered adequate if the gold standard is going to be used only to estimate the overall performance of a system.21 In our study, the actual generalizability coefficient that took the three raters into account (equation 3) was above 0.7 for all the pneumonia concepts. The lowest generalizability coefficient was on the infiltrate concept, 0.68 per rater (equation 2) and 0.86 when the three raters (equation 3) were taken into account. We were not surprised by this lower coefficient, because deciding whether a particular infiltrate is compatible with acute bacterial pneumonia is sometimes difficult. The generalizability coefficient per rater (equation 2) on the acute bacterial pneumonia inference was 0.72 and correlates well with the 0.70 coefficient published by Hripcsak et al.21
Having good reliability measures does not reflect absence of disagreement within the gold standard. Disagreement between gold standard physicians averaged 17 percent for the concept infiltrate and 16 percent for the inference on acute bacterial pneumonia. Similar disagreement measures were found in another NLP evaluation study11 and in studies for which physicians performed other diagnostic tasks.24,25
A limitation of this study is the small number of clinical conditions that were evaluated. Although SymText was developed to code most of the conditions on a chest x-ray report, we limited this evaluation of SymText to one clinical condition (acute bacterial pneumonia). The level of performance may differ for other diseases, such as congestive heart failure, neoplasms, pneumothorax, and atelectasis.
Another limitation is the prevalence of pneumonia in our sample of reports. To increase the prevalence of acute bacterial pneumonia, we enriched our data set with 75 reports from patients with a primary discharge diagnosis of pneumonia. We do not know how well the performance measures will generalize to a population of reports with the actual disease prevalence. There is contradiction in the literature on how performance measures such as recall, precision, and specificity vary with the prevalence of conditions. Traditionally, recall (sensitivity) and specificity have generally been thought of as being independent of disease prevalence. In contrast, precision (positive predictive value) is highly dependent on disease prevalence.26 A recent study using simulation of these performance measures actually demonstrated that they all vary with prevalence.27
A third limitation of this study is that we did not compare the NLP performance with the average physician performance. In our methodology the scores used for the McNemar test are dichotomous, and there is no way to compute averages. As far as we know, the only outcome metric in NLP evaluation that allows computation of averages is a distance metric.11 This metric was used in an evaluation study for six clinical conditions and was not applicable in our study. However, we did compare the NLP system with each of the physicians and found that the system was different from some physicians and not different from others.
Depending on the context of the application, one performance measure might be more important than another. For the antibiotic assistant program, recall is more important than precision and specificity. However, other applications may require higher precision and tolerate lower sensitivity. Therefore, when developing and evaluating generally applicable NLP systems, a system that yields good performance in all the measures (recall, precision and specificity) is preferred.
SymText is more sensitive than AAKS without significant loss of specificity and precision. Therefore, we are now planning to use SymText to store the coded pneumonia data in the electronic medical record of the HELP system. Using SymText potentially increases the detection of pneumonia and may improve the overall recommendations of the antibiotic assistant for pneumonia patients. This potential improvement needs to be demonstrated in the clinical context of the real-time recommendations of the antibiotic assistant.
Other applications may benefit from having pneumonia-related concepts coded in the hospital information system. Radiographic information from chest x-ray reports is required for clinical pneumonia guidelines.28 Natural language processing systems can provide, from chest x-ray reports, coded data that support real-time computerization of pneumonia guidelines. However, pneumonia guidelines require information about localization and severity of pneumonia findings. To drive computerized pneumonia guidelines, NLP systems will have to be evaluated not only for the ability to detect clinical condition but also for the ability to localize and determine extension of the disease.
Quality assurance initiatives in the radiology department may benefit from having coded data in the hospital information system. A quality assurance study on diagnostic interpretations of radiologists could compare pneumonia interpretations with outcome data from other sources, such as the discharge diagnosis. Quality assurance studies in diagnostic interpretations of the radiologists usually require double readings of films. However, double reading of films is expensive and time consuming. Comparing coded interpretations from chest x-ray reports with outcome measures like the discharge diagnoses may reduce the number of films that require the traditional double reading process.29
Automatic systems need coded data. However, a large quantity of information is stored in free-text format. Natural language processing systems are an appealing method of encoding free-text reports and unlocking the content of those reports for a variety of applications. Evaluation studies are be needed to reveal whether NLP systems can fulfill the needs of those applications. However, issues such as implementation in a hospital information system, extensibility to different types of reports, and portability to other institutions must be addressed before NLP systems attain widespread use.
Conclusion
We have shown that the performance of an NLP system was similar to the performance of physicians and superior to the performance of lay persons and keyword searches in the extraction of pneumonia-related concepts from chest x-ray reports. The encoded pneumonia information has the potential to support several pneumonia-related applications, such as the antibiotic assistant, computerized clinical protocols for pneumonia, and quality assurance applications in the radiology department.
Acknowledgments
The authors thank the physicians who read the reports: Alexandra Pagel Eidelwein, MD, Gustavo Oderich, MD, Kenneth Zollo, MD, Ann Giulliano, MD, Bruce Bray, MD, Gregory Patton, MD, and Chuck Mullet, MD. They also thank Lee Christensen, David Chapman, Shauna Bruun, and Becky Seegmiller for their collaboration in this project.
This work was supported in part by grant LM-06539 from the National Library of Medicine and grant HL-53427 from the National Heart, Lung and Blood Institute of the National Institutes of Health. Dr. Fiszman is supported by a scholarship (FAPESP 96/3922-9) from the Research Foundation of the State of Sao Paulo, Brazil.
References
- 1.Auble TE, Yealy DM, Fine MJ. Assessing prognosis and selecting an initial site of care for adults with community-acquired pneumonia. Infect Dis Clin North Am. 1998;12(3):741–59. [DOI] [PubMed] [Google Scholar]
- 2.Aronsky D, Haug PJ. Diagnosing community-acquired pneumonia with a Bayesian network. Proc AMIA Symp. 1998:632–6. [PMC free article] [PubMed]
- 3.Evans RS, Pestotnik SL, Classen DC, Burke JP. Development of an automated antibiotic consultant. MD Comput. 1993;10(1):17–22. [PubMed] [Google Scholar]
- 4.Evans RS, Pestotnik SL, Classen DC, et al. A computer-assisted management program for antibiotics and other anti-infective agents. N Engl J Med. 1998;338(4):232–8. [DOI] [PubMed] [Google Scholar]
- 5.Sager N. Medical Language Processing: Computer Management of Narrative Data. New York: Springer-Verlag, 1997.
- 6.Friedman C, Alderson PO, Austin JH, Cimino JJ, Johnson SB. A general natural-language text processor for clinical radiology. J Am Med Inform Assoc. 1994;1:161–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sager N, Lyman M, Bucknall C, Nhan N, Tick LJ. Natural language processing and the representation of clinical data. J Am Med Inform Assoc. 1994;1:142–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zingmond D, Lenert LA. Monitoring free-text data using medical language processing. Comput Biomed Res. 1993;26(5):467–81. [DOI] [PubMed] [Google Scholar]
- 9.Haug P, Koehler S, Lau LM, Wang P, Rocha R, Huff S. A natural language understanding system combining syntactic and semantic techniques. Proc Annu Symp Comput Appl Med Care. 1994:247–51. [PMC free article] [PubMed]
- 10.Haug PJ, Koehler S, Lau LM, Wang P, Rocha R, Huff SM. Experience with a mixed semantic/syntactic parser. Proc Annu Symp Comput Appl Med Care. 1995:284–8. [PMC free article] [PubMed]
- 11.Hripcsak G, Friedman C, Alderson PO, DuMouchel W, Johnson SB, Clayton PD. Unlocking clinical data from narrative reports: a study of natural language processing. Ann Intern Med. 1995;122(9):681–8. [DOI] [PubMed] [Google Scholar]
- 12.Pryor TA. The HELP system. The HELP medical record system. MD Comput. 1988;5(5):22–33. [PubMed] [Google Scholar]
- 13.Gundersen ML, Haug PJ, Pryor TA, et al. Development and evaluation of a computerized admission diagnoses encoding system. Comput Biomed Res. 1996;29(5):351–72. [DOI] [PubMed] [Google Scholar]
- 14.Fiszman M, Haug PJ, Frederick PR. Automatic extraction of pioped interpretations from ventilation/perfusion lung scan reports. Proc AMIA Symp. 1998:860–4. [PMC free article] [PubMed]
- 15.Allen J. Natural Language Understanding. Redwood City, Calif: Benjamin Cummings, 1994.
- 16.Pearl J. Probabilistic rasoning in intelligent systems: networks of plausible inference. San Francisco, Calif: Morgan Kaufmann, 1988.
- 17.Wilcox A, Hripcsak G. Classification algorithms applied to narrative reports. Proc AMIA Symp. 1999:455–9. [PMC free article] [PubMed]
- 18.Chapman WW, Haug PJ. Comparing expert systems for identifying chest x-ray reports that support pneumonia. Proc AMIA Symp. 1999:216–20. [PMC free article] [PubMed]
- 19.Friedman CP, Wyatt JC. Evaluation Methods in Medical Informatics. New York: Springer-Verlag, 1997.
- 20.Shavelson RJ, Webb NM. Generalizability Theory: A Primer. Newbury Park, Calif: Sage, 1991.
- 21.Hripcsak G, Kuperman GJ, Friedman C, Heitjan DF. A reliability study for evaluating information extraction from radiology reports. J Am Med Inform Assoc. 1999;6:143–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Whitehurst GJ. Interrater agreement for journal manuscript reviews. Am Psychol. 1984; 39(1):22–8. [Google Scholar]
- 23.Zar JH. Biostatistical Analysis. Englewood Cliffs, N.J.: Prentice Hall, 1974.
- 24.Herman PG, Gerson DE, Hessel SJ, et al. Disagreements in chest roentgen interpretation. Chest. 1975;68(3):278–82. [DOI] [PubMed] [Google Scholar]
- 25.Koran LM. The reliability of clinical methods, data and judgments. N Engl J Med. 1975;293(14):695–701. [DOI] [PubMed] [Google Scholar]
- 26.Kramer MS. Clinical Epidemiology and Biostatistics: A Primer for Clinical Investigators and Decision Makers. New York: Spinger-Verlag, 1988.
- 27.Brenner H, Gefeller O. Variation of sensitivity, specificity, likelihood ratios and predictive values with disease prevalence. Stat Med. 1997;16(9):981–91. [DOI] [PubMed] [Google Scholar]
- 28.Niederman MS, Bass JB Jr, Campbell GD, et al. Guidelines for the initial management of adults with community-acquired pneumonia: diagnosis, assessment of severity, and initial antimicrobial therapy. American Thoracic Society. Medical Section of the American Lung Association. Am Rev Respir Dis. 1993;148(5):1418–26. [DOI] [PubMed] [Google Scholar]
- 29.Haug PJ, Frederick PR, Tocino I. Quality control in a medical information system. Med Decis Making. 1991;11(4 suppl):S57–60. [PubMed] [Google Scholar]


