Abstract
In herpes simplex virus, lytic replication is initiated by the viral transactivator VP16 acting with cellular cofactors Oct-1 and HCF-1. Although this activator complex has been studied in detail, the role of HCF-1 remains elusive. Here, we show that HCF-1 contains an activation domain (HCF-1AD) required for maximal transactivation by VP16 and its cellular counterpart LZIP. Expression of the VP16 cofactor p300 augments HCF-1AD activity, suggesting a mechanism of synergy. Infection of cells lacking the HCF-1AD leads to reduced viral immediate-early gene expression and lowered viral titers. These findings underscore the importance of HCF-1 to herpes simplex virus replication and VP16 transactivation.
Keywords: p300‖transcriptional activation‖herpes simplex virus (HSV)
Herpes simplex virus (HSV) serves as a paradigm for understanding the regulation of RNA polymerase II-mediated transcription in mammalian cells. HSV lytic replication follows a cascade of gene expression beginning with the immediate-early (IE) genes (reviewed in refs. 1 and 2). At low multiplicities of infection, IE gene expression depends on VP16 (Vmw65 or αTIF), which directs the assembly of a multiprotein–DNA complex that includes two cellular proteins, HCF-1 and Oct-1, and a VP16-responsive DNA sequence (the TAATGARAT element) found in every IE gene promoter (reviewed in refs. 3–5). Transactivation by the VP16-induced complex requires the well characterized C-terminal acidic activation domain of VP16 (VP16AD), which has been shown to interact with numerous targets (reviewed in refs. 1, 6, and 7).
How HCF-1 contributes to transcriptional activation by the VP16-induced complex is poorly understood. HCF-1 is a ubiquitous chromatin-associated protein required for cell proliferation (8–10). Analysis of tsBN67 cells, a hamster cell line that expresses a temperature-sensitive (ts) version of HCF-1, showed that HCF-1 is essential for progression through G1 (11). Transactivation by VP16 is equally sensitive to the ts mutation and reflects loss of VP16-induced complex formation (11–13).
HCF-1 is synthesized as a 2,035-aa precursor that undergoes proteolytic processing at reiterated sites (HCFPRO repeats) near the center of the polypeptide (14–16). The resulting N- and C-terminal subunits form a stable complex mediated by matched pairs of self-association sequences termed SAS1N-SAS1C and SAS2N-SAS2C (17). Association with VP16 requires an N-terminal domain consisting of six kelch repeats that form a β-propeller structure (13, 18–20). In addition to VP16, the HCF-1 β-propeller interacts with the cellular basic leucine zipper proteins LZIP (Luman) and Zhangfei (12, 21, 22). HCF-1 binds to the activation domain of LZIP and mutations that disrupt this interaction significantly reduce LZIP-dependent activation (23, 24). Other N-terminal regions of HCF-1 interact with the cellular transcription factors Sp1 and GABP (25, 26). Overexpression of the domain of HCF-1 required for GABP interaction inhibits GABP-dependent transcriptional activation (26). Thus, the N terminus of HCF-1 contains multiple modules capable of interacting with cellular and viral transcription factors.
Here we show that HCF-1 contains an activation domain (HCF-1AD) located in the C-terminal subunit. The HCF-1AD was not required for association of the N- and C-terminal subunits. Deletion of the HCF-1AD severely reduced transactivation by VP16 and LZIP. The functional interaction between the VP16AD and HCF-1AD is not restricted to the VP16-induced complex because the isolated activation domains were capable of synergistically activating transcription. Expression of p300, a known coactivator for VP16 (6, 27), enhanced the HCF-1AD function, implying a mechanism for synergy. The strong dependence on the HCF-1AD is highly specific because unrelated activators that are not known to recruit HCF-1 were unaffected by the HCF-1AD deletion. Furthermore, HSV lytic replication and transcription of the ICP0 and ICP4 genes were significantly reduced in the absence of the HCF-1AD. These observations prompt a change in our view of HCF-1, from a simple complex assembly factor to a more active component in the transactivation process.
Materials and Methods
Plasmid Constructs.
Human HCF-1 fragments were generated by PCR and subcloned into expression vectors pCGT (adds T7-epitope tag) or pCGNGal4 [adds hemagglutinin (HA)-epitope and Gal4DBD (28)]. LexA-HCF-1AD was generated by inserting the HCF-1AD into pCDNA3 plasmid, modified with the LexADBD. HCF-1ΔrepΔAD was generated by fusing pCGNHCF-1N1011 with HCF-1CΔAD through an engineered BglII site. LZIPN228 was generated by PCR amplification from full-length LZIP. GFP fusion proteins were generated by subcloning into pEGFP-C2 (CLONTECH). Plasmids encoding Gal4-Sp1, Gal4-c-Jun, Gal4-CREB, p5xGal4-E1B-luc, and pCMV-lacZ have been described (24). Plasmids encoding VP16FL, VP16ΔC, and Gal4-VP16AD have been described (15). The p300 plasmid was a gift of Lisa Daily, New York University. Luciferase reporters 1xTAAT/GA-tk-luc and 6xTAAT-tk-luc were made by inserting one copy (1x) of the sequence 5′-CCGGAACGGAAGCGGAAACCGCCGGATCGGGCGGTAATGAGATGCCATGCGGGC-3′ or six copies (6x) of the sequence 5′-CTCGAGGGCGGTAATGAGATACGACTCGAG-3′ (TAATGARAT in bold) into a modified version of pGL3-luciferase (Promega) containing the HSV tk promoter (gift of Heather Harding and David Ron, New York University). The cAMP response element (CRE) reporter contains four cAMP-response elements (gift of Ron Prywes, Columbia University, New York). The ERE-luc reporter and pCMV-ER were gifts of Michael Garabedian, New York University (29). The 2xGal4-2xLexA-E1B-luc reporter has been described (30).
Cell Culture and Protein Expression Assays.
HeLa, 293T, and tsBN67-derived cells were transfected by electroporation. Luciferase reporter assays, immunoprecipitations, and gel-mobility shift assays were performed as described (15, 24). Luciferase assays were normalized to β-galactosidase. For fluorescence microscopy, transfected HeLa cells were fixed in 0.2% formaldehyde, stained with Hoechst 33258 (Sigma), and observed with a Zeiss Axioplan 2 microscope.
Stable tsBN67-Derived Cells.
Subconfluent tsBN67 cells were transfected with 1 μg of a T7 epitope-tagged expression vector encoding either HCF-1Δrep WT or ΔAD and 500 ng pSV2neo by using Lipofectamine 2000 (Life Technologies, Grand Island, NY). After 24 h, Geneticin (0.8 mg/ml) was included to select for stable transfectants. After 24 h, cells were transferred to 39.5°C and cultured until individual colonies could be isolated and expanded.
Virus Growth Assays.
Infections were carried out with wild-type HSV-1 Patton strain (gift of Ian Mohr, New York University). Before infection, R-tsBN67 HCF-1Δrep WT or ΔAD cells were seeded at a density of 5 × 105 cells per dish and incubated at 33.5°C or 39.5°C. After 24 h, cells were infected at a multiplicity of infection (moi) of 0.001 and maintained at 33.5°C or 39.5°C in DMEM supplemented with 2% FBS for 5 days, with daily rocking to distribute the virus. At 5 days post infection, cells were lysed by freeze/thaw and total internal and external virus were collected and released by sonication for 1 min. Viral yield was measured by serial dilution on 1 × 106 Vero cells. After 4 days at 37°C, cells were fixed with 10% trichloroacetic acid for 10 min at room temperature and stained with 0.5% crystal violet in 80% methanol.
Preparation of RNA and RT-PCR.
Before infection, R-tsBN67 HCF-1Δrep WT or ΔAD cells were seeded at a density of 5 × 105 cells per dish and incubated at 39.5°C. After 24 h, cells were infected at a moi of 0.001 and maintained at 39.5°C in DMEM supplemented with 2% FBS for 4 h. Total RNA was extracted using the Ultraspec RNA Isolation System (Biotecx Laboratories, Houston) and analyzed using the Access RT-PCR system (Promega) with the following primers: β-actin, 5′-AACCCCAAGGCCAACCGTGAAAAGATGACC-3′ (sense) and 5′-GGTCCCTCCTTCTCCTACGCCG-3′ (antisense); ICP0, 5′-TTCGGTCTCCGCCTGAGAGT-3′ (sense) and 5′-GACCCTCCAGCCGCATACGA-3′ (antisense) (31); ICP4, 5′-CGACACGGATCCACGACCC-3′ (sense) and 5′-GATCCCCCTCCCGCGCTTCGTCCG-3′ (antisense) (32). Amplification products were fractionated on a 2.2% agarose gel and visualized by ethidium bromide staining.
Results
HCF-1 Contains a C-Terminal Activation Domain.
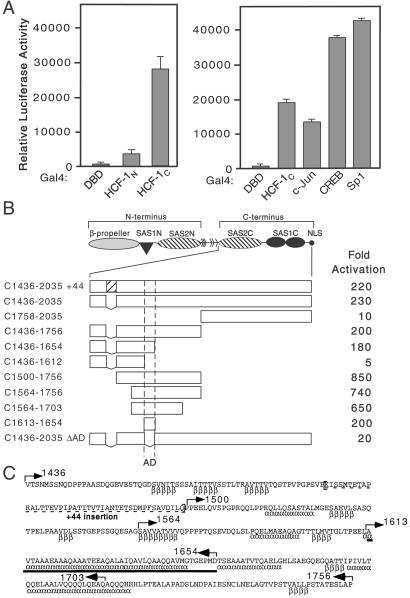
To understand how HCF-1 augments VP16 and LZIP dependent transactivation, we tested the N- and C-terminal subunits of HCF-1 for intrinsic activation. The N terminus (residues 1–1011, HCF-1N) and the C terminus of HCF-1 (residues 1436–2035, HCF-1C) were fused to the Gal4 DNA binding-domain (Gal4DBD) and expressed in 293T cells with a Gal4-responsive reporter (Fig. 1A). Gal4-HCF-1N gave only a modest stimulation (5-fold) compared with Gal4DBD alone, whereas Gal4-HCF-1C increased transcription 200-fold. Activation by Gal4-HCF-1C was slightly higher than Gal4-c-Jun and only 2-fold weaker than Gal4-CREB and Gal4-Sp1, other well characterized activators.
Figure 1.
The C terminus of HCF-1 contains an activation domain. (A) 293T cells were transfected with expression plasmids (250 ng) encoding Gal4DBD, Gal4-HCF-1N, Gal4-HCF-1C, or the activation domains of Gal4-c-Jun, Gal4-CREB, and Gal4-Sp1, and assayed for transactivation of a 5xGal4-E1B-luc reporter (500 ng). Extracts were prepared after 40 h and assayed for luciferase activity. Values represent means and standard deviations of three independent transfections. (B) The structure of the HCF-1 precursor is shown. The boundaries of the activation domain were delineated using N-terminal, C-terminal, or internal deletions fused to the Gal4DBD. Luciferase activity is scored as fold activation above Gal4DBD alone. (C) Sequence of human HCF-1 (residues 1436–1756) encompassing the HCF-1AD (solid bar). Alternative splicing removes 44 amino acids (dashed underline). Secondary structure predictions [α (α-helix) and β (β-sheet)] were derived using the Frishman and Argos method (50). Arrows indicate the boundaries of truncations used in B.
The C terminus of HCF-1 can be divided into a region rich in acidic amino acid residues (residues 1530–1735) and a pair of fibronectin type 3 (Fn3) repeats (residues 1758–2035) (15, 17). To further map the activation function, we assayed each region individually (Fig. 1B). A fragment corresponding to residues 1436–1756 activated transcription 200-fold, similar to HCF-1C, whereas residues 1758 to 2035 activated transcription only 10-fold. Thus, the activation domain of HCF-1 (termed HCF-1AD) lies within the acidic region, and does not require the Fn3 repeats. A splice variant of the C terminus that results in a 44-aa insertion had no effect on activation (Fig. 1B).
Fig. 1C shows the sequence of residues 1436–1756 from human HCF-1. Secondary structure predictions indicate several potential β-sheets and long α-helices (indicated below sequence as β and α). To further delineate the boundaries of the HCF-1AD, we assayed a series of additional truncations for transactivation. Deletion of N-terminal sequences (residues 1436–1499) led to a 4-fold increase in activity (compare C1436–1756 with C1500–1756), whereas truncations from the C terminus (deleting residues 1613–1756) abolished activity. A region spanning residues 1613–1654 was common to all strongly activating fragments and thus defines the HCF-1AD (bold underline in Fig. 1C). We tested this directly by fusing the 42-aa peptide to the Gal4DBD (C1613–1654), and also by deleting this sequence from the C terminus (C1436–2035 ΔAD). The 42-aa peptide activated the reporter as strongly as HCF-1C, confirming that this peptide contains the full activation function. Accordingly, activation was abolished when these residues were deleted from HCF-1C, indicating that there were no additional activating sequences.
The HCF-1AD Is Not Required for Self-Association or Nuclear Localization.
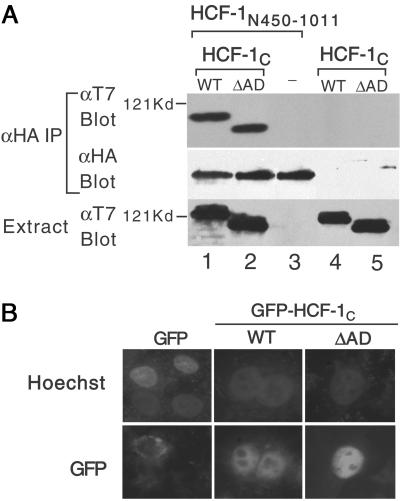
The SAS2C domain lies between residues 1436 and 1758 (17), and it is possible that the HCF-1AD deletion interferes with subunit association. We addressed this using a coimmunoprecipitation assay. Extracts were prepared from cells expressing HA-tagged HCF-1N450–1011, which contains only the SAS2N element and either WT or ΔAD versions of T7-tagged HCF-1C (Fig. 2A). HCF-1N450–1011 associated equally well with WT and ΔAD versions of HCF-1C (lanes 1 and 2), indicating that the removal of HCF-1AD does not interfere with SAS2C function. We also examined nuclear localization by fusing the WT and ΔAD versions of HCF-1C to green fluorescence protein (GFP). Both versions of GFP-HCF-1C showed similar patterns of nuclear localization (Fig. 2B), indicating that the HCF-1AD deletion does not interfere with subcellular localization.
Figure 2.
Deletion of HCF-1AD does not affect association with the N terminus or nuclear localization. (A) HA-tagged HCF-1N450–1011 and T7-tagged HCF-1C WT or ΔAD were coexpressed in transfected 293T cells and association assayed by immunoprecipitation with αHA antibody followed by immunoblotting with the αT7 antibody. The HA-tagged HCF-1N450–1011 is in lanes 1–3 and the T7-tagged HCF-1C WT and ΔAD in lanes 1 and 4 and lanes 2 and 5, respectively. (B) HeLa cells were transfected with expression plasmids (500 ng) encoding GFP or GFP-tagged HCF-1C WT or ΔAD. DNA is visualized with Hoechst 33258 and GFP by fluorescence microscopy.
Transactivation by VP16 and LZIP Is Compromised in Cells Stably Expressing a Version of HCF-1 Lacking the HCF-1AD.
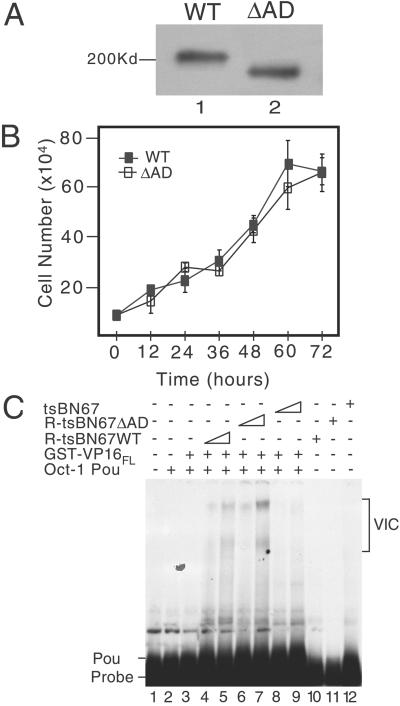
To investigate the functional relevance of deleting the HCF-1AD, we took advantage of the tsBN67 cell line, which contains a ts mutation in the N-terminal subunit of HCF-1 (11, 13). At the nonpermissive temperature, tsBN67 HCF-1 is functionally inactivated but can be complemented by expression of recombinant N terminus (13). To prevent exchange of subunits between recombinant and endogenous HCF-1 proteins, we used a derivative, HCF-1Δrep, which does not undergo proteolytic processing (16). Cells were transfected with HCF-1Δrep WT or ΔAD and selected for growth at 39.5°C. Colonies expressing WT and ΔAD proteins arose at similar frequency, indicating that HCF-1AD is not required for complementation and is consistent with previous studies showing that replacement of the mutant N-terminal subunit (residues 1–902) is sufficient (13). From multiple stable isolates, we selected and expanded two representative lines (R-tsBN67 HCF-1ΔrepWT and R-tsBN67 HCF-1ΔrepΔAD). Both cell lines expressed similar levels of WT or ΔAD HCF-1Δrep (Fig. 3A , lanes 1 and 2), proliferated with similar kinetics (Fig. 3B), and showed no obvious morphological differences (data not shown).
Figure 3.
Characterization of R-tsBN67 HCF-1Δrep WT and ΔAD cells. (A) Immunoblot comparing tsBN67-derived cell lines stably expressing T7-tagged HCF-1Δrep WT and ΔAD. Protein extracts were prepared from equal numbers of R-tsBN67-HCF-1Δrep WT (lane 1) and ΔAD (lane 2) cells, resolved on a SDS-7% polyacrylamide gel and immunoblotted with the αT7 antibody. (B) Analysis of cell proliferation. R-tsBN67-HCF-1Δrep WT (■) and ΔAD (□) cells were seeded at a density of 1 × 105 cells per 10-cm dish and incubated at 39.5°C for 3 days. At the indicated times, three separate dishes were harvested and counted. (C) Protein extracts were prepared from an equal number of tsBN67, R-tsBN67-HCF-1ΔrepWT, and HCF-1ΔrepΔAD cells and assayed for the ability to support VP16-induced complex formation in a gel-mobility shift assay using a 32P-labeled ICP0 TAATGARAT element as a probe. Lane 1 contains probe alone, lanes 2–9 contain probe with Oct-1 POU domain, and lanes 3–9 contain probe, Oct-1 POU domain, and VP16FL fused to GST. Two amounts of each cell extract (a 3-fold difference) were incubated with VP16 and Oct-1 POU: WT extract (lanes 4 and 5), ΔAD extract (lanes 6 and 7), and tsBN67 extract (lanes 8 and 9). In lanes 10, 11, and 12, the probe was incubated with the highest amount of WT, ΔAD, and tsBN67 extracts.
Although the HCF-1 β-propeller is necessary and sufficient for binding to VP16 and complex assembly (13, 33), it is possible that deletion of the HCF-1AD affects VP16-induced complex formation. To address this, we prepared protein extracts from equal numbers of R-tsBN67 HCF-1Δrep WT and ΔAD cells, as well as the parental tsBN67 line, cultured at 39.5°C. VP16-induced complex formation was examined by gel-mobility shift assay (Fig. 3C), using a TAATGARAT-containing probe and supplemented with recombinant Oct-1 POU domain and full-length GST-VP16 (15). As shown previously, the extract from tsBN67 cells failed to promote VP16-induced complex formation (Fig. 3C, lanes 8 and 9), confirming that the endogenous HCF-1 protein was inactive for complex formation at 39.5°C (11). In marked contrast, extracts from the WT (lanes 4 and 5) and ΔAD (lanes 6 and 7) cell lines gave rise to VP16-induced complexes (labeled VIC). Similar levels of complex forming activity were observed in the rescued lines, indicating that deletion of the HCF-1AD does not prevent VP16-induced complex assembly.
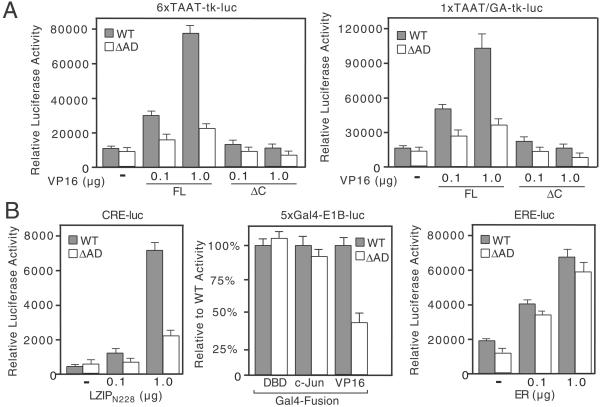
Using these cell lines, we examined transcriptional activation by the VP16-induced complex (Fig. 4A). Equal numbers of R-tsBN67 HCF-1Δrep WT and ΔAD cells were cotransfected with 0.1 or 1.0 μg of a full-length VP16 expression plasmid (VP16FL) and one of two VP16-responsive reporter genes: 6xTAAT-tk-luc, containing six copies of a TAATGARAT motif from the HSV-1 ICP0 promoter, or 1xTAAT/GA-tk-luc, containing a single TAATGARAT and adjacent CGGAAR sequence from the ICP4 promoter (34). In WT cells (shaded bars), VP16FL activated both reporters in a dose-dependent manner. In striking contrast, VP16FL was unable to stimulate either reporter as effectively in ΔAD cells (open bars), implying an important role for the HCF-1AD in activation by the VP16-induced complex. Deletion of the VP16 activation domain (VP16ΔC) abolished transactivation in both cell lines, indicating an absolute requirement for the VP16AD.
Figure 4.
Transactivation by VP16 and LZIP is severely compromised in tsBN67-derived cells expressing HCF-1ΔrepΔAD. (A) R-tsBN67-HCF-1ΔrepWT and R-tsBN67-HCF-1ΔrepΔAD cells were transiently transfected with 6xTAAT-tk-luc (500 ng) or 1xTAAT/GA-tk-luc (1 μg) reporters together with 0.1 or 1.0 μg of the VP16FL or VP16ΔC expression plasmids and assayed for luciferase activity after incubation at 39.5°C for 40 h. (B) As in A, except cells were transfected with: (Left) a CRE-luciferase reporter (CRE-luc, 500 ng) and an expression plasmid encoding LZIPN228; (Center) a Gal4-responsive reporter (5xGal4-E1B-luc reporter, 500 ng) and 500 ng of either Gal4DBD, Gal4-c-Jun, or Gal4-VP16AD; or (Right) an estrogen response element reporter (ERE-luc, 500 ng) and plasmid encoding the human estrogen receptor (ER).
We also examined activation by LZIP using a cAMP-response element (CRE)-containing promoter (Fig. 4B). Expression of a constitutively active version (LZIPN228), lacking the membrane tethering domain, stimulated the CRE-reporter up to 35-fold in WT cells but gave no more than 10-fold stimulation in ΔAD cells (Fig. 4B Left). Thus the HCF-1AD stimulates LZIP-mediated activation. Interestingly, activation by the isolated VP16AD (Gal4-VP16) was also reduced in the ΔAD cells, indicating a role for the HCF-1AD beyond the context of VP16-induced complex. As controls, we tested Gal4-c-Jun (Fig. 4B Center) and the human estrogen receptor (ER; Fig. 4B Right), two activators that do not require HCF-1 (ref. 24 and data not shown). In both cases, the relative level of activation was similar in the two cell lines and argues that the lack of activation by VP16 and LZIPN228 in ΔAD cells is a direct consequence of the HCF-1AD deletion.
Synergy Between HCF-1AD and VP16AD.
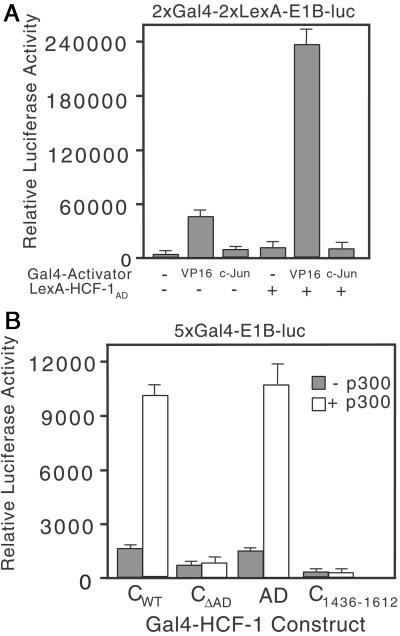
To investigate the relationship between the HCF-1AD and the VP16AD independent of the VP16-induced complex, we assayed a synthetic promoter containing two Gal4 sites and two LexA sites (Fig. 5A). We transfected just enough Gal4-VP16 (100 ng) and LexA-HCF-1AD (10 ng) expression plasmid to obtain modest activation when each fusion was assayed alone. A greater than additive increase was achieved by simultaneous expression of the two activators, suggesting the HCF-1AD and VP16AD activate in a synergistic manner. In contrast, coexpression of Gal4-c-Jun and LexA-HCF-1AD were no more than additive.
Figure 5.
The HCF-1AD synergizes with VP16 and p300. (A) 293T cells were transfected with 2xGal4-2xLexA-E1B-luc (200 ng) together with Gal4 activator (VP16AD or c-Jun, 100 ng) and LexA-HCF-1AD (10 ng). Extracts were prepared after 40 h and assayed for luciferase activity. (B) 293T cells were cotransfected with expression plasmids (500 ng) encoding Gal4-HCF-1CWT or ΔAD, Gal4-HCF-1C1436–1612, or Gal4-HCF-1AD with the 5xGal4-E1B-luc reporter gene (500 ng) and with (open bars) or without (filled bars) an expression plasmid (6 μg) encoding full-length p300.
To identify potential targets of the HCF-1AD we tested p300, a known coactivator of VP16 (6, 27). Expression of full-length p300 led to a 4- to 5-fold increase in transactivation by fragments containing the HCF-1AD (Fig. 5B). Although this experiment does not distinguish between a direct and indirect interaction, it suggests that the HCF-1AD functions through recruitment of coactivators such as p300.
Role of the HCF-1AD in Infection of tsBN67 Cells by HSV.
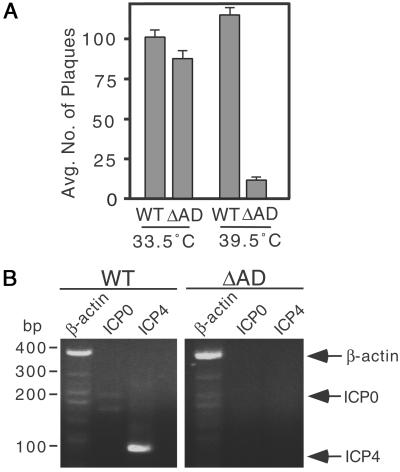
Although VP16 is not essential for IE gene activation, it has been shown to increase the probability that a cell infected with a single virus particle will enter the lytic cycle. Thus at low-multiplicities of infection (low moi), viruses carrying mutations in VP16 that interfere with transcriptional activation exhibit reduced levels of IE gene expression and a greatly increased particle/plaque-forming units ratio (35–37). To address the role the HCF-1AD in lytic replication, R-tsBN67 HCF-1Δrep WT and ΔAD cells were infected with wild-type HSV at a dose of 0.001 plaque forming units per cell. Parallel infections were performed at 33.5 and 39.5°C. After 5 days, total virus was recovered and levels of replication quantitated by measuring plaque formation by serial dilution on Vero cells (Fig. 6A). In multiple independent assays, infection of the two cell lines at 33.5°C gave similar viral yields. At 39.5°C, viral titers were increased slightly for WT cells, showing that this virus stock was able to replicate similarly at high and low temperatures, but were reduced 10-fold in ΔAD cells. This result shows that at low multiplicities, the HCF-1AD is required for efficient HSV replication.
Figure 6.
The HCF-1AD is involved in HSV lytic replication. (A) Wild-type HSV-1 at moi of 0.001 was used to infect R-tsBN67-HCF-1Δrep WT and ΔAD cells maintained at 33.5°C and 39.5°C. After 5 days, cells were harvested, sonicated, and serial dilutions plated on Vero cells. After 4 days, cells were fixed and stained with crystal violet to visualize viral plaques. Graph shows the mean number of plaques at 10−6 the dilution based on four independent assays. (B) Wild-type HSV-1 at moi of 0.001 was used to infect R-tsBN67-HCF-1Δrep WT and ΔAD cells maintained at 39.5°C. RNA was isolated after 4 h and assayed by RT-PCR. Specific amplification products from hamster β-actin (lanes 1 and 4, amplification fragment, 360 bp), ICP0 (lanes 2 and 5, amplification fragment, 157 bp), and ICP4 (lanes 3 and 6 amplification fragment, 101 bp) are indicated.
To demonstrate that reduced viral yield was due to a defect in VP16 transactivation, we used RT-PCR to examine HSV IE gene expression. Parallel cultures of R-tsBN67 HCF-1Δrep WT and ΔAD cells were maintained at 39.5°C and infected with wild-type HSV at a dose of 0.001 plaque forming units per cell. After 4 h, total RNA was isolated and analyzed by RT-PCR to detect ICP0 and ICP4 IE gene transcripts, as well as cellular β-actin as a positive control. As shown in Fig. 6B, viral ICP4 and ICP0 transcripts were readily detectable in R-tsBN67 HCF-1ΔrepWT cells but not ΔAD cells (compare lanes 2 and 3 with lanes 5 and 6). Hamster β-actin RNA was expressed at relatively equal levels in both cell lines (compare lanes 1 and 4). This result indicates that the decrease in viral replication is most likely a direct consequence of reduced transactivation by VP16.
Discussion
Prevailing models of the VP16-induced complex view HCF-1 as an assembly factor that promotes the association of VP16 with the Oct-1 POU domain and with DNA (33, 38, 39). Biochemical studies have shown that the N-terminal subunit of HCF-1, specifically the β-propeller domain (residues 1–380), is sufficient for complex formation, although sequences in the C-terminal subunit further stabilize the association with DNA (13, 18–20). Here we report a second critical role for HCF-1, namely its intrinsic activation function.
Deletion of the HCF-1AD leads to a profound loss of transactivation by the VP16-induced complex but does not affect complex formation, self-association, or nuclear localization. On face value, this new function is unexpected. In yeast the HCF-1 β-propeller is sufficient to reconstitute VP16-dependent transcription (13). Also, numerous studies have shown that the isolated VP16AD can act as a strong activator when tethered to promoters through a heterologous DNA-binding domain and implies that the VP16AD can activate transcription in most promoter contexts (40–42). Indeed, in the assay conditions used here, deletion of the VP16AD was more severe than deletion of the HCF-1AD, reducing activation to almost that of the reporter alone. The strict dependence on the VP16AD is consistent with previous studies and implies a modulatory role for the HCF-1AD. Further biochemical and genetic studies are required to elucidate the mechanism underlying this functional relationship between the two activation domains. Synergy might involve the recruitment of coactivators such as p300 or unmasking the VP16AD by other domains within the VP16-induced complex. Requirement for the HCF-1AD was observed when using a minimal TAATGARAT sequence (6xTAAT-tk-luc), and in this context it seems unlikely that the HCF-1AD functions through interactions with other DNA-binding proteins, such as GABP or Sp1, which bind to sites flanking the TAATGARAT elements in the HSV IE promoters (43). Studies by Schaffner and colleagues (44) have shown that the VP16-induced complex is a relatively poor activator when placed at a distance from the core promoter, in marked contrast to Gal4-VP16AD. We suggest that VP16 may exist in different conformations, which depend on proximity to the general transcription machinery. Perhaps the HCF-1AD facilitates the transition to the more active conformation or recruits cofactors that promote such a rearrangement. It is worth noting that when fused to the Gal4 DNA-binding domain, the VP16AD is present in two copies on each binding site. Perhaps the HCF-1AD cooperates with the VP16AD, providing activation domain synergy.
Our study provides additional support for the importance of HCF-1 in HSV lytic replication (45). We show that at a low moi, virion production was reduced by at least an order of magnitude in R-tsBN67 HCF-1ΔrepΔAD cells when compared with cells complemented with wild-type HCF-1Δrep. At low particle-to-cell ratios, viruses carrying transactivation-deficient mutants of VP16 show reduced yields of infectious virions, reflecting a requirement for VP16 to boost initial synthesis of the IE proteins (35–37, 46). Although VP16 performs multiple roles during HSV infection, including regulation of the virion host shutoff protein and other necessary steps in virion maturation and egress (46–48), our data shows that there is a significant reduction in IE gene expression in ΔAD cells.
Functional studies of the cellular transcription factors LZIP and GABP have established a role for HCF-1 as a coactivator (24, 26). In both examples, HCF-1 has been shown to interact directly with the activation domain, albeit through two separate mechanisms, and this association is required for optimal transactivation. HCF-1 is comparatively abundant in proliferating cells and therefore has the capacity to interact with a large number of cellular transcription factors (10, 22, 25, 26). Whether the HCF-1AD is important in other cases remains to be determined. Interestingly, complementation of the tsBN67 cell proliferation defect did not show a requirement for the HCF-1AD. This is consistent with prior complementation studies which established that the first 902 amino acids of human HCF-1 spanning the β-propeller and adjacent SAS1N and SAS1C domains were sufficient to restore growth at the nonpermissive temperature (13). The underlying cause of the tsBN67 cell cycle defect is not known and conceivably might reflect a nontranscriptional function of HCF-1.
In conclusion, our results reveal a critical role for HCF-1C in allowing transactivation of IE promoters by the VP16-induced complex and in providing coactivator activity used by cellular transcription factors such as LZIP. Although the mechanism of synergy between activation domains of VP16 and HCF-1 remain to be determined, it is clear that HCF-1 has the potential to control both VP16 promoter targeting and activity. The strong dependence on a cellular cofactor may be of particular relevance during establishment or reactivation from latency in sensory neurons. In unstimulated neurons, which are the primary reservoir of latent HSV, HCF-1 is sequestered in the cytoplasm, where it is presumably unavailable as a transcription factor (49). Stimuli that trigger reactivation of latent virus also lead to a rapid translocation of HCF-1 into the nucleus, suggesting that HCF-1 might play a direct role in regulating the initiation of the lytic cascade. Compartmentalization of HCF-1 provides an elegant mechanism to ensure minimal expression of the IE genes during latency, which may last for decades, and at the same time allow for rapid activation in response to physiological stress conditions.
Acknowledgments
We thank Rich Freiman, Michael Garabedian, Ian Mohr, and Naoko Tanese for constructive comments on the manuscript. Michael Garabedian, Shahana Mahajan, Matt Mulvey, Ron Prywes, Herb Samuels, and Naoko Tanese provided valuable discussion, reagents, or advice. This work was supported by National Science Foundation Grant MCB-98-16856 and National Institutes of Health Grants GM61139 and AI07180-21.
Abbreviations
- IE
immediate-early
- HSV
herpes simplex virus
- moi
multiplicity of infection
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Flint J, Shenk T. Annu Rev Genet. 1997;31:177–212. doi: 10.1146/annurev.genet.31.1.177. [DOI] [PubMed] [Google Scholar]
- 2.Weir J P. Gene. 2001;271:117–130. doi: 10.1016/s0378-1119(01)00512-1. [DOI] [PubMed] [Google Scholar]
- 3.O'Hare P. Semin Virol. 1993;4:145–155. [Google Scholar]
- 4.Herr W. Cold Spring Harbor Symp Quant Biol. 1998;63:599–607. doi: 10.1101/sqb.1998.63.599. [DOI] [PubMed] [Google Scholar]
- 5.Wilson A C, Cleary M A, Lai J S, LaMarco K, Peterson M G, Herr W. Cold Spring Harbor Symp Quant Biol. 1993;58:167–178. doi: 10.1101/sqb.1993.058.01.021. [DOI] [PubMed] [Google Scholar]
- 6.Kundu T K, Palhan V B, Wang Z, An W, Cole P A, Roeder R G. Mol Cell. 2000;6:551–561. doi: 10.1016/s1097-2765(00)00054-x. [DOI] [PubMed] [Google Scholar]
- 7.Utley R T, Ikeda K, Grant P A, Cote J, Steger D J, Eberharter A, John S, Workman J L. Nature. 1998;394:498–502. doi: 10.1038/28886. [DOI] [PubMed] [Google Scholar]
- 8.Kristie T M, Sharp P A. J Biol Chem. 1993;268:6525–6534. [PubMed] [Google Scholar]
- 9.Wilson A C, Parrish J E, Massa H F, Nelson D L, Trask B J, Herr W. Genomics. 1995;25:462–468. doi: 10.1016/0888-7543(95)80046-o. [DOI] [PubMed] [Google Scholar]
- 10.Wysocka J, Reilly P T, Herr W. Mol Cell Biol. 2001;21:3820–3829. doi: 10.1128/MCB.21.11.3820-3829.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Goto H, Motomura S, Wilson A C, Freiman R N, Nakabeppu Y, Fukushima K, Fujishima M, Herr W, Nishimoto T. Genes Dev. 1997;11:726–737. doi: 10.1101/gad.11.6.726. [DOI] [PubMed] [Google Scholar]
- 12.Freiman R N, Herr W. Genes Dev. 1997;11:3122–3127. doi: 10.1101/gad.11.23.3122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wilson A C, Freiman R N, Goto H, Nishimoto T, Herr W. Mol Cell Biol. 1997;17:6139–6146. doi: 10.1128/mcb.17.10.6139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kristie T M, Pomerantz J L, Twomey T C, Parent S A, Sharp P A. J Biol Chem. 1995;270:4387–4394. doi: 10.1074/jbc.270.9.4387. [DOI] [PubMed] [Google Scholar]
- 15.Wilson A C, LaMarco K, Peterson M G, Herr W. Cell. 1993;74:115–125. doi: 10.1016/0092-8674(93)90299-6. [DOI] [PubMed] [Google Scholar]
- 16.Wilson A C, Peterson M G, Herr W. Genes Dev. 1995;9:2445–2458. doi: 10.1101/gad.9.20.2445. [DOI] [PubMed] [Google Scholar]
- 17.Wilson A C, Boutros M, Johnson K M, Herr W. Mol Cell Biol. 2000;20:6721–6730. doi: 10.1128/mcb.20.18.6721-6730.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hughes T A, La Boissière S, O'Hare P. J Biol Chem. 1999;274:16437–16443. doi: 10.1074/jbc.274.23.16437. [DOI] [PubMed] [Google Scholar]
- 19.La Boissière S, Walker S, O'Hare P. Mol Cell Biol. 1997;17:7108–7118. doi: 10.1128/mcb.17.12.7108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Simmen K A, Newell A, Robinson M, Mills J S, Canning G, Handa R, Parkes K, Borkakoti N, Jupp R. J Virol. 1997;71:3886–3894. doi: 10.1128/jvi.71.5.3886-3894.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lu R, Yang P, O'Hare P, Misra V. Mol Cell Biol. 1997;17:5117–5126. doi: 10.1128/mcb.17.9.5117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lu R, Misra V. Nucleic Acids Res. 2000;28:2446–2454. doi: 10.1093/nar/28.12.2446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lu R, Yang P, Padmakumar S, Misra V. J Virol. 1998;72:6291–6297. doi: 10.1128/jvi.72.8.6291-6297.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Luciano R L, Wilson A C. Proc Natl Acad Sci USA. 2000;97:10757–10762. doi: 10.1073/pnas.190062797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gunther M, Laithier M, Brison O. Mol Cell Biochem. 2000;210:131–142. doi: 10.1023/a:1007177623283. [DOI] [PubMed] [Google Scholar]
- 26.Vogel J L, Kristie T M. EMBO J. 2000;19:683–690. doi: 10.1093/emboj/19.4.683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kraus W L, Manning E T, Kadonaga J T. Mol Cell Biol. 1999;19:8123–8135. doi: 10.1128/mcb.19.12.8123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tanaka M, Herr W. Cell. 1990;60:375–386. doi: 10.1016/0092-8674(90)90589-7. [DOI] [PubMed] [Google Scholar]
- 29.Su L F, Knoblauch R, Garabedian M J. J Biol Chem. 2001;276:3231–3237. doi: 10.1074/jbc.M005547200. [DOI] [PubMed] [Google Scholar]
- 30.Dorris D R, Struhl K. Mol Cell Biol. 2000;20:4350–4358. doi: 10.1128/mcb.20.12.4350-4358.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chen X, Li J, Mata M, Goss J, Wolfe D, Glorioso J C, Fink D J. J Virol. 2000;74:10132–10141. doi: 10.1128/jvi.74.21.10132-10141.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kramer M F, Coen D M. J Virol. 1995;69:1389–1399. doi: 10.1128/jvi.69.3.1389-1399.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Babb R, Huang C C, Aufiero D J, Herr W. Mol Cell Biol. 2001;21:4700–4712. doi: 10.1128/MCB.21.14.4700-4712.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Douville P, Hagmann M, Georgiev O, Schaffner W. Virology. 1995;207:107–116. doi: 10.1006/viro.1995.1056. [DOI] [PubMed] [Google Scholar]
- 35.Ace C I, McKee T A, Ryan J M, Cameron J M, Preston C M. J Virol. 1989;63:2260–2269. doi: 10.1128/jvi.63.5.2260-2269.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Smiley J R, Duncan J. J Virol. 1997;71:6191–6193. doi: 10.1128/jvi.71.8.6191-6193.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tal-Singer R, Pichyangkura R, Chung E, Lasner T M, Randazzo B P, Trojanowski J Q, Fraser N W, Triezenberg S J. Virology. 1999;259:20–33. doi: 10.1006/viro.1999.9756. [DOI] [PubMed] [Google Scholar]
- 38.Kristie T M, Sharp P A. Genes Dev. 1990;4:2383–2396. doi: 10.1101/gad.4.12b.2383. [DOI] [PubMed] [Google Scholar]
- 39.Stern S, Herr W. Genes Dev. 1991;5:2555–2566. doi: 10.1101/gad.5.12b.2555. [DOI] [PubMed] [Google Scholar]
- 40.Cousens D J, Greaves R, Goding C R, O'Hare P. EMBO J. 1989;8:2337–2342. doi: 10.1002/j.1460-2075.1989.tb08361.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sadowski I, Ma J, Triezenberg S, Ptashne M. Nature. 1988;335:563–564. doi: 10.1038/335563a0. [DOI] [PubMed] [Google Scholar]
- 42.Triezenberg S J, Kingsbury R C, McKnight S L. Genes Dev. 1988;2:718–729. doi: 10.1101/gad.2.6.718. [DOI] [PubMed] [Google Scholar]
- 43.Thompson C C, McKnight S L. Trends Genet. 1992;8:232–236. [Google Scholar]
- 44.Hagmann M, Georgiev O, Schaffner W. J Virol. 1997;71:5952–5962. doi: 10.1128/jvi.71.8.5952-5962.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.La Boissière S, O'Hare P. J Virol. 2000;74:99–109. doi: 10.1128/jvi.74.1.99-109.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mossman K L, Smiley J R. J Virol. 1999;73:9726–9733. doi: 10.1128/jvi.73.12.9726-9733.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lam Q, Smibert C A, Koop K E, Lavery C, Capone J P, Weinheimer S P, Smiley J R. EMBO J. 1996;15:2575–2581. [PMC free article] [PubMed] [Google Scholar]
- 48.Weinheimer S P, Boyd B A, Durham S K, Resnick J L, O'Boyle D R., Jr J Virol. 1992;66:258–269. doi: 10.1128/jvi.66.1.258-269.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kristie T M, Vogel J L, Sears A E. Proc Natl Acad Sci USA. 1999;96:1229–1233. doi: 10.1073/pnas.96.4.1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Frishman D, Argos P. Proteins. 1997;27:329–335. doi: 10.1002/(sici)1097-0134(199703)27:3<329::aid-prot1>3.0.co;2-8. [DOI] [PubMed] [Google Scholar]