Abstract
Histone deacetylases (HDACs) are thought to function as critical mediators of transcriptional repression. However, the physiological targets and posttranslational modifications of the class II HDACs are largely unknown. Here we show that the C terminus of HDAC 6 is both necessary and sufficient for specific association with polyubiquitin. This region contains a putative zinc finger but lacks significant similarity to other known ubiquitin binding domains. Thus, we have designated this region as a PAZ domain, for Polyubiquitin Associated Zinc finger. Although the PAZ domain possesses homology with the zinc finger of deubiquitinating enzymes, it is dispensable for the deubiquitinating activity we find associated with HDAC6 following immunopurification. We also show that both HDAC 5 and HDAC 6 are ubiquitinated in vitro and in vivo. However, both of these proteins are stable in vivo and do not appear to be targeted for rapid degradation by the proteasome. Thus, HDAC6 is linked to the ubiquitin system via ubiquitin conjugation, polyubiquitin binding, and copurification with deubiquitinating enzymes.
Transcriptional activation and repression are processes dependent on DNA accessibility regulated by chromatin remodeling, nucleosomal positioning, and posttranslational modification of histones (reviewed in ref. 1). Sequence-specific transcriptional activators frequently recruit ATP-dependent DNA chromatin remodeling complexes and histone acetyltransferases (HATs) enabling engagement of the basal transcriptional machinery. Gene activation correlates with phosphorylation and subsequent acetylation of the N-terminal tails of histones H3 and H4 (reviewed in ref. 2). Transcriptionally active chromatin has also been correlated with polyubiquitination of the C-terminal tails of histone H2A (3) and H2B (4, 5). Recent evidence suggests that monoubiquitination of the linker histone, H1 mediated by TAFII250 (6), may also be required for transcriptional activation.
Many repressor proteins, like transcriptional activators, also bind specific DNA sequences; however, they initiate a reciprocal cascade of events including deacetylation, dephoshorylation, methylation, and quite possibily deubiquitination of histones (reviewed in refs. 7 and 8). These changes lead to localized chromatin structural modifications which apparently block engagement of the general transcriptional apparatus. Thus, it is not surprising that purified repressor complexes contain chromatin remodeling proteins, methylases, and histone deacetylases (HDACs).
To date, 11 different mammalian histone deacetylases have been identified. They are grouped into three classes according to their homology to the yeast deacetylases, rpd3 (for mammalian class I), hda1 (class II), and the NAD-dependent sir2 (class III) (reviewed in refs. 9 and 10). In vitro the class I HDACs [1, 2, 3, 8, 11] and II HDACs [4, 5, 6, 7, 9, 10] are capable of deacetylating all of the core histones. However, their substrate specificity or redundancy in vivo is not well defined. Several class II HDACs have been reported to interact with corepressors including SMRT (11), N-Cor (12), CtBP (13, 14), B-Cor (15), TR2 (16), ETO-2 (17), and Bcl6 (18). However, binding partners for HDAC 6, 9, and 10 are largely unknown.
The intracellular localization and likely functions of several class II HDACs are dynamically regulated. HDAC 4, 5, and presumably HDAC7, have nuclear localization signals (NLSs) that direct them to the nucleus when bound to the MEF2 family of transcription factors. In addition, sumoylation of HDAC4 may be important for its nuclear import (19). Site specific phosphorylation of HDAC 4, 5, and 7 by CaM kinases I or IV releases them from MEF2 and unmasks nuclear export signals (NESs) leading to cytoplasmic sequestration apparently mediated by 14–3-3 proteins (reviewed in refs. 9 and 20). Sumoylation and phosphorylation, however, are the only examples of posttranslational modifications of the class II HDACs to date.
HDAC6, a novel deacetylase in that it contains two functional catalytic domains, also displays nucleo-cytoplasmic shuttling capablities. HDAC6 contains three NES signals, the most N-terminal of which is responsible for the enzyme's cytoplasmic localization in rapidly dividing cells (21). This observation raises the intriguing possibility that this deacetylase has unique cytoplasmic, nontranscriptionally related targets. In fact, cytoplasmic HDAC6 from testis extracts copurifies with mammalian homologues of ubiquitin-fusion degradation protein (UFD3), as well as cdc48p, an ATPase involved in protein trafficking from endoplasmic reticulum to cytoplasm (22). In addition, HDAC6 has been shown to deacetylate α-tubulin in polymerized microtubules thus potentially enhancing chemotactic cell motility (23). HDAC6 has recently been reported to be sumoylated, although the biological consequence of this modification is not known (19). Although a cytoplasmic enzyme, HDAC6 can deacetylate all of the core histones in vitro (24) and may specifically regulate transcription in response to signals that induce differentiation or arrest proliferation, because it accumulates in the nucleus after sodium butyrate treatment and serum withdrawal (21, 25). Thus, extracellular signals are likely to control HDAC6 localization and its interaction with target proteins.
Signaling cascades have long been recognized to modulate transcriptional activators, most often via phosphorylation, as a means to control gene expression. Phosphorylation not only affects transcription factor localization, DNA binding, and interaction with other factors, it frequently also directs ubiquitination, traditionally considered as the penultimate step preceeding proteasomal–mediated degradation. Indeed, many transcription factors are short-lived proteins and are degraded by the ubiquitin proteasome pathway (reviewed in ref. 26), although recent evidence shows that ubiquitination may alter the activity of the targeted factor without modifying its half life (27, 28) (reviewed in refs. 29 and 30). Interestingly, the transactivation domains of several transcription factors overlap sequences involved in ubiquitin-mediated degradation (31, 32) and ubiquitination is suggested to increase transcriptional activation. Even ubiquitin itself, when fused to the VP16 activator, markedly stimulates transcriptional potency (31). Thus, ubiquitination of transcriptional activators, and presumably also repressors, can directly influence their activity.
Several intriguing recent reports indicate that proteasome subunits are involved in transcriptional regulation. Yeast strains mutant for two 19S ATPases have defects consistent with impaired transcriptional elongation by RNA polymerase II. In vivo, these proteins interact with the transcriptional elongation factor Cdc68 (33). In addition, a specific subset of the 19S ATPases are recruited to activated promoters, and their presence is required for transcriptional elongation in a proteolytic-independent manner (34).
In light of studies linking transcriptional activation, chromatin structure, and ubiquitination, we elected to address whether repressor proteins, namely the class II HDACs, are linked to the ubiquitin system. We took a two pronged approach by first addressing whether the class II HDACs were ubiquitinated and degraded and second, performing genetic screens to identify relevant HDAC binding proteins.
Materials and Methods
Reagents.
Flag antibodies and ubiquitin were purchased from Sigma. Ubiquitin antibodies were from Babco (Richmond, CA). HA antibodies and Fugene were from Roche. MG-132, ubiquitin-aldehyde (Ub-al), and polyubiquitin chains were obtained from Calbiochem, Boston Biochemicals, and Affiniti, respectively.
In Vitro Conjugation of HDACs.
HDAC5 and HDAC6 were subcloned into the pCS2+MT vector and were translated in vitro using TNT-reticulocyte lysate (Promega) in the presence of [35S]methionine. Conjugation was performed as described (35). As indicated, reactions contained either Ub-al (0.5 μg), a specific inhibitor of deubiquitinating enzymes (DUBs) (36) and the proteasome inhibitor MG-132 (50 μM), or 10 mM 2-deoxyglucose and 0.2 μg hexokinase. Reactions were resolved on SDS/6.5% PAGE and visualized by PhosphoImager.
Pulse–Chase Experiments.
HDACs were transiently transfected into Cos-1 cells, and 40 h later, degradation was assessed by pulse–chase labeling and immunoprecipitation. Briefly, cells were labeled with [35S]methionine (≈150 μCi/ml; 1 Ci = 37 GBq) for 30 min (pulse), washed extensively with PBS, and either harvested immediately (0 time for “chase”) or further incubated for the indicated periods of “chase” time in the presence of 50× excess unlabeled methionine. Cells were lysed in boiling lysis buffer containing 1× PBS, 1% SDS, 1 mM EDTA, 2 mM sodium orthovanadate, 80 μM MG-132, and protease inhibitors. Equal amounts of trichloroacetic acid-precipitable cpms were immunoprecipitated in buffer containing 1× PBS, 1% Triton X, 0.25% deoxycholate, 0.5% SDS, 1 mM EDTA, 0.5% BSA, 2 mM sodium orthovanadate, and protease inhibitors. After SDS/PAGE, proteins were visualized by autoradiograms.
In Vivo Ubiquitination.
293 cells were transfected with 8 μg of HDAC and 6 μg of hemagglutinin-tagged ubiquitin (HA-ub). Approximately 46 h after transfection, cells were treated with 80 μM MG-132 for 2 h before harvesting. Cells were lysed as described for pulse–chase experiments with the exception that the lysis and immunoprecipitation buffers contained 3 μM Ub-al. Flag-immunoprecipitates were analyzed by SDS/PAGE and Western blots using HA-antibodies.
Two-Hybrid Screens.
Full-length human HDAC6 or the catalytically inactive HDAC6 (H216A/H611A) (24) were used as baits in two-hybrid screens as described (37, 38). Each screen yielded 3 × 106 primary transformants, of those 44 (HDAC6) and 62 (H216A/H611A) expressed high levels of β-galactosidase activity. These clones were cured of their original bait plasmids and were then mated with Amr70 strains containing either the original bait plasmids or lamin B. Nearly all library clones interacted strongly with HDAC6 and negligibly with lamin. HDAC6Δzf was used as a bait to screen 6 × 106 primary yeast transformants; however, no specific interactors were identified.
GST Pull-Down Assays.
293 cells were mock transfected or transfected with flag-HDAC6 or flag-HDAC6Δzf. Forty-eight hours later, cells were snap frozen on dry ice/ethanol and thawed in 5 ml buffer containing: 0.4 M KCl, 20 mM Hepes (pH 7.4), 1 mM DTT, and 20% glycerol. Equal amounts of cell lysates were added to equal amounts of purified GST, GST-polyubiquitin clone 6.3, or GST-monoubiquitin, and incubated 3 h at 4°C in buffer containing 150 mM NaCl, 1 mM EDTA, 20 mM Tris (pH 8.0), 0.5% Nonidet P-40, and 10% glycerol. Pull-downs were electrophoresed on SDS/8% PAGE and subjected to anti-flag Western blots.
In Vitro DUB Assays.
GST-HDAC6 or immunopurified HDAC6 was assayed for 1 h at 37°C in a buffer containing 50 mM Tris (pH 8.0), 5 mM MgCl2, 5 mM DTT, and 3 μg K48-linked polyubiquitin chains in the absence or presence of 6 μM Ub-al. Reactions were terminated by the addition of SDS sample buffer. Reaction mixtures were electrophoresed on SDS/15% PAGE and blotted with anti-ubiquitin antibodies.
Results
HDAC5 and HDAC6 Are Ubiquitinated in Vivo and in Vitro.
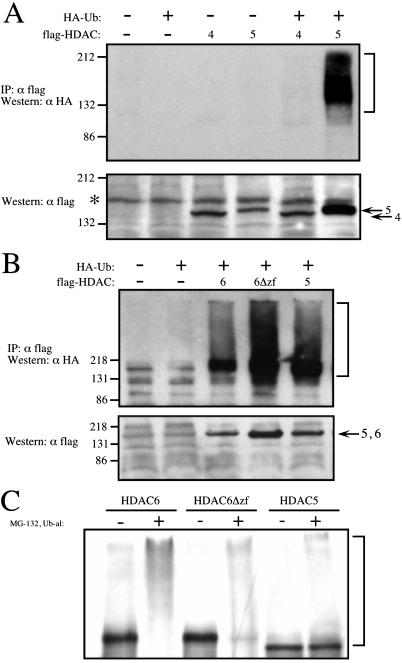
We first examined the ubiquitination status of HDAC4, HDAC5, and HDAC6. Flag-tagged class II HDACs were transfected into 293 cells with or without HA-Ub. To increase the steady-state level of ubiquitin conjugates, the cells were incubated in the presence of the proteasome inhibitor, MG-132, and lysed with boiling lysis buffer. HDACs were subsequently immunoprecipitated with anti-flag antibody, subjected to SDS/PAGE, and immunoblotted for ubiquitination using HA antibodies. As shown in Fig. 1 A and B, HDAC5 and HDAC6 were highly ubiquitinated in vivo in the presence of HA-Ub, whereas HDAC4 was not.
Figure 1.
Ubiquitination of HDAC4, HDAC5, and HDAC6. (A and B) 293 cells were transfected with flag-tagged HDAC4, HDAC5, HDAC6, or HDAC6Δzf (comprising amino acids 1–1111) with or without HA-Ub. Before harvesting, cells were treated with 80 μM proteasome inhibitor, MG-132, for 2 h. Cell lysates were immunoprecipitated with flag antibody and blotted for HA. Brackets denote ubiquitin conjugated HDACs (Upper). The corresponding flag Western blots of whole cell lysates (5% immunoprecipitation input). Asterisk denotes a nonspecific band that cross reacts with the flag antibody (Lower). A and B are run on SDS/8% and 10% PAGE, respectively. (C) In vitro ubiquitination of HDAC5 and HDAC6. [35S]methionine-labeled HDACs were incubated with HeLa nuclear extract in the absence and presence of the proteasome inhibitor, MG-132, and the DUB inhibitor, Ub-al. Reactions were electrophoresed on SDS/6.5% PAGE and analyzed by PhosphoImager.
Because we established that HDAC5 and HDAC6 were ubiquitinated in vivo, we wanted to confirm these results in vitro. As depicted in Fig. 1C, ubiquitin conjugated intermediates of in vitro translated HDAC5 and HDAC6 were detected in the presence MG-132 and the DUB inhibitor, Ub-al. Thus, these proteins are ubiquitinated both in vivo and in vitro.
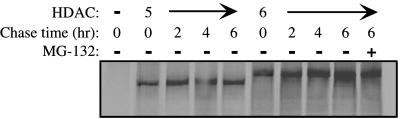
Because HDAC5 and HDAC6 are ubiquitinated, we determined whether they had short half lives in vivo. To test this, we performed pulse–chase experiments and found that both HDAC5 and HDAC6 are relatively stable proteins in vivo (Fig. 2). In addition, continued exposure to MG-132 during the chase had no significant effect on HDAC5 (data not shown) or HDAC6 levels (Fig. 2). In contrast, c-Myc, a protein known to be rapidly degraded, displayed a short half life under similar conditions (data not shown). Thus, although HDAC5 and HDAC6 are ubiquitinated, they are stable proteins in vivo.
Figure 2.
Pulse–chase analysis of HDAC5 and HDAC6. Cos-1 cells expressing flag-HDAC5 or HDAC6 were pulse labeled with [35S]methionine for 30 min and chased for the indicated times with media containing an excess of cold methionine. HDACs were immunoprecipitated with flag antibody and analyzed on SDS/8% PAGE by autoradiography.
The Zinc Finger of HDAC6 Associates with Polyubiquitin.
To search for potential binding partners for HDAC6 we performed two-hybrid screens (39) using the full-length catalytically active and inactive HDAC6 (24) as baits. The inactive HDAC6 contains point mutations within each of the deacetylase domains of active site histidine residues (H216A/H611A). We screened 3 × 106 transformants for each bait, and found that the only positive interacting clones for HDAC6 and H216A/H611A contained cDNA inserts encoding polyubiquitin or ubiquitin B. Both of these genes encode several tandem repeats of ubiquitin (10 for polyubiquitin and 4 for ubiquitin B).
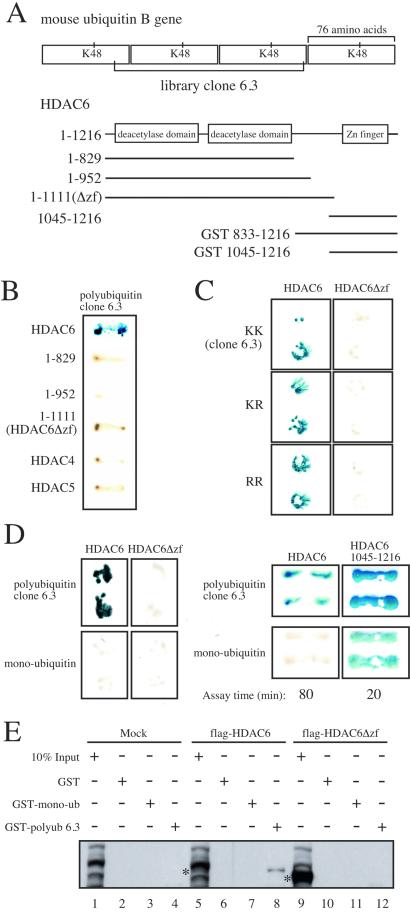
To characterize the interaction of HDAC6 and polyubiquitin, we created several variants of a polyubiquitin clone (6.3) containing 2.5 ubiquitin repeats (Fig. 3A Upper). We wanted to assess whether branched polyubiquitin chains were required for the interaction with HDAC6. Typically, once a substrate is monoubiquitinated, the successive ubiquitin moieties are attached through K48 linkages. Therefore, we mutated all K48 residues to R within the VP16-polyubiquitin clone so as to disrupt potential K48-linked polyubiquitination. As shown in Fig. 3C, HDAC6 interacted with wild-type polyubiquitin (KK) as well as polyubiquitin containing one K48R mutation or both K48R mutations. HDAC6, however, failed to interact with VP16-monoubiquitin (Fig. 3D). Thus, HDAC6 noncovalantly binds tandem repeats of polyubiquitin independent of R48 and fails to interact with a single ubiquitin moiety. In addition, these data substantiate the argument that HDAC6 is not being mono- or polyubiquitinated with the VP16 fusion proteins in the directed two-hybrid assay.
Figure 3.
The C terminus of HDAC6 interacts with polyubiquitin in yeast two-hybrid and GST pull-down assays. (A) Schematic diagram of the HDAC6 baits and polyubiquitin library clone 6.3. (B) Interaction of polyubiquitin clone 6.3 (as indicated by blue color) with different regions of HDAC6, HDAC4, or HDAC 5. (C) Yeast strains expressing mutations in residue K48 within the 6.3 library clone were mated to yeast expressing HDAC6 or HDAC6Δzf (in duplicate) and tested for β-galactosidase activity. (D) Interaction of HDAC6 and the isolated zinc finger domain (amino acids 1045–1216) with polyubiquitin clone 6.3 and monoubiquitin (in duplicate). β-galactosidase assay with amino acids 1045–1216 were terminated after 20 min, whereas the assay with full-length HDAC6 was carried out for 80 min. (E) HDAC6 interacts with GST-polyubiquitin in vitro. GST, GST-monoubiquitin, and GST-polyubiquitin were purified on glutathione Sepharose beads and incubated with 293 lysates from cells mock transfected, transfected with flag-HDAC6, or flag-HDAC6Δzf. Proteins binding to the GST fusion proteins were run on SDS/8% PAGE and blotted with flag antibodies. Asterisks denote HDAC 6 or HDAC6Δzf in whole cell lysates. Several bands in the mock input lanes nonspecifically react with the flag antibodies.
The C-terminal region of HDAC6 (amino acids 1134–1192) possesses significant homology to the zinc finger found in some members of both classes of DUBs, the ubiquitin C-terminal hydrolases (UCHs) and ubiquitin-specific processing proteases (UBPs) (Fig. 4, see Discussion). Although the role of the zinc finger in these enzymes has not been characterized, we tested whether the zinc finger of HDAC6 was responsible for binding polyubiquitin. In a directed yeast two-hybrid assay, truncation mutants (Fig. 3A) comprising amino acids 1–830, 1–953, and 1–1111 all failed to bind polyubiquitin (Fig. 3B). Thus, the C-terminal 105 aa of HDAC6, those encompassing the zinc finger, are necessary for interaction with polyubiquitin.
Figure 4.
Amino acid alignment of the zinc finger of HDAC6 with human DUBs and BRCA1 associated proteins. Asterisks denote conserved C and H residues. USPs are members of the UBP family.
Having established that the zinc finger in HDAC6 is necessary for interaction with polyubiquitin, we wanted to determine whether this region was sufficient. Surprisingly, in directed two-hybrid analysis, the isolated zinc finger interacted with polyubiquitin more efficiently than full-length HDAC6 and also interacted with monoubiquitin, albeit more weakly (Fig. 3D). These results indicate that although the zinc finger is sufficient to interact with polyubiquitin, there are likely to be specificity determinants within full-length HDAC6 which prevent interaction with monoubiquitin.
Some proteins containing ubiquitin binding regions require these domains for efficient ubiquitination (40). Thus, we tested whether the zinc finger was required for HDAC6 ubiquitination. HDAC6Δzf, like its full-length counterpart, was highly ubiquitinated in vivo and in vitro (Fig. 1 B and C). Thus, the HDAC6 zinc finger is dispensable for ubiquitination. The HDAC6 interaction with polyubiquitin is specific, as HDAC4 and HDAC5 fail to bind polyubiquitin in the two-hybrid assay (Fig. 3B). Our data using mammalian cells indicates that both HDAC5 and HDAC6Δzf are highly ubiquitinated (Fig. 1B) and yet do not interact with polyubiquitin in yeast (Fig. 3 B, C, and D). Thus, it is unlikely that the HDAC6-polyubiquitin interaction in yeast is driven by ubiquitination of HDAC6 with the VP16-polyubiquitin clone.
To further explore the interaction of HDAC6 with polyubiquitin, we performed GST pull-down assays. GST, GST-monoubiquitin, and GST-polyubiquitin (clone 6.3) were incubated with 293 cell lysates transfected with empty vector, HDAC6, or HDAC6Δzf. As shown in Fig. 3E, a GST-polyubiquitin interacts with HDAC6 (lane 8) but not HDAC6Δzf (lane 12) from lysates expressing considerably more HDAC6Δzf (lane 9) as compared with HDAC6 (lane 5). In contrast, GST-monoubiquitin could not bind either HDAC6 (lane 7) or HDAC6Δzf (lane 11). These data confirm our yeast two-hybrid results in showing that HDAC6 interacts specifically with a polyubiquitin tandem repeat through its zinc finger.
HDAC6 Associates with DUB Activity.
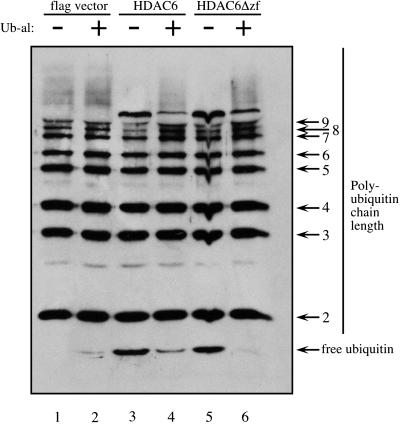
Because HDAC6 binds tandem repeats of polyubiquitin and has homology to DUBs, we wanted to determine whether HDAC6 was able to deubiquitinate a branched polyubiquitin chain. Purified GST fusions encoding HDAC6 amino acids 833-1216 and 1045–1216 (schematic Fig. 3A) showed no observable DUB activity toward K48-linked polyubiquitin chains (data not shown). However, both HDAC6 and HDAC6Δzf purified from cells contained substantial DUB activity toward high molecular weight polyubiquitin chains. We expressed flag-tagged HDAC6 or HDAC6Δzf in 293 cells and immunopurified them on flag antibody columns. Equal amounts of eluted HDAC6 and HDAC6Δzf (as assessed by Western blots, data not shown) were incubated with the same substrate to assess DUB activity. Fractions from flag-vector transfected 293 cells that were immunopurified contained no observable DUB activity inhibitable by Ub-al (Fig. 5 compare lanes 1 and 2). In the absence of Ub-al, immunopurified HDAC6 and HDAC6Δzf showed reduced amounts of branched polyubiquitin chains of 7 or more ubiquitin moieties and a concomitant increase in free ubiquitin as compared with reactions in the presence of Ub-al (Fig. 5, compare lanes 3 and 5 to 4 and 6). We therefore conclude that the DUB activity of immunopurified HDAC6 probably comes from associated DUB enzymes. Thus, although HDAC6 may not itself be a DUB enzyme, it associates with endogenous DUB enzymes and this association is independent of the HDAC6 zinc finger.
Figure 5.
Deubiquitinating activity copurifies with full-length HDAC6 and HDAC6Δzf. Flag HDAC6 and HDAC6Δzf were immunopurified from 293 cells and were assayed for DUB activity in vitro. Reaction mixtures were resolved on SDS/15% PAGE and Western blotted with anti-ubiquitin antibodies.
Discussion
Because ubiquitination as a posttranslational modification has been demonstrated to be important not only for protein stability but also transcriptional activity, we decided to study the role of ubiquitination of the class II HDAC members 4, 5, and 6. We have shown that HDAC5 is ubiquitin-conjugated both in vitro and in vivo but is a relatively stable protein (Fig. 1 and 2) whose turnover is not affected by proteasome inhibitor. Although the biological significance of HDAC5 ubiquitination is unknown, it is worth pointing out that several proteins involved in inhibition of differentiation, as is HDAC5 (41), are specifically degraded upon induction of differentiation (42–44). Thus, in some way ubiquitination of HDAC5 might “prime” it for rapid elimination in response to specific signals. Indeed, in a two-hybrid screen we found that HDAC5 associated with the S12/rpn8 subunit, known to localize within the lid of the 26S proteasome (data not shown).
Another possibility is that ubiquitination of HDAC5 and/or HDAC6 may facilitate interaction with other proteins or directly affect deacetylase activity. Because we have shown that HDAC6 associates with DUB activity (Fig. 5), it will be interesting to determine whether this association depends on the ubiquitination status of HDAC6. In support of this hypothesis, HDAC6 has been reported to be in a complex with cdc48p (22), a chaperone that specifically interacts with polyubiquitinated proteins and is involved in transport from the endoplasmic reticulum to the cytoplasm (reviewed in ref. 45).
To elucidate binding partners of HDAC6, we conducted two-hybrid screens with the full-length, catalytically active and inactive HDAC6 as baits. All of the positive interactors contained different regions of polyubiquitin or ubiquitin B genes. By homology search, we found that HDAC6 contains a C-terminal zinc finger with strong homology to the zinc fingers of DUBs (reviewed in refs. 46 and 47). Here, we have shown that the zinc finger from HDAC6 interacts specifically with polyubiquitin and not monoubiquitin in yeast and in in vitro binding assays (Fig. 3). Perhaps the zinc finger within the DUBs also serves to recognize polyubiquitinated proteins.
The first characterized polyubiquitin-binding sequences were two short ≈20-aa regions within the S5A subunit of the 26S proteasome (48). Homology searches along with Hidden Markov Models based on these sequences have defined a UIM (ubiquitin-interaction motif) domain contained in many types of proteins (49) including some UBPs. The UIM motif defined by four acidic amino acids followed by Φ-X-X-Ala-X-X-X-Ser-X-X-Ac, where Φ is a large hydrophobic residue and Ac is an acidic residue (49). This motif is not itself a domain, but likely forms an α-helix that is incorporated into different protein folds. The UIM from the S5a subunit of the proteasome prefers to recognize polyubiquitin chains of 4 or more ubiquitin moieties (50). Recently, UIMs from four proteins that mediate receptor endocytosis (eps15, eps15R, epsins, and Hrs) have been characterized (reviewed in ref. 51). The UIMs from these proteins, like that of the S5a subunit of the proteasome, display preferences for binding polyubiquitin over monoubiquitin (40). Intriguingly, the UIM is required for these proteins to be monoubiquitinated, but the motif itself is not ubiquitinated (40). These results are in contrast to HDAC6 where the zinc finger is dispensable for polyubiquitination (Fig. 1).
Recently, much attention has focused on another ubiquitin-binding domain, the ubiquitin-associated (UBA) domain (reviewed in ref. 52). One of the first characterizations of this domain was through a two-hybrid screen with p62, a phosphotyrosine independent interactor of the SH2 domain of p56lck. The vast majority of cDNAs isolated from the p62 interaction screen encoded polyubiquitin, diubiquitin, or a ubiquitin fusion protein, UBA52 (rUB) (53). In yeast and in vitro the p62 UBA domain interacts with monomeric or oligomeric ubiquitin. UBA domains within other proteins such as ubiquitin conjugating enzymes, UBPs, enzymes involved in nucleotide excision repair, and several kinases have been reported to facilitate mono- and diubiquitin binding (54) as well as mediate protein-protein interaction (55). This domain has recently been shown to preferentially bind K48-linked multiubiquitin chains as opposed to monoubiquitin (56–58).
The UBA domain and UIM motif, although both capable of binding polyubiquitin, do not show any sequence similarity to the zinc finger of HDAC6. As mentioned, the UIM is predicted to form a single alpha helix. The 45–55 aa UBA domain forms a three helix bundle with the helices at 45° angles to one another (59). On the surface of the bundle is a hydrophobic patch that is thought to be responsible for interaction with ubiquitin moieties. The structure of the HDAC6 zinc finger is currently unknown. However, all of the crystal and NMR structures of zinc finger proteins (reviewed in ref. 60) differ considerably from the UBA structure. Thus, we have likely identified a unique polyubiquitin-binding domain which we call a PAZ domain, for Polyubiquitin Associated Zinc finger. This domain is also found in many UBPs and in the BRCA1 associated proteins 1 and 2 (Fig. 4). We would predict then that BRCA1 associated proteins might also bind polyubiquitin. Careful sequence analysis similar to Hofmann's approach (49, 52) could identify unrecognized PAZ domains in other proteins.
While this study was underway, Seigneurin-Berny et al. (22) linked HDAC6 with the ubiquitin system. They found that the zinc finger motif within HDAC6 was associated with monoubiquitin. The authors state that the isolated C terminus of HDAC6 binds monoubiquitin better than does the full-length HDAC6. In our study, we have failed to detect substantial interactions between HDAC6 and monoubiquitin in both the yeast two-hybrid and GST pull-down assays (Fig. 3). We have, however, also observed that the isolated zinc finger interacts strongly with both polyubiquitin and monoubiquitin. Seigneurin-Berny et al. (22) purified ubiquitin-binding proteins from testis extracts using ubiquitin-agarose and isolated 9 proteins including HDAC6 and three DUBs, ubiquitin C-terminal hydrolase 5, UCH-L1, and UCH-L3. It is possible that these particular DUBs interact directly with HDAC6 and the activity we see derives from one or more of these enzymes.
What are the ubiquitinated binding targets of HDAC6? We attempted to address this question by conducting a two-hybrid screen with HDAC6Δzf as the bait. We reasoned that by removing the polyubiquitin-binding domain, we might now uncover other weaker interactors. However, of 6 × 106 transformants, no specific interactors of HDAC6Δzf were identified. Although we have addressed possible connections between the ubiquitin machinery and HDAC6, we are still uncertain about the biological roles of these phenomena.
HDAC6 may provide a link between deacetylation and deubiquitination. Surprisingly, in the literature, no connection has been reported between deacetylation and deubiquitination or between polyubiquitination and transcriptional repression. It is well-documented that ubiquitinated histone H1, H2A, and H2B correlate with transcriptionally active chromatin regions (3–6, 61), and that acetylated histones H3 and H4 correlate with increased transcriptional activation (reviewed in ref. 62). The nucleosome structure suggests that both ubiquitination and acetylation would destabilize tight packing between histones (63). Ubiquitination of H2A-H2B heterodimers does destabilize interaction with H3–H4 tetramers within chromatin (64). Transcriptional repression would then be achieved through both deacetylation and deubiquitination. HDAC6 might bind ubiquitinated histones, deacetylate them, and recruit DUBs for deubiquitination. Alternatively, because HDAC6 is reported to have a role in the cytoplasm (23), it is possible that binding polyubiquitin may be more critical in this subcellular compartment than in the nucleus.
Acknowledgments
We thank Drs. C. M. Grozinger and S. L. Schreiber for the mammalian expression vectors for HDAC 4, 5, 6, and HDAC6 (H216A/H611A). We thank Drs. A. Ciechanover, D. E. Gottschling, and E. Seto for comments on the manuscript. We are indebted to David J. Elzi and Jason J. Yada for help with the two-hybrid screens. This work was supported by a National Institutes of Health/National Cancer Institute Grant R01CA57138 (to R.N.E.). S.S.H. is supported by the National Institutes of Health, and A.O. is supported by the Human Frontiers in Science Program. R.N.E. is an American Cancer Society Research Professor.
Abbreviations
- HDAC
histone deacetylase
- DUB
deubiquitinating enzyme
- HA-Ub
hemagglutinin-tagged ubiquitin
- Ub-al
ubiquitin-aldehyde
- UIM
ubiquitin-interaction motif, UBA, ubiquitin-associated domain
- UBP
ubiquitin-specific processing protease
References
- 1.Jenuwein T, Allis C D. Science. 2001;293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- 2.Cheung P, Allis C D, Sassone-Corsi P. Cell. 2000;103:263–271. doi: 10.1016/s0092-8674(00)00118-5. [DOI] [PubMed] [Google Scholar]
- 3.Levinger L, Varshavsky A. Cell. 1982;28:375–385. doi: 10.1016/0092-8674(82)90355-5. [DOI] [PubMed] [Google Scholar]
- 4.Davie J R, Murphy L C. Biochemistry. 1990;29:4752–4757. doi: 10.1021/bi00472a002. [DOI] [PubMed] [Google Scholar]
- 5.Nickel B E, Allis C D, Davie J R. Biochemistry. 1989;28:958–963. doi: 10.1021/bi00429a006. [DOI] [PubMed] [Google Scholar]
- 6.Pham A D, Sauer F. Science. 2000;289:2357–2360. doi: 10.1126/science.289.5488.2357. [DOI] [PubMed] [Google Scholar]
- 7.Narlikar G J, Fan H-Y, Kingston R E. Cell. 2002;108:475–487. doi: 10.1016/s0092-8674(02)00654-2. [DOI] [PubMed] [Google Scholar]
- 8.Spencer V A, Davie J R. Gene. 1999;240:1–12. doi: 10.1016/s0378-1119(99)00405-9. [DOI] [PubMed] [Google Scholar]
- 9.Bertos N, Wang A, Yang X. Biochem Cell Biol. 2001;79:243–252. [PubMed] [Google Scholar]
- 10.Gray S G, Eckstrom T J. Exp Cell Res. 2001;262:75–83. doi: 10.1006/excr.2000.5080. [DOI] [PubMed] [Google Scholar]
- 11.Kao H Y, Downes M, Ordentlich P, Evans R M. Genes Dev. 2000;14:55–66. [PMC free article] [PubMed] [Google Scholar]
- 12.Huang E Y, Zhang J, Miska E A, Guenther M G, Kouzarides T, Lazar M A. Genes Dev. 2000;14:45–54. [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang C L, McKinsey T A, Lu J R, Olson E N. J Biol Chem. 2001;276:35–39. doi: 10.1074/jbc.M007364200. [DOI] [PubMed] [Google Scholar]
- 14.Dressel U, Bailey P J, Wang S C, Downes M, Evans R M, Muscat G E. J Biol Chem. 2001;276:17007–17013. doi: 10.1074/jbc.M101508200. [DOI] [PubMed] [Google Scholar]
- 15.Huynh K D, Fischle W, Verdin E, Bardwell V J. Genes Dev. 2000;14:1810–1823. [PMC free article] [PubMed] [Google Scholar]
- 16.Franco P J, Farooqui M, Seto E, Wei L N. Mol Endocrinol. 2001;15:1318–1328. doi: 10.1210/mend.15.8.0682. [DOI] [PubMed] [Google Scholar]
- 17.Amann J M, Nip J, Strom D K, Lutterbach B, Harada H, Lenny N, Downing J R, Meyers S, Hiebert S W. Mol Cell Biol. 2001;21:6470–6483. doi: 10.1128/MCB.21.19.6470-6483.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lemercier C, Brocard M P, Puvion-Dutilleul F, Kao H Y, Albagli O, Khochbin S. J Biol Chem. 2002;277:22045–22052. doi: 10.1074/jbc.M201736200. [DOI] [PubMed] [Google Scholar]
- 19.Kirsh O, Seeler J-S, Pichler A, Gast A, Müller S, Miska E A, Mathieu M, Harel-Bellan A, Kouzarides T, Melchior F, Dejean A. EMBO J. 2002;21:2682–2691. doi: 10.1093/emboj/21.11.2682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Fischle W, Kiermer V, Dequiedt F, Verdin E. Biochem Cell Biol. 2001;79:337–348. [PubMed] [Google Scholar]
- 21.Verdel A, Curtet S, Brocard M P, Rousseaux S, Lemercier C, Yoshida M, Khochbin S. Curr Biol. 2000;10:747–749. doi: 10.1016/s0960-9822(00)00542-x. [DOI] [PubMed] [Google Scholar]
- 22.Seigneurin-Berny D, Verdel A, Curtet S, Lemercier C, Garin J, Rousseaux S, Khochbin S. Mol Cell Biol. 2001;21:8035–8044. doi: 10.1128/MCB.21.23.8035-8044.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hubbert C, Guardiola A, Shao R, Kawaguchi Y, Ito A, Nixon A, Yoshida M, Wang X-F, Yao T-P. Nature. 2002;417:455–458. doi: 10.1038/417455a. [DOI] [PubMed] [Google Scholar]
- 24.Grozinger C M, Hassig C A, Schreiber S L. Proc Natl Acad Sci USA. 1999;96:4868–4873. doi: 10.1073/pnas.96.9.4868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lai A, Kennedy B K, Barbie D A, Bertos N R, Yang X J, Theberge M-C, Tsai S-C, Seto E, Zhang Y, Kuzmichev A, et al. Mol Cell Biol. 2001;21:2918–2932. doi: 10.1128/MCB.21.8.2918-2932.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ciechanover A, Orian A, Schwartz A L. BioEssays. 2000;22:442–451. doi: 10.1002/(SICI)1521-1878(200005)22:5<442::AID-BIES6>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- 27.Deng L, Wang C, Spencer E, Yang L, Braun A, You J, Slaughter C, Pickart C, Chen Z J. Cell. 2000;103:351–361. doi: 10.1016/s0092-8674(00)00126-4. [DOI] [PubMed] [Google Scholar]
- 28.Kaiser P, Flick K, Wittenberg C, Reed S I. Cell. 2000;102:303–314. doi: 10.1016/s0092-8674(00)00036-2. [DOI] [PubMed] [Google Scholar]
- 29.Conaway R C, Brower C S, Weliky Conaway J. Science. 2002;296:1254–1258. doi: 10.1126/science.1067466. [DOI] [PubMed] [Google Scholar]
- 30.Hicke L. Nat Rev Mol Cell Biol. 2001;2:195–201. doi: 10.1038/35056583. [DOI] [PubMed] [Google Scholar]
- 31.Salghetti S E, Caudy A A, Chenoweth J G, Tansey W P. Science. 2001;293:1651–1653. doi: 10.1126/science.1062079. [DOI] [PubMed] [Google Scholar]
- 32.Salghetti S E, Muratani M, Wijnen H, Futcher B, Tansey W P. Proc Natl Acad Sci USA. 2000;97:3118–3123. doi: 10.1073/pnas.050007597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ferdous A, Gonzalez F, Sun L, Kodadek T, Johnston S A. Mol Cell. 2002;7:981–991. doi: 10.1016/s1097-2765(01)00250-7. [DOI] [PubMed] [Google Scholar]
- 34.Gonzalez F, Delahodde A, Kodadek T, Johnston S A. Science. 2002;296:548–550. doi: 10.1126/science.1069490. [DOI] [PubMed] [Google Scholar]
- 35.Orian A, Whiteside S, Israel A, Stancovski I, Schwartz A L, Ciechanover A. J Biol Chem. 1995;270:21707–21714. doi: 10.1074/jbc.270.37.21707. [DOI] [PubMed] [Google Scholar]
- 36.Hershko A, Rose I A. Proc Natl Acad Sci USA. 1987;84:1829–1833. doi: 10.1073/pnas.84.7.1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vojtek A B, Hollenberg S M, Cooper J A. Cell. 1993;74:205–214. doi: 10.1016/0092-8674(93)90307-c. [DOI] [PubMed] [Google Scholar]
- 38.Hollenberg S M, Sternglanz R, Cheng P F, Weintraub H. Mol Cell Biol. 1995;15:3813–3822. doi: 10.1128/mcb.15.7.3813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chien C T, Bartel P L, Sternglanz R, Fields S. Proc Natl Acad Sci USA. 1991;88:9578–9582. doi: 10.1073/pnas.88.21.9578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Polo S, Sigismund S, Faretta M, Guidi M, Capua M R, Bossi G, Chen H, De Camilli P, Di Fiore P P. Nature. 2002;416:451–455. doi: 10.1038/416451a. [DOI] [PubMed] [Google Scholar]
- 41.McKinsey T A, Zhang C L, Olson E N. Proc Natl Acad Sci USA. 2000;97:14400–14405. doi: 10.1073/pnas.260501497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bonni S, Wang H R, Causing C G, Kavsak P, Stroschein S L, Luo K, Wrana J L. Nat Cell Biol. 2001;3:587–595. doi: 10.1038/35078562. [DOI] [PubMed] [Google Scholar]
- 43.Stroschein S L, Bonni S, Wrana J L, Luo K. Genes Dev. 2001;15:2822–2836. doi: 10.1101/gad.912901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Miyake S, Sellers W R, Safran M, Li X, Zhao W, Grossman S R, Gan J, DeCaprio J A, Adams P D, Kaelin W G., Jr Mol Cell Biol. 2000;20:8889–8902. doi: 10.1128/mcb.20.23.8889-8902.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bays N W, Hampton R Y. Curr Biol. 2002;12:366–371. doi: 10.1016/s0960-9822(02)00862-x. [DOI] [PubMed] [Google Scholar]
- 46.Wilkinson K D. FASEB J. 1997;11:1245–1256. doi: 10.1096/fasebj.11.14.9409543. [DOI] [PubMed] [Google Scholar]
- 47.Chung C H, Baek S H. Biochem Biophys Res Commun. 1999;266:633–640. doi: 10.1006/bbrc.1999.1880. [DOI] [PubMed] [Google Scholar]
- 48.Young P, Deveraux Q, Beal R E, Pickart C M, Rechsteiner M. J Biol Chem. 1998;273:5461–5467. doi: 10.1074/jbc.273.10.5461. [DOI] [PubMed] [Google Scholar]
- 49.Hofmann K, Falquet L. Trends Biochem Sci. 2001;26:347–350. doi: 10.1016/s0968-0004(01)01835-7. [DOI] [PubMed] [Google Scholar]
- 50.Thrower J S, Hoffman L, Rechsteiner M, Pickart C M. EMBO J. 2000;19:94–102. doi: 10.1093/emboj/19.1.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Riezman H. Nature. 2002;416:381–383. doi: 10.1038/416381a. [DOI] [PubMed] [Google Scholar]
- 52.Hofmann K, Bucher P. Trends Biochem Sci. 1996;21:172–173. [PubMed] [Google Scholar]
- 53.Vadlamudi R K, Joung I, Strominger J L, Shin J. J Biol Chem. 1996;271:20235–20237. doi: 10.1074/jbc.271.34.20235. [DOI] [PubMed] [Google Scholar]
- 54.Chen L, Shinde U, Ortolan T G, Madura K. EMBO Rep. 2001;2:933–938. doi: 10.1093/embo-reports/kve203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bertolaet B L, Clarke D J, Wolff M, Watson M H, Henze M, Divita G, Reed S I. J Mol Biol. 2001;313:955–963. doi: 10.1006/jmbi.2001.5105. [DOI] [PubMed] [Google Scholar]
- 56.Wilkinson C R, Seeger M, Hartmann-Petersen R, Stone M, Wallace M, Semple C, Gordon C. Nat Cell Biol. 2001;3:939–943. doi: 10.1038/ncb1001-939. [DOI] [PubMed] [Google Scholar]
- 57.Funakoshi M, Sasaki T, Nishimoto T, Kobayashi H. Proc Natl Acad Sci USA. 2002;99:745–750. doi: 10.1073/pnas.012585199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rao H, Sastry A. J Biol Chem. 2002;277:11691–11695. doi: 10.1074/jbc.M200245200. [DOI] [PubMed] [Google Scholar]
- 59.Dieckmann T, Withers-Ward E S, Jarosinski M A, Liu C-F, Chen I S Y, Feigon J. Nat Struct Biol. 1998;5:1042–1047. doi: 10.1038/4220. [DOI] [PubMed] [Google Scholar]
- 60.Laity J H, Lee B M, Wright P E. Curr Opin Struct Biol. 2001;11:39–46. doi: 10.1016/s0959-440x(00)00167-6. [DOI] [PubMed] [Google Scholar]
- 61.Davie J R, Murphy L C. Biochem Biophys Res Commun. 1994;203:344–350. doi: 10.1006/bbrc.1994.2188. [DOI] [PubMed] [Google Scholar]
- 62.Grunstein M. Nature. 1997;389:349–352. doi: 10.1038/38664. [DOI] [PubMed] [Google Scholar]
- 63.Luger K, Mader A W, Richmond R K, Sargent D F, Richmond T J. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 64.Li W, Nagaraja S, Delcuve G P, Hendzel M J, Davie J R. Biochem J. 1993;296:737–744. doi: 10.1042/bj2960737. [DOI] [PMC free article] [PubMed] [Google Scholar]