Abstract
The 5′ UTR of turnip yellow mosaic virus RNA contains two conserved hairpins with internal loops consisting of C⋅C and C⋅A mismatches. In this article, evidence is presented indicating that the 5′ proximal hairpin functions as an encapsidation initiation signal. Extensive mutagenesis studies on this hairpin and sequencing of virus progeny showed a clear preference for C⋅C and C⋅A mismatches within the internal loop. The importance of these mismatches lies in their pH-dependent protonation and stable base pair formation. Encapsidation efficiency was found to be severely affected for several mutants lacking the protonatable mismatches in the internal loop of the 5′ proximal hairpin. Furthermore, gel mobility-shift assays were performed with various RNA hairpins and empty capsids with a hole. Protonatable hairpins containing C⋅C and/or C⋅A pairs were found to bind specifically to the interior of the protein shell under acidic conditions (pH 4.5) in the presence of spermidine. Based on these results we propose that this binding of protonated cytosines to the coat protein of turnip yellow mosaic virus may represent a new motif in RNA–protein interactions.
Turnip yellow mosaic virus (TYMV) is a nonenveloped plant virus and the type member of the genus Tymovirus. The virion is an icosahedral particle with T = 3 symmetry, and the protein shell consists of 180 identical subunits (1). The genomic RNA is 6,318 nt long and has an unusually high cytosine content (38%) and a rather low guanine content (18%). The genome carries three ORFs coding for the overlapping protein, which is thought to be involved in cell-to-cell transport, the RNA-dependent RNA polymerase polyprotein, and the coat protein, which is expressed from a subgenomic messenger RNA. The 3′ UTR has been shown to contain a tRNA-like structure whose function is still unclear (2).
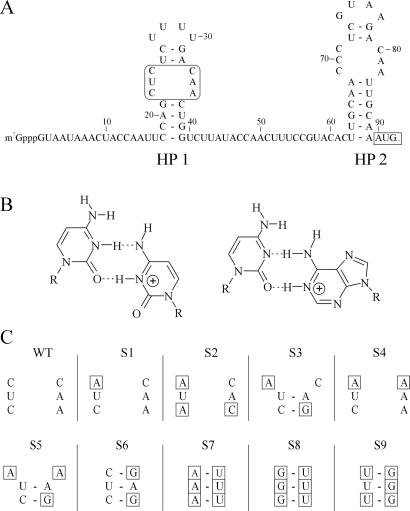
The 5′ UTR of TYMV RNA is 90 nt long and has been shown, by means of structure probing and sequence comparison, to harbor two simple hairpins containing symmetric internal loops, which consist of C⋅C and C⋅A mismatches (ref. 3; see Fig. 1A). Under slightly acidic conditions these mismatches can form rather stable base pairs. NMR and UV melting experiments have confirmed that protonation of the C⋅C and C⋅A mismatches in hairpin 2 (HP2) stabilizes the structure when the pH is lowered from 7 to 5 (refs. 3 and 4; see Fig. 1B). A preliminary study of the possible function of the protonatable cytosines in the two 5′ UTR hairpins suggested a dependence of TYMV on these nucleotides to establish a successful infection (5). A role for these hairpins in encapsidation was proposed. However, conclusions about the apparent requirement of C⋅C and/or C⋅A mismatches were preliminary, as this study described one single mutant for which only a few revertants were recovered after passaging in Chinese cabbage. Moreover, this construct could not solve the question of whether an A⋅C pair instead of C⋅A was also allowed, because the chosen mutation rather forced the recovery of C⋅C and C⋅A mismatches through single step reversions. A⋅C mismatches would require two reversions, which may be the reason they were not observed. Also, mutants in which the symmetric internal loop lacks C⋅C and C⋅A mismatches were not tested.
Figure 1.
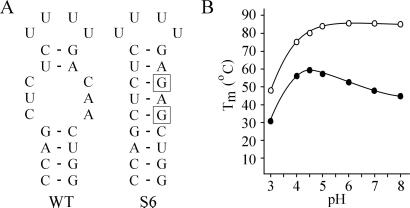
Overview of the substitution mutations made in the internal loop of the 5′ proximal hairpin of TYMV RNA. (A) The 5′ UTR of TYMV RNA including the two hairpins containing the protonatable internal loops (HP1 and HP2). The mutagenesis site and the start codon of movement protein (MP) are boxed. (B) Proposed structure of a protonated C⋅C and C⋅A mismatch. Note the identical positioning of the two exocyclic amino groups in the deep groove. (C) Overview of the substitution mutants S1–S9 used in this study. Base substitutions in the internal loop, as compared with WT, are boxed.
Here we describe a mutagenesis study of the 5′ proximal hairpin of TYMV RNA [hairpin 1 (HP1), see Fig. 1A] to delimit the requirement of C⋅C, C⋅A, and/or A⋅C mismatches in the internal loop. Also, data on packaging efficiency linking the absence of C⋅C, C⋅A, and/or A⋅C mismatches in the 5′ proximal hairpin with altered ratios of filled and empty capsids as compared with the WT situation are presented. The influence of C⋅C and C⋅A pairs on the stability of HP1 was studied as well as the binding of this hairpin to the coat protein capsid. Data are presented showing the increased affinity of protonatable hairpins in binding to TYMV coat protein under acidic conditions. From these results it is concluded that the protonatable 5′ proximal hairpin may function as an encapsidation initiation signal, and that this property is probably shared by all members of the genus Tymovirus. A novel type of RNA–protein interaction that is accompanied by the base pairing of protonatable bases under acidic conditions is proposed.
Materials and Methods
Construction of cDNA Mutants.
To obtain mutant TYMV RNA, an infectious cDNA clone of the Blue Lake strain of TYMV, pBL16 (6), was used. All mutants were constructed by PCR. PCR was performed by using mutagenesis primers together with the oligonucleotide 5′-GGAAT GGACC ATGGG TAGGTC (NCO), followed by template extension using oligonucleotide 5′-GGATC CAAGC TTAAT AACGA CTCAC TATAG GTAAT CAAAT ACCAA TT (HIN). Primers used for RT-PCR were PCR-5′ (5′-biotin-GTAAT CAAAT ACCAA TT) and PCR-3′ (5′-CAATG CTAAT TGGAA GG). Primer PCR-3′ was also used for sequencing purposes. The sequences of the mutagenesis primers used are: mutant S1 (5′-CAAAT ACCAA TTCCA GCTAT CTTTT GACAA CTGGT CTTAT ACCAA C), mutant S2 (5′-CAAAT ACCAA TTCCA GATAT CTTTT GACAC CTGGT CTTAT ACCAA C), mutant S3 (5′-CAAAT ACCAA TTCCA GCTAT CTTTT GACAG CTGGT CTTAT ACCAA C), mutant S4 (5′-CAAAT ACCAA TTCCA GCTAT CTTTT GAAAA CTGGT CTTAT ACCAA C), mutant S5 (5′-CAAAT ACCAA TTCCA GCTAT CTTTT GAAAG CTGGT CTTAT ACCAA C), mutant S7 (5′-CAAAT ACCAA TTCCA GAAAT CTTTT GATTT CTGGT CTTAT ACCAA C), mutant S8 (5′-CAAATA CCAA TTCCA GGGGT CTTTT GATTT CTGGT CTTAT ACCAA C), and mutant S9 (5′-CAAAT ACCAA TTCCA GTTTT CTTTT GAGGG CTGGT CTTAT ACCAA C). The resulting PCR products, containing the T7 promoter sequence as well as the 5′ part of the viral genome, were digested with HindIII and NcoI and ligated in pUC21. Sequence analysis was performed by using a T7 Sequencing kit (Amersham Pharmacia). These fragments were then cloned into pBL16, followed again by sequencing. All primers were synthesized by Eurogentec, Brussels.
RNA Fragments.
RNA fragments corresponding to the WT HP1 (5′-CCAGC UCUCU UUUGA CAACU GG) and S6 (5′-CCAGC UCUCU UUUGA GAGCU GG) were chemically synthesized (BioSyn group, Leiden Institute of Chemistry). RNA fragment Y (5′-CUUUU GCAAA AGCAA AAGC) was chemically synthesized by NAPS Nucleinsäuresynthesen, Göttingen, Germany (a gift of J. Nagel). UV melting experiments were performed as described (3). In the binding assays, labeling of RNA was performed with T4 polynucleotide kinase (United States Biochemical) and [γ-32P]-ATP (Amersham Pharmacia).
Plants.
Chinese cabbage (Brassica pekinensis) was grown under an 8- to 16-h light-dark regime. Standard conditions in the growth chambers include a light intensity of 5,000–6,000 lux, a humidity of 70%, and a constant temperature of 23°C. Three-week-old plants were used for infection with RNA.
RNA Preparation and Plant Inoculation.
Plasmid DNA of the various clones was linearized with NdeI. Transcription with T7 RNA polymerase (Amersham Pharmacia), and subsequent plant inoculation was performed according to ref. 5. On average 2 μg of RNA was applied per leaf. Symptom development was followed for about 4 weeks after inoculation, and only leaves showing systemic symptoms were used either for further analysis of the 5′ UTR sequence, passaging of the progeny onto new plants, or capsid analysis.
Extraction of the Progeny Virus from Plants.
Virus was isolated after each round of infection by using the bentonite procedure, as described for TYMV (7). For inoculation of new plants the supernatant of the first centrifugation step was used. At this stage of the virus purification virus samples were also taken for RNA sequence analysis by RT-PCR.
Analysis of the Capsid Composition.
Virus was isolated according to the bentonite procedure (7), and the pellet was dissolved in 10 mM Na acetate, pH 6.0. Bottom component (B component) and natural top component (NTC) of TYMV were separated by using agarose gel electrophoresis as described (5). The gels were stained with Coomassie brilliant blue, according to Maniatis et al. (8). Quantitative analysis was performed by scanning of the stained gels using the GelDoc system of BioRad including the quantity one software package. Samples for electron microscopy were applied on copper grids (mesh 100) with a carbon-coated Formvar support film and glow-discharged. For negative staining 16% ammonium molybdate, pH 7.0, was used.
Analysis of the Viral RNA.
RNA was isolated from the virus by phenol/chloroform extraction, followed by RT-PCR using primers HIN and NCO. At this stage the PCR products were either cloned into pUC21 for sequencing of individual clones or biotinylated primers were used. The biotinylated PCR fragments were sequenced after separation and purification of the strands with Dynabeads (Dynal).
Calculation of RNA Secondary Structure Stability.
The influence of the mutations on RNA secondary structure formation was calculated by using the efn-server running mfold version 3.0 (9).
Gel Mobility-Shift Assay.
For the preparation of artificial top component (ATC), B component was separated from NTC by CsCl-gradient centrifugation. The initial density of the CsCl solution was 1.45 g⋅ml−1, and centrifugation was for 18 h at 60,000 rpm at 10°C. B component and the NTC were individually removed from the centrifuge tubes and dialyzed against 10 mM Tris⋅HCl, pH 7.0 and 10 mM Na acetate buffer, pH 6.0, respectively. Conditions of the freeze-thaw procedure were as described (10) and include 8–10 mg⋅ml−1 of the B component in 30 mM NaCl, 10 mM Tris⋅HCl buffer, pH 7.0. A CsCl-gradient centrifugation step (initial density 1.29 g⋅ml−1) was performed to separate the ATC preparation from the released RNA and intact B component. The ATC was dialyzed against 10 mM Na acetate, pH 6.0 and used for the binding experiments.
Binding experiments were performed as follows: One volume containing a fixed amount of 32P-labeled RNA (final concentration ≈10 nM) was mixed with an equal volume containing an ATC protein preparation in varying concentrations (16–2,000 nM in 10 mM Na acetate, pH 6.0). The RNA solution contained the components needed to vary the binding conditions (spermidine, pH). Incubation of the final solution was for 1 h at 20°C. After incubation, 1/6th volume of 30% glycerol was added, and the sample was directly loaded and electrophoresed on a 5% native polyacrylamide gel to separate ATC from unbound RNA. The gels were run at 4°C for 2–3 h. ATC particles were visualized by staining with Coomassie brilliant blue.
Results
Design and Analysis of Mutants of HP1.
This mutagenesis study focuses on the 5′ proximal hairpin (HP1) in the 5′ UTR of TYMV RNA (Fig. 1A). The substitution mutants S1–S9 (Fig. 1C) were designed to test the C⋅C, C⋅A, and A⋅C mismatch requirement for TYMV infection. The influence of these mutations on the secondary structure was predicted by using the Zuker mfold program. The introduction of three consecutive G residues (S8 and S9) could have profound effects on the RNA folding in a C-rich genome. No alternative structures, however, were predicted that would change the proposed folding of the 5′ UTR.
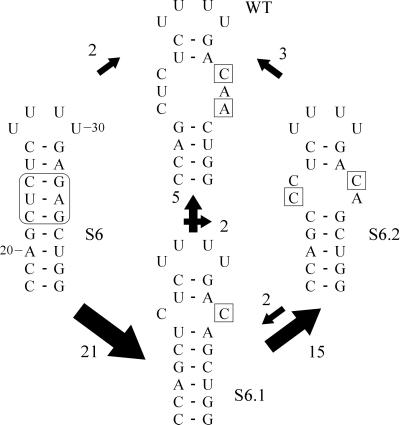
To extend the findings previously reported (see ref. 5), first mutant S6 was screened for the occurrence of reversions in the progeny. The original mutation was never recovered in the progeny of S6 (see also pBL16e in ref. 5). The changes in sequence of the 5′ UTR were monitored during four rounds of infection. Fig. 2 shows in a schematic representation a summary of the reversion pathways observed based on 134 individual sequence determinations. All revertants were found to include the introduction of one or more C⋅C and/or C⋅A mismatches. Additional mismatches, such as A⋅G or U⋅G, were found in only 7% of the recovered progeny, in some cases located outside the internal loop. However, the latter mismatches were always accompanied by the formation of C⋅A and/or C⋅C pairs at the position of the internal loop (not shown). Starting from revertant S6.1, the preferred introduction of the C⋅A mismatch in S6.2 at a non-WT position is of great interest because the formation of the WT sequence, also by means of one reversion only, is expected to be at least equally viable. From both S6.1 and S6.2 a minority of the progeny reverted to the WT sequence. After four rounds of infection the majority of the progeny still contained the double mismatch (S6.2).
Figure 2.
Reversion pathway for mutant S6. Changes in the 5′ UTR sequence were monitored over four rounds of infection. Differences in sequence as compared with the original mutant are boxed. The arrows and their size indicate the direction and frequency of accumulation of changes in the internal loop region. After the first round of infection 21 of 25 cases first generated S6.1, having one C⋅C mismatch at the same position as the WT. In the successive rounds of infection the majority of revertants S6.1 (75%) evolved to form S6.2, including a C⋅C as well as a C⋅A mismatch. Numbering of the sequence is according to the nucleotide position in the viral RNA genome.
Next, mutants S1, S2, and S3 were made to investigate whether only C⋅C and C⋅A mismatches are allowed or whether A⋅C mismatches also are permitted in the internal loop region. Because TYMV is able to generate reversions in its RNA genome during infection, we have also constructed mutants S4 and S5 to force the virus to introduce either C⋅A or A⋅C mismatches by single site reversion pathways. Mutants S1–S5 did not reveal any changes in the progeny after one round of infection. Therefore, these mutants were not analyzed further, although reversions in succeeding rounds of infection could not be excluded. The introduction of A⋅C mismatches (mutants S1, S2, and S3) apparently did not have a major influence on TYMV viability during infection. More surprisingly, the introduction of an A⋅A mismatch with or without the presence of a C⋅A mismatch did not force TYMV to introduce reversions in the first round of infection (mutants S4 and S5, respectively).
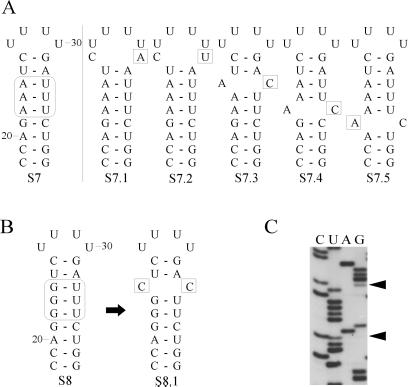
Unlike S6, mutant S7 excludes C⋅C and C⋅A mismatch introduction by a single substitution. In S7 only A⋅C mismatches can be introduced in the internal loop region in this fashion. The introduction of three A-U base pairs in the internal loop region (S7; Fig. 3A) did not always lead to reversions as the original mutation often was found unchanged after sequence analysis. After three rounds of infection the majority of the progeny with altered sequences contained a C⋅A or A⋅C mismatch (S7.1 and S7.3–5). Interestingly, the mismatches were not restricted to the internal loop region (S7.1, S7.2, and S7.5) as if reducing the stability of the hairpin is important as well.
Figure 3.
Observed reversion for mutant S7 (A) and S8 (B). Differences in sequence as compared with the original mutation are boxed. (C) Autoradiogram showing that the progeny of mutant S8 contains a mixed sequence at the positions indicated by arrowheads.
Finally, mutants S8 and S9 were made to exclude all possibilities of protonatable mismatch formation by means of single base substitutions within the defined internal loop region. The mutants designed to force the virus to create a double reversion for introduction of a C⋅C, C⋅A, or A⋅C mismatch took three rounds of infection before any change occurred. In only one case, progeny of the triple G⋅U mutant (S8; Fig. 3B) was found to contain a mixed sequence (Fig. 3C). The introduction of a C⋅C mismatch (S8.1) was confirmed after cloning of the 5′ UTR PCR fragment into pUC21, followed by single colony sequencing. The U⋅G mutant (S9) was not yet altered after three rounds of infection, and further screening was terminated.
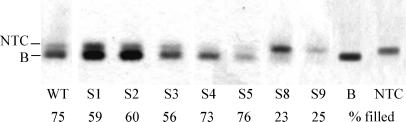
Packaging Efficiency.
TYMV preparations typically contain empty capsids (NTC) besides filled virions, containing the RNA genome (B component). During agarose gel electrophoresis, B component migrates slightly faster than the empty capsids (5), and the percentage of NTC particles and B component was determined to analyze the packaging efficiency. Only those mutants of which it was found that no reversions had taken place (Fig. 4) were analyzed. In the WT preparation about 75% of the particles are filled. As shown in Fig. 4, the packaging efficiency of mutants S4 and S5 did not seem to be affected. The percentage of filled particles for mutants S1, S2, and S3 was slightly lower (approximately 55–60%). Remarkably, the G⋅U and U⋅G mutants (S8 and S9, respectively) were down to as little as 25% of B component present in the virus preparation. To test the reliability of this technique the WT preparation was also analyzed with electron microscopy after negative staining. Counting more than 1,000 particles from WT preparations resulted in the same percentage of filled particles (75% ± 5%). Analysis of mutant S8 by electron microscopy yielded a ratio of 30% ± 8% for filled particles (>500 particles).
Figure 4.
Determination of the ratio of filled and empty particles of WT and various mutant virus preparations by means of agarose gel electrophoresis. Lane B represents purified B component. The values indicate the average percentage of filled particles for every mutant.
Protonatable Cytosines in HP1.
The frequent reintroduction of C⋅C, C⋅A, and/or A⋅C mismatches in the 5′ proximal hairpin raises the question of how and under what conditions these mismatches could play a functional role in the virus life cycle. For this purpose RNA fragments corresponding to the 5′ proximal HP1 of the WT and of mutant S6 were synthesized chemically (Fig. 5A). UV melting curves determined at a range of pH values revealed a remarkable difference in behavior of the two hairpins (Fig. 5B). The substitution of only two residues in the WT hairpin leads to the formation of a perfect Watson–Crick hairpin stem and gives rise to a maximum Tm of around 85°C at neutral pH. When the pH is lowered a decrease in Tm is observed for hairpin S6, which corresponds to the destabilization of the helix by protonation of C and A residues involved in Watson–Crick base pairing. In contrast, the WT HP1 revealed a very different pH dependence, showing an increase in Tm at more acidic conditions, reaching a maximum of 58°C at pH 4.5. The increase in Tm is 11°C going from pH 7.0 to pH 4.5.
Figure 5.
UV melting as a function of pH. (A) RNA fragments used. The boxes indicate substitutions in mutant S6 as compared with WT. (B) Influence of pH on midpoint temperature Tm. ●, WT 5′ proximal hairpin; ○, mutant S6 hairpin.
Binding Studies.
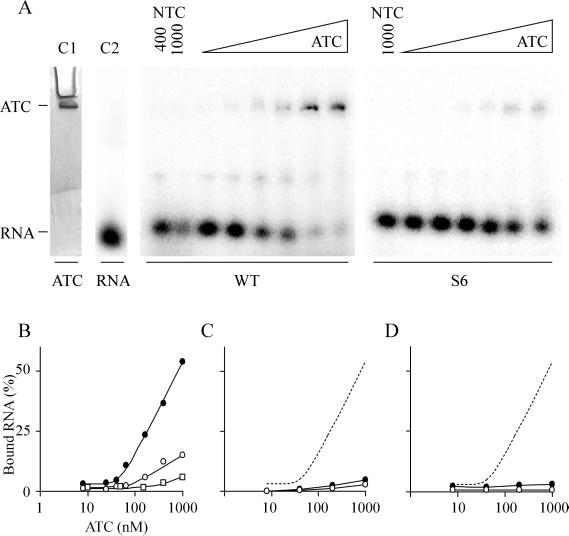
The reintroduction of protonatable mismatches after infection with mutant RNA, the strong influence on packaging efficiency, and the stabilization of these hairpins under acidic conditions all suggested a role for HP1 in encapsidation initiation. Gel mobility-shift assays were performed to study the possible role of the protonatable hairpins in binding to TYMV coat protein under acidic conditions. For this purpose so-called ATC was prepared by using the freeze-thaw method as described by Katouzian-Safadi et al. (10). Coat protein monomers, to be preferred in this type of binding studies, cannot be obtained or kept under physiological conditions (11). ATC particles consist of a TYMV coat protein shell containing a hole corresponding to the lack of one or two capsomers, caused by the escape of the viral RNA during the freeze-thaw treatment.
RNA fragments corresponding to the WT HP1 and mutant S6 hairpin were incubated with ATC under varying conditions and analyzed by native PAGE. Because of the large size of the capsid particle the band corresponding to the protein shell only just migrates into the gel (Fig. 6A, lane C1). The RNA fragments migrate either at the position of free RNA (Fig. 6A, lane C2) or at the height of ATC, indicative of complex formation. The specific binding conditions in this experiment included 50 mM Na acetate, pH 4.5, 3 mM spermidine and incubation for 1 h at 20°C. As shown in Fig. 6A, increasing amounts of RNA were bound with increasing amounts of protein (ranging from 8 to 1,000 nM of ATC). It is clear that the WT HP1 is complexed with the ATC particle with higher affinity than the S6 hairpin. The result is summarized in Fig. 6B. With 1,000 nM of ATC 50% of the WT RNA could be shifted in the gel (Fig. 6B, filled circles), whereas mutant S6 showed an approximate 4-fold lower binding efficiency (Fig. 6B, open circles). Control shift assays were also performed by using NTC particles (without a hole in the capsid) at concentrations of either 400 or 1,000 nM (Fig. 6A, lanes NTC). No binding of the RNA fragments to the NTC particles was observed. Because hairpin S6 differs from the WT HP1 by two substitutions, hairpin Y, also containing Watson–Crick base pairs only, was used as a control (for sequence see Materials and Methods). Hairpin Y consists of a stem of 7 bp, but the sequence of the stem and the 4-nt loop is completely unrelated to hairpin S6. Fig. 6B shows that there is almost no binding of hairpin Y to ATC (open squares).
Figure 6.
Gel mobility-shift assay. (A) Binding experiments using WT HP1 and mutant hairpin S6 at different concentrations of ATC, ranging from 8 to 1,000 nM. Binding conditions include 50 mM Na acetate, pH 4.5, and 3 mM spermidine. The control lane (C1) containing ATC only was stained separately with Coomassie brilliant blue. In lane C2 32P-labeled WT HP1 was loaded. The control lanes NTC contain either 1,000 and 400 nM, or 1,000 nM only of NTC in the binding reactions with HP1 and hairpin S6, respectively. (B) Plot of the relative amounts of bound RNA as shown in A for the WT HP1 (●) and mutant S6 (○). Hairpin Y (□), consisting of Watson–Crick base pairs but with a different sequence, was used as a control. (C) Binding conditions: 50 mM Na acetate, pH 4.5. The dashed line is the WT HP1 shift as found in B. (D) Binding conditions: 50 mM Na cacodylate, pH 7.0, and 3 mM spermidine.
The conditions for binding the RNA to the coat protein were very specific. In the absence of spermidine no binding was observed (Fig. 6C). Also, complex formation at pH 7.0, in the presence of spermidine, did not occur (Fig. 6D). Other binding conditions, including high salt conditions or the addition of magnesium, were not successful either (not shown).
Discussion
In this article, evidence is presented that the 5′ proximal HP1 of the 5′ UTR of TYMV RNA may function as an encapsidation signal. Forced evolution experiments show that the presence of C⋅A and/or C⋅C oppositions in HP1 is important for optimal viability of the virus. Besides C⋅C and C⋅A pairs, A⋅C “mismatches” were found to be functional. Replacement of the C⋅C and C⋅A base pairs by other ones have a clear effect on encapsidation efficiency as concluded from an altered ratio of RNA-containing particles to empty capsids. Analysis of the influence of C⋅C and C⋅A pairs on the stability of hairpin HP1, using UV melting experiments, revealed an increase of the Tm on lowering the pH, whereas a hairpin containing only Watson–Crick base pairs shows a decrease in Tm under acidic conditions. Furthermore, HP1 can be specifically bound to the interior of an empty viral capsid at low pH, in the presence of spermidine.
Structural Requirements of HP1.
Initially, it was reported that only C⋅C and/or C⋅A mismatches were allowed in the 5′ proximal hairpin of TYMV RNA (5). In this study we show that A⋅C mismatches are virtually equivalent. Also, an additional type of revertants was found occasionally, generating changes outside the internal loop. By far the majority of the revertants have reintroduced C⋅C, C⋅A, or A⋅C pairs. However, a role for relieving the hairpin stability per se cannot be excluded as, for instance, the rather stable mutant S6 hairpin might have an inhibitory effect on translation or replication. From the frequency of reversions that were observed it may be concluded that on stabilization of HP1 the need for compensating mutations becomes more apparent. From a statistical point of view destabilization of the helix of HP1 can be reached by 54 different substitutions, whereas in S6 and S7, only 15 and 13 of the possible 54 reversions, respectively, lead to the introduction of a C⋅C, C⋅A, or A⋅C mismatch. Here, it is clear that when stability relaxing reversions are observed they consistently correspond to C⋅C, C⋅A, and/or A⋅C mismatches, often at the same position as in WT.
HP1 and HP2 represent a repeated motif, which in other tymoviruses even occurs in three or four copies (3). It is therefore not surprising to see that neither deletion of HP1 (unpublished results) nor of HP2 alone (5) is detrimental to the virus. Deletion of both 5′ UTR hairpins, however, abolishes infection of secondary leaves (systemic symptoms), indicating a defect in encapsidation (5). The deletion or mutation of either hairpin can therefore be compensated by the other. It allows mutants containing substitutions to remain unchanged for a certain period until a selective advantage is generated through the introduction of a C⋅C, C⋅A, and/or A⋅C mismatch. This is especially clear for the G⋅U pair containing mutants (S8 and S9), which need a double substitution to revert to WT structure. Most likely the selection pressure on these mutants can be raised by deletion of HP2.
Packaging Efficiency.
Another indication for an important role of the internal loop of HP1 for encapsidation came from the drop in relative amount of filled particles after replacement of the internal loop of HP1 by other sequences. This finding confirms the results of Hellendoorn et al. (5). The ratio of only 25% of filled particles for certain mutants was not a consequence of the methods used, because the WT preparation always contained 70–80% of this B component, which is a well-documented property of TYMV. Why mutant S5 behaves like WT, despite the absence of a protonatable pair, is unclear. Interestingly, one of the possible A⋅A oppositions (the single hydrogen-bonded AA-N7 amino) presents two exocyclic amino groups in the deep groove comparable to the situation in the C⋅C and C⋅A pair (Fig. 1B) and therefore might function in a pH-independent manner. However, a gel mobility-shift assay with an RNA hairpin containing the A⋅A mismatch did not reveal detectable binding at pH 7.0 (results not shown). For all mutants, it is acknowledged that differences in the NTC/B component ratio might be influenced by a compromise in levels of replication or translation of mutant viral RNA.
Protonation of C⋅C and C⋅A Mismatches.
The results of the UV melting experiments are in agreement with those obtained for HP2 (3), for which one-dimensional NMR measurements were performed, supporting the formation of protonated C⋅C and C⋅A pairs at pH 5.0. How this type of protonated base pairs is positioned in the RNA double helix of HP1 is not yet known and would require a more extensive analysis with NMR or x-ray diffraction techniques. However, a few examples of other stems containing C⋅C and/or C⋅A mismatches studied with these techniques have been reported, and it was shown that a single C⋅A mismatch causes a severe distortion of the RNA double-helical axis with a bend of 20° as well as an expansion of the major groove, whereas two adjoining mismatches showed only a mild helical distortion (12, 13). A functional C⋅A pair has been described to occur in the small nuclear RNA U6, where it exists as a protonated base pair playing a role in the regulation of RNA splicing. It is interesting to see that in trypanosomatids this pair is replaced by the isosteric C⋅C pair (14).
Requirements for Binding of RNA to TYMV Capsids.
Specific binding of HP1 to empty capsids of TYMV was shown by using a gel mobility-shift assay and appears to require both acidic conditions and the presence of spermidine. An interaction of the protonated HP1 with the natural empty capsid, lacking a hole in the protein shell, was not observed, indicating that binding occurs at the interior of the protein shell. No (or reduced) binding was observed when magnesium ions were used instead of spermidine, despite the fact that magnesium is an important component of the TYMV virion (15).
Amounts of the polyamine spermidine up to as many as 400–500 molecules per virion (15) are believed to function in neutralizing the negative charge of the RNA phosphate residues. Furthermore, this polyamine was suggested to be instrumental in TYMV assembly (16), which is supported by our results.
The reduced binding efficiency of the control hairpin S6 demonstrated the requirement of the protonatable C⋅C and C⋅A mismatches for optimal binding. However, because a certain level of binding was observed for this control hairpin as well, it is suggested that additional interactions between the viral coat protein and the RNA take place. In our view, hairpin S6 was the best possible control in this study as only two base substitutions were introduced. The change in primary structure as well as in overall secondary structure was thereby minimized so that the influence of the protonatable C⋅C and C⋅A pairs could be studied. The results suggest that the internal loop sequence is dominantly involved in protein binding. This notion is confirmed by the binding at background levels of a hairpin consisting of a different stem and loop sequence (hairpin Y).
The conditions established for the interaction of the 5′ proximal hairpin of TYMV RNA with the coat protein are in agreement with results obtained with so-called dissociation–reassociation experiments involving genomic RNA of TYMV. It was found that ATC associates specifically with TYMV RNA under acidic conditions in the presence of spermidine. No apparent affinity was detected for other plant viral RNAs like tobacco mosaic virus RNA and alfalfa mosaic virus RNA, which are not particularly C rich (17). Based on these studies a role for cytosines in single-stranded regions in protein binding was proposed (18). Furthermore, the protonation of the 5′ proximal hairpins, necessary for their possible function as encapsidation initiation signals, is in good agreement with the notion that TYMV assembly is driven by a low pH generated in the chloroplasts by active photosynthesis (as reviewed in ref. 19).
A Novel Type of RNA–Protein Interaction?
Encapsidation signals on the RNA level have been described for a large number of spherical viruses. Mostly, however, the main principles underlying the RNA–protein interactions in spherical viruses involve the binding of basic amino acids such as lysines and arginines to the phosphate backbone of the RNA (20–22). Establishing the crystal structure of TYMV, including the finding of some ordered regions of RNA within the capsid, did not provide an understanding of molecular details of encapsidation for this virus (1).
Based on the results described in this article and as far as encapsidation initiation is concerned, we propose that TYMV uses protonated C⋅C and C⋅A base pairs in binding to the coat protein. Whether the excess of cytosines, as found mainly within single-stranded regions all over the genomic RNA (23), are cooperating in the same fashion in packaging remains to be investigated. A preliminary analysis of the C-rich regions has shown that similar hairpins, containing C⋅A and C⋅C pairs in stem regions, may exist (4).
The amino acids in the TYMV coat protein functioning as binding partners for the RNA still remain to be identified, although several candidates have been put forward on the basis of UV crosslinking and crystallography (1, 24). It should be very interesting to visualize the interaction between an RNA hairpin containing the protonated C⋅C and C⋅A pairs, e.g., HP1, and the ATC empty capsid to test the interaction proposed above. A direct interaction of protonated C⋅C or C⋅A pairs with protein has, to the best of our knowledge, not been described before and therefore may represent a novel type of RNA–protein interaction.
Acknowledgments
We thank Drs. R. Olsthoorn and M. de Smit for critically reading the manuscript. This work was supported by the European Commission (Project BIO4-98-0189).
Abbreviations
- TYMV
turnip yellow mosaic virus
- NTC
natural top component
- ATC
artificial top component
- B component
bottom component
- HP1
hairpin 1
- HP2
hairpin 2
- Tm
melting temperature
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Canady M A, Larson S B, Day J, McPherson A. Nat Struct Biol. 1996;3:771–781. doi: 10.1038/nsb0996-771. [DOI] [PubMed] [Google Scholar]
- 2.Giegé R. Proc Natl Acad Sci USA. 1996;93:12078–12081. doi: 10.1073/pnas.93.22.12078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hellendoorn K, Michiels P J A, Buitenhuis R, Pleij C W A. Nucleic Acids Res. 1996;24:4910–4917. doi: 10.1093/nar/24.24.4910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hellendoorn K. Ph.D. thesis. Leiden, The Netherlands: Leiden University; 1997. [Google Scholar]
- 5.Hellendoorn K, Verlaan P W G, Pleij C W A. J Virol. 1997;71:8774–8779. doi: 10.1128/jvi.71.11.8774-8779.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Skotnicki M L, Mackenzie A M, Gibbs A J. Arch Virol. 1992;127:25–35. doi: 10.1007/BF01309572. [DOI] [PubMed] [Google Scholar]
- 7.Dunn D B, Hitchborn J H. Virology. 1965;172:555–563. [Google Scholar]
- 8.Maniatis T, Sambrook J, Fritsch E F. Molecular Cloning: A Laboratory Manual. 2nd Ed. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 9.Mathews D H, Sabina J, Zuker M, Turner D H. J Mol Biol. 1999;288:911–940. doi: 10.1006/jmbi.1999.2700. [DOI] [PubMed] [Google Scholar]
- 10.Katouzian-Safadi M, Favre A, Haenni A L. Eur J Biochem. 1980;112:479–486. doi: 10.1111/j.1432-1033.1980.tb06110.x. [DOI] [PubMed] [Google Scholar]
- 11.Matthews R E F. Plant Virology. 3rd Ed. New York: Academic; 1991. pp. 209–227. [Google Scholar]
- 12.Jang S B, Hung L-W, Holbrook E L, Carter R J, Holbrook S R. Biochemistry. 1998;37:11726–11731. doi: 10.1021/bi980758j. [DOI] [PubMed] [Google Scholar]
- 13.Pan B, Mitra S N, Sundaralingam M. J Mol Biol. 1998;283:977–984. doi: 10.1006/jmbi.1998.2140. [DOI] [PubMed] [Google Scholar]
- 14.Huppler A, Nikstad L J, Allman A M, Brow D A, Butcher S E. Nat Struct Biol. 2002;9:431–435. doi: 10.1038/nsb800. [DOI] [PubMed] [Google Scholar]
- 15.Eecen H G, Van Dierendonck J H, Pleij C W A, Mandel M, Bosch L. Biochemistry. 1985;24:3610–3617. [Google Scholar]
- 16.Balint R, Cohen S S. Virology. 1985;144:181–193. doi: 10.1016/0042-6822(85)90316-2. [DOI] [PubMed] [Google Scholar]
- 17.Bouley J P, Briand J P, Jonard G, Witz J, Hirth L. Virology. 1975;63:312–319. doi: 10.1016/0042-6822(75)90305-0. [DOI] [PubMed] [Google Scholar]
- 18.Jonard G, Briand J P, Bouley J P, Witz J, Hirth L. Philos Trans R Soc London Ser B. 1976;276:123–129. doi: 10.1098/rstb.1976.0103. [DOI] [PubMed] [Google Scholar]
- 19.Rohozinski J, Hancock J M. J Gen Virol. 1996;77:163–165. doi: 10.1099/0022-1317-77-2-163. [DOI] [PubMed] [Google Scholar]
- 20.Jones T A, Liljas L. J Mol Biol. 1984;177:735–767. doi: 10.1016/0022-2836(84)90047-0. [DOI] [PubMed] [Google Scholar]
- 21.Larson S B, Koszelak S, Day J, Greenwood A, Dodds J A, McPherson A. Nature. 1993;361:179–182. doi: 10.1038/361179a0. [DOI] [PubMed] [Google Scholar]
- 22.Schneemann A, Reddy V S, Johnson J E. Adv Virus Res. 1998;50:381–446. doi: 10.1016/s0065-3527(08)60812-x. [DOI] [PubMed] [Google Scholar]
- 23.Hellendoorn K, Mat A W, Gultyaev A P, Pleij C W A. Virology. 1996;224:43–54. doi: 10.1006/viro.1996.0505. [DOI] [PubMed] [Google Scholar]
- 24.Ehresmann B, Briand J P, Reinbolt J, Witz J. Eur J Biochem. 1980;108:123–129. doi: 10.1111/j.1432-1033.1980.tb04702.x. [DOI] [PubMed] [Google Scholar]