Abstract
Electron microscopy of the RSC chromatin-remodeling complex reveals a ring of protein densities around a central cavity. The size and shape of the cavity correspond closely to those of a nucleosome. Results of nuclease protection analysis are consistent with nucleosome binding in the cavity. Such binding could explain the ability of RSC to expose nucleosomal DNA in the presence of ATP without loss of associated histones.
The wrapping of DNA around a histone octamer in the nucleosome is an impediment to many DNA transactions, including transcription (1, 2). The impediment may be removed by the combined action of chromatin-remodeling and histone-modifying complexes (3, 4). The first chromatin-remodeling complex to be described, SWI/SNF from yeast, is important for the transcription of ≈1% of all genes and is not required for cell viability (5). A similar complex, termed RSC, is more abundant in yeast and is required for cell viability (6). Human cells contain two such chromatin-remodeling complexes as well, one of which appears to be a homolog of SWI/SNF and the other a homolog of RSC (7).
Chromatin-remodeling complexes perturb nucleosome structure in an ATP-dependent manner. SWI/SNF, RSC, and related complexes expose nucleosomal DNA to attack by nucleases, catalyze histone octamer “sliding” and transfer between DNA molecules, and reduce the supercoiling of DNA circles bearing nucleosomes (6, 8–13). The exposure of nucleosomal DNA occurs without loss of histones, which is paradoxical, because the DNA behaves as if it were free and bound at the same time. Here we report on structural studies of RSC and RSC–nucleosome complexes that provide a possible basis for this seemingly contradictory behavior and for understanding the remodeling process.
Materials and Methods
RSC was prepared as described (6). Nucleosomes were formed from 154-bp DNA, uniformly labeled with [α-32P]dATP, and rat liver histones as described (10). Reactions of RSC with nucleosomes were in 15 μl containing 15 mM Hepes (pH. 7.5), 3 mM MgCl2, 10 mM potassium acetate, and 75 μg/ml BSA, for 20 min at 30°C.
To prepare samples for electron microscopy, about 3 μl of protein solution (≈30 μg/ml) was applied to freshly glow-discharged (in the presence of amylamine), carbon-coated Maxtaform, 300-mesh Cu/Rh grids (Ted Pella, Inc., Redding, CA), and preserved by negative staining in uranyl acetate. All images were collected under low-dose conditions on Kodak SO-163 film. Images were recorded at 0.3 μm underdefocus and ×60,000 ±1% magnification, using a Philips CM120 (FEI/Philips) microscope equipped with a LaB6 filament, operating at an accelerating voltage of 100 kV. Micrographs were digitized on a Zeiss SCAI flat-bed scanning densitometer (ZI/Zeiss) with a step size of 7 μm. Digitized images of particles preserved in negative stain were 3-fold pixel-averaged, which resulted in a pixel size of 3.312 Å on the object scale. All image processing was carried out by using the spider software package (14).
Results
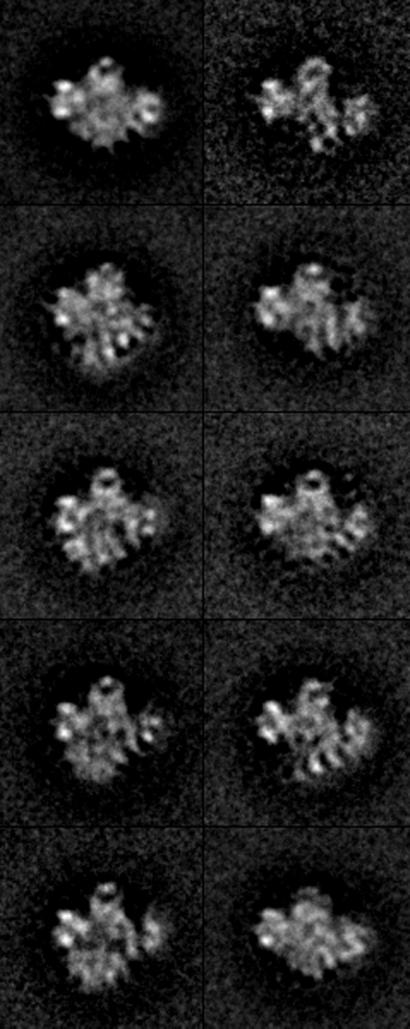
RSC is a 15-subunit complex with a total mass of ≈106 Da (6), large enough for meaningful 3D reconstruction from electron micrographs of individual particles. To this end, images of particles preserved in negative stain were recorded at tilt angles of 0° and 55°. Images from the zero-tilt micrographs were subjected to reference-free alignment and averaging (15). The results were indicative of a predominant orientation of the particles on the electron microscope grid, but with considerable variation in particle structure or conformation (not shown). The images were therefore processed by hierarchical ascendant classification (16), which divided them into comparatively homogeneous groups. At the limited resolution of this analysis (≈25 Å) and in projection, the top half of the RSC structure appeared fairly constant, whereas the bottom part varied in position, and was in some cases entirely absent (Fig. 1).
Figure 1.
Structure of RSC in projection. A total of 5,880 RSC particles preserved in uranyl acetate were computationally aligned and sorted into homogeneous classes (according to their conformation) using Hierarchical Ascendant Classification (16). A central area of lower density is apparent in several of the class averages shown (≈45% of particles). Most of the variation in RSC conformation is related to a domain forming the bottom part of the structure, which is either missing (≈35% of particles) or collapsed against the top of the structure (≈22% of particles).
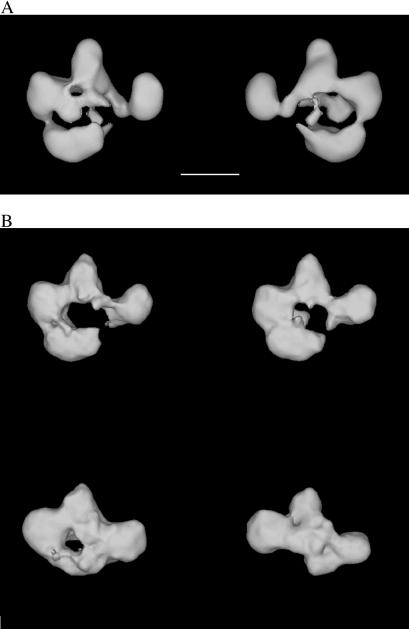
Particles in tilted micrographs corresponding to those in the zero-tilt micrographs were used to calculate 3D reconstructions. The resolution of the reconstructions was estimated at ≈26–30 Å by the Fourier Shell correlation method (17, 18). RSC was seen to consist of four modules surrounding a central cavity (Fig. 2A). Differences in the 3D reconstructions from different homogeneous groups defined in projection revealed only small changes in the conformation and relative position of three of the modules (defining the top portion of the central cavity), but revealed significant variation in the position of the fourth module (Fig. 2B).
Figure 2.
The RSC chromatin-remodeling complex. (A) RSC consists of four modules that define a central cavity. Two views of the structure (front and back) are shown. Scale bar corresponds to 100 Å. (B) The most significant variation in RSC conformation was caused by the collapse (Upper) or absence (Lower) of a module that forms the lower part of the RSC structure.
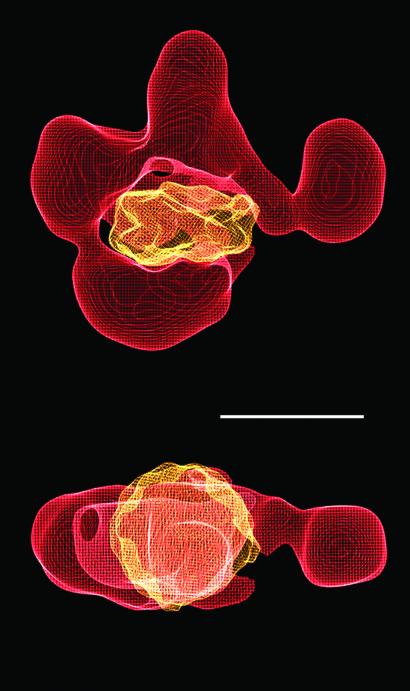
Biochemical studies have shown that RSC binds tightly to a nucleosome (Ka ≈ 10−8 M−1), forming an apparent 1:1 complex (19). In the presence of ATP, RSC converts the bound nucleosome to an “activated” state, characterized by an enhanced susceptibility of the DNA to nuclease attack (6). The size and shape of the central cavity in the RSC reconstruction are appropriate for binding a nucleosome: there is a snug fit of the x-ray structure of a nucleosome core particle, with no steric clash (Fig. 3).
Figure 3.
Possible mode of RSC–nucleosome interaction. An x-ray structure of the nucleosome (20) was filtered to 25 Å and manually fitted in the central cavity of the RSC structure by using the program o (21). The close fit between the nucleosome and the RSC cavity is apparent in two different views (front and top). Scale bar corresponds to 100 Å.
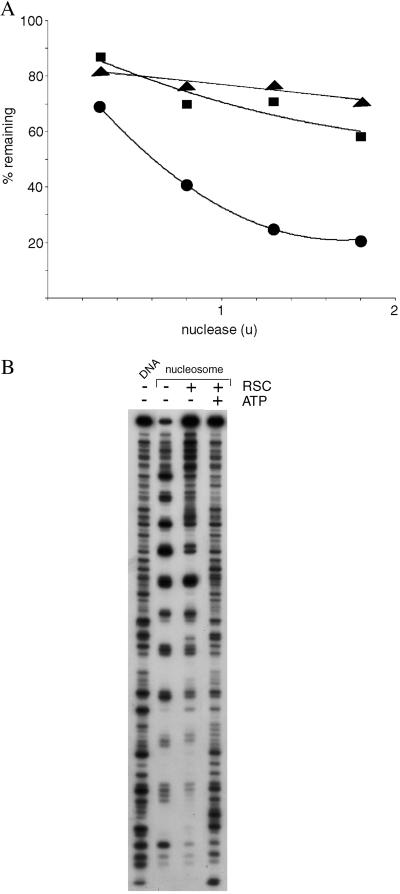
This mode of RSC–nucleosome interaction is supported by results of nuclease digestion analysis. Despite the well known capacity of RSC and related chromatin-remodeling complexes to expose nucleosomal DNA to nuclease attack in an ATP-dependent manner, we report here that RSC confers protection against digestion. The rate of double-strand cleavage of nucleosomal DNA by micrococcal nuclease was diminished by RSC, both in the presence and absence of ATP (Fig. 4A). The rate of single-strand cleavage by DNase I was also much reduced, both in the presence and absence of ATP (Fig. 4B, compare intensities of uppermost bands, caused by uncut DNA). Finally, the pattern of cutting by DNase I was affected by RSC, even in the absence of ATP: the nucleosome alone was maximally exposed to digestion every 10 residues, as expected; addition of RSC produced a “footprint,” with a diminished frequency of digestion at major sites outside the central region. These findings are readily explained by nucleosome binding in the RSC cavity. General protection against nuclease attack and a RSC footprint over much of the nucleosomal DNA are indicative of RSC enveloping the nucleosome. Persistent exposure of the DNA in the central region of the nucleosome is also consistent with binding in the RSC cavity: the central region, near the midpoint (dyad) of the two turns of DNA around the histone octamer, is located between the DNA entering and leaving the nucleosome; DNA can only enter and leave through the opening of the RSC cavity; the central region of the nucleosome will therefore be exposed at the opening of the cavity.
Figure 4.
Nuclease protection analysis of RSC–nucleosome complexes. (A) Micrococcal nuclease digestion. Nucleosomes (25 ng DNA) were treated with RSC (125 ng, ■ and ▴) or no RSC (●), and with ATP (0.5 mM, ▴) or no ATP (● and ■). Micrococcal nuclease was then added in the amounts indicated (u, units) for 5 min at 37°C, followed by 15 μl of 50 mM Tris⋅Cl, pH 8/20 mM EDTA/0.5 M NaCl/1% SDS. DNA was recovered by digestion with proteinase K and phenol-chloroform extraction, followed by electrophoresis in a 7% polyacrylamide gel in TBE buffer. The percentage of the DNA remaining in the band due to the full-length 154-bp fragment was determined with a PhosphorImager. (B) DNase I footprinting. Nucleosomes containing 3′-end-labeled DNA, or the DNA alone, were treated with RSC and ATP or not as indicated, followed by DNase I digestion and gel electrophoresis, all as described (6).
It is noteworthy that addition of ATP to the RSC–nucleosome complex did not alter the overall susceptibility of the DNA to digestion; the amount of uncut DNA remained about the same (Fig. 4). Rather, the addition of ATP affected the cutting pattern, exposing the entire DNA, as noted (8). Such exposure might occur if the nucleosome were to rotate, revealing all sites in the opening of the RSC cavity.
Discussion
The possibility of nucleosome binding in the RSC cavity can be tested by structure determination of a RSC–nucleosome complex. Images of RSC–nucleosome complexes preserved in negative stain were more homogeneous than those of RSC alone, as shown by hierarchical ascendant classification, indicative of reduced conformational variability of RSC due to interaction with the nucleosome. A preliminary 3D reconstruction was consistent with binding in the cavity, but the extra density was less than expected for a nucleosome (data not shown). The deficiency could reflect incomplete occupancy of the cavity (though there was other evidence of significant RSC–nucleosome interaction), or may have resulted from an artifact of staining.
Although binding of a nucleosome in the RSC cavity therefore remains speculative, it is in keeping with biochemical evidence for a 1:1 RSC–nucleosome complex and with a high affinity of RSC for naked DNA. It could also explain the central paradox of RSC action: the DNA in the activated state of the nucleosome is exposed along its entire length, and yet the nucleosome remains intact. The sequestration of the nucleosome in the RSC cavity might permit a partial separation without loss of association of the histones and DNA.
Acknowledgments
This research was supported by National Institutes of Health Grants GM-36659 and AI-21144 (to R.D.K.), and by a Human Frontier Science Program grant (to F.J.A. and R.D.K.).
References
- 1.Lorch Y, LaPointe J W, Kornberg R D. Cell. 1987;49:203–210. doi: 10.1016/0092-8674(87)90561-7. [DOI] [PubMed] [Google Scholar]
- 2.Knezetic J A, Luse D S. Cell. 1986;45:95–104. doi: 10.1016/0092-8674(86)90541-6. [DOI] [PubMed] [Google Scholar]
- 3.Wu J, Grunstein M. Trends Biochem Sci. 2000;25:619–623. doi: 10.1016/s0968-0004(00)01718-7. [DOI] [PubMed] [Google Scholar]
- 4.Becker P B, Horz W. Annu Rev Biochem. 2002;71:247–273. doi: 10.1146/annurev.biochem.71.110601.135400. [DOI] [PubMed] [Google Scholar]
- 5.Sudarsanam P, Iyer V R, Brown P O, Winston F. Proc Natl Acad Sci USA. 2000;97:3364–3369. doi: 10.1073/pnas.050407197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cairns B R, Lorch Y, Li Y, Lacomis L, Erdjument-Bromage H, Tempst P, Laurent B, Kornberg R D. Cell. 1996;87:1249–1260. doi: 10.1016/s0092-8674(00)81820-6. [DOI] [PubMed] [Google Scholar]
- 7.Xue Y, Canman J C, Lee C S, Nie Z, Yang D, Moreno G T, Young M K, Salmon E D, Wang W. Proc Natl Acad Sci USA. 2000;97:13015–13020. doi: 10.1073/pnas.240208597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Imbalzano A N, Kwon H, Green M R, Kingston R E. Nature. 1994;370:481–485. doi: 10.1038/370481a0. [DOI] [PubMed] [Google Scholar]
- 9.Cote J, Quinn J, Workman J L, Peterson C L. Science. 1994;265:53–60. doi: 10.1126/science.8016655. [DOI] [PubMed] [Google Scholar]
- 10.Lorch Y, Zhang M, Kornberg R D. Cell. 1999;96:389–392. doi: 10.1016/s0092-8674(00)80551-6. [DOI] [PubMed] [Google Scholar]
- 11.Whitehouse I, Flaus A, Cairns B R, White M F, Workman J L, Owen-Hughes T. Nature. 1999;400:784–787. doi: 10.1038/23506. [DOI] [PubMed] [Google Scholar]
- 12.Jaskelioff M, Gavin I M, Peterson C L, Logie C. Mol Cell Biol. 2000;20:3058–3068. doi: 10.1128/mcb.20.9.3058-3068.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lorch Y, Zhang M, Kornberg R D. Mol Cell. 2001;7:89–95. doi: 10.1016/s1097-2765(01)00157-5. [DOI] [PubMed] [Google Scholar]
- 14.Frank J, Radermacher M, Penczek P, Zhu J, Li Y, Ladjadj M, Leith A. J Struct Biol. 1996;116:190–199. doi: 10.1006/jsbi.1996.0030. [DOI] [PubMed] [Google Scholar]
- 15.Penczek P, Grassucci R A, Frank J. Ultramicroscopy. 1994;53:251–270. doi: 10.1016/0304-3991(94)90038-8. [DOI] [PubMed] [Google Scholar]
- 16.Frank J. Three-Dimensional Electron Microscopy of Macromolecular Assemblies. New York: Academic; 1996. [Google Scholar]
- 17.Saxton W O, Baumeister W. J Microsc. 1982;127:127–138. doi: 10.1111/j.1365-2818.1982.tb00405.x. [DOI] [PubMed] [Google Scholar]
- 18.van Heel M. Ultramicroscopy. 1987;21:95–100. doi: 10.1016/0304-3991(87)90078-7. [DOI] [PubMed] [Google Scholar]
- 19.Lorch Y, Cairns B R, Zhang M, Kornberg R D. Cell. 1998;94:29–34. doi: 10.1016/s0092-8674(00)81218-0. [DOI] [PubMed] [Google Scholar]
- 20.Luger K, Mader A W, Richmond R K, Sargent D F, Richmond T J. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 21.Jones T A, Zou J Y, Cowan S W, Kjeldgaard Acta Crystallogr A. 1991;47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]