Abstract
Calcineurin (Cn), a Ca2+/calmodulin-dependent Ser/Thr protein phosphatase, is an important participant in signaling pathways that activate T cells. It is the target of the immunosuppressive drugs cyclosporin A (CsA) and FK506. These drugs bind proteins known as cyclophilin (Cyp) and FK506-binding protein, respectively, and the drug–protein complexes in turn inhibit Cn. We report the crystal structure of a Cyp/CsA/Cn ternary complex, determined to a resolution of 3.1 Å. Residues 3–9 of CsA, particularly N-methyl leucines 4 and 6, and Trp-121 of Cyp form a composite surface for interaction with Cn. The hydrophobic interface buries two hydrogen bonds. The structure accounts clearly for the effects of mutations in Cn on CsA-resistance and for the way modifications of CsA alter immunosuppressive activity.
Calcineurin (Cn), also known as protein phosphatase 2B, is a Ca2+/calmodulin-dependent Ser/Thr protein phosphatase. It participates in a number of cellular regulatory pathways, of which the best studied is the activation and nuclear translocation of the transcriptional regulator, nuclear factor of activated T cells (NFAT). Triggered by an increase in intracellular Ca2+ concentration, Cn dephosphorylates NFAT and exposes a nuclear localization signal. The immunosuppressive drugs, cyclosporin A (CsA) and FK506, work by complexing with an “immunophilin” [cyclophilin (Cyp) and FK506-binding protein (FKBP), respectively], in such a way that the drug/immunophilin complex can associate with Cn and inhibit dephosphorylation of NFAT and other substrates.
Cn has two subunits: a catalytic subunit, CnA, and a Ca2+-binding regulatory subunit, CnB. CnA comprises four regions: a catalytic domain (residues 20–340), a CnB-binding segment (residues 349–372), a calmodulin-binding segment (residues 390–414), and a C-terminal autoinhibitory helix (residues 469–486). CnB resembles calmodulin, and like calmodulin, it binds to an extended α-helix on its partner, in this case, CnA (1–3). The CnB-binding segment of CnA then projects as a 5-turn α-helix away from the body of the catalytic domain; binding of CnB renders this helix stable in its projecting conformation (Fig. 1) (2, 3). Calmodulin itself, bearing four Ca2+ ions, binds the segment between the CnB site and the autoinhibitory helix; it is thought to stiffen that segment into an α-helical conformation and thereby pull the autoinhibitory helix away from the catalytic site.
Fig 1.
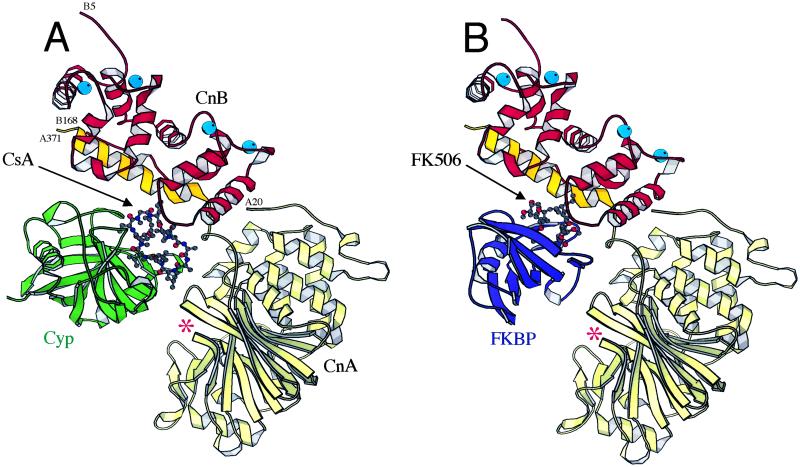
Ribbon diagrams of the Cyp/CsA/Cn ternary complex (A), and the FKBP/FK506/Cn complex (B) (2, 3). CsA and FK506 are shown in ball-and-stick representations; CnA is in yellow (with the CnB-binding segment in dark yellow); CnB, red; Cyp, green; FKBP, blue; and Ca2+ ions, cyan balls. The Cn active-site cleft is indicated with an asterisk.
CsA is a cyclic peptide of 11 amino acid residues (Fig. 2B) (4). Structures of Cyp/CsA binary complexes show that one side of the CsA ring (comprising residues 1–3 and 9–11) fits snugly into a groove on Cyp, leaving the opposite side (the “effector domain”) free to interact with Cn (5–7). Cyp is a cis-trans prolyl isomerase, but its enzymatic activity is distinct from its role in Cn inhibition (8). Thus, CsA appears to have “hijacked” Cyp, to create an intracellular inhibitory complex much larger than CsA itself. The mechanism of inhibition has been inferred from structures of a ternary FKBP/FK506/Cn complex (2, 3), because no Cyp/CsA/Cn crystals have hitherto been obtained. FK506 is a macrocyclic molecule unrelated to CsA. Its cognate immunophilin, FKBP, is, like Cyp, a prolyl isomerase. The ternary-complex structure shows that FK506 is effectively an adaptor that mediates tight association of the two proteins (Fig. 1B). Neither FK506 nor FKBP contacts the Cn active site directly, but the immunophilin is so positioned that protein and polypeptide substrates are unable to gain access to it.
Fig 2.
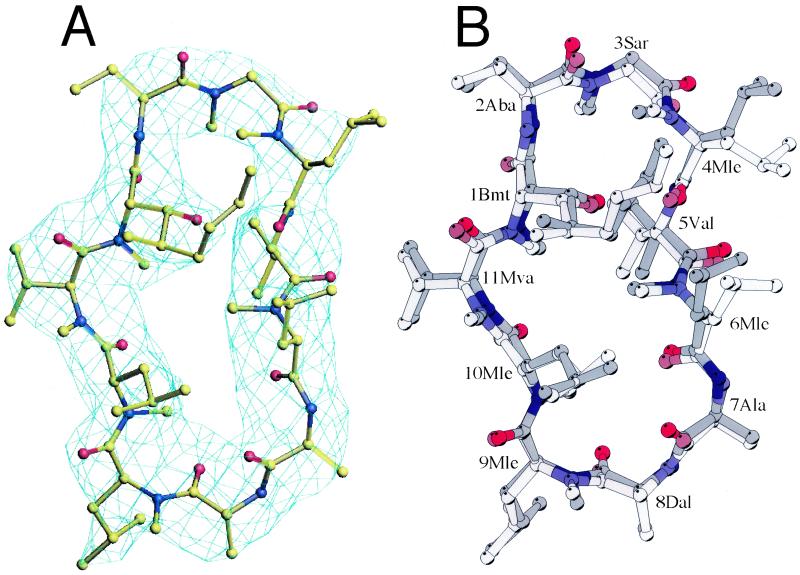
(A) Electron density for CsA, from a 2Fo − Fc map calculated before CsA was added to the model. Contour is at 1 σ. The final CsA model is shown in the density. (B) CsA conformation before and after binding to Cn. Superposition of Cyp-Cn-bound CsA (dark color) onto Cyp-bound CsA (lighter color, PDB code ) based on the least-squares fit of the two Cyp molecules. (Note that there are 10 virtually identical CsA/Cyp complexes in one asymmetric unit in this crystal form, we chose the first one for structural comparison.) The following abbreviations are used for the modified amino acids of CsA. Bmt, 4-[(E)-2-butenyl]-4,N-dimethyl threonine; Aba, α-amino butyric acid; Sar, sarcosine; Ala, l-alanine; Dal, d-alanine; Mle, N-methyl leucine; Mva, N-methyl valine.
We have determined the structure of a Cyp/CsA/Cn ternary complex from crystals obtained by suitably “trimming” the Cn component. As anticipated, CsA and Cyp form a composite interface for tight association with Cn. Residues 3–9 of CsA, the presumed “effector domain,” create a hydrophobic interface with Cn, containing two buried hydrogen bonds. Cn undergoes a modest, hinge-like closure when it binds the inhibitor, but the Cyp/CsA interaction is essentially identical to that seen in the binary complex (5, 6). The overall geometry of the Cyp/CsA/Cn complex resembles that of FKBP/FK506/Cn. Two N-methyl leucine (Mle) residues, CsA4Mle and CsA6Mle, make particularly important interactions with Cn, as predicted from structure-activity relationships.
Materials and Methods
Protein Expression and Purification.
N-terminally His-tagged, full-length human Cnα was expressed in Escherichia coli (with an expression plasmid supplied by J. O. Liu) and purified according to ref. 9, with the modification that neither the pBB131 plasmid (encoding the Saccharomyces cerevisiae myristoyl-CoA:protein N-myristoyltransferase) nor myristic acid was included during expression. The N terminus of the CnB subunit was therefore not myristoylated. An additional step of ion-exchange chromatography (Q-Sepharose HP, Amersham Pharmacia) was included. Human CypA was expressed in E. coli with an expression plasmid supplied by Wesley I. Sundquist, purified as described (10), and concentrated in 10 mM Tris⋅HCl, pH 7.5/50 mM NaCl/1 mM CaCl2.
Complex Formation and Limited Trypsinization.
The ternary complex of Cyp/CsA/Cn was made by mixing Cn with a slight excess of Cyp and CsA in 10 mM Tris⋅HCl, pH 7.5/50 mM NaCl/5 mM CaCl2. We were unable to obtain crystals of the intact complex, despite extensive effort. We therefore chose to remove flexible segments of Cn in the ternary complex by limited proteolysis (11). For preparative proteolysis, 20 mg of the ternary complex at ≈1 mg/ml in 10 mM Tris⋅HCl/50 mM NaCl/5 mM CaCl2, pH 7.5, were mixed with 120 μl of 10 mg/ml trypsin in 1 mM HCl. The solution was incubated on ice for 3.5 h. PMSF dissolved in isopropanol was then added to inactivate the trypsin. This procedure resulted in a truncated CnA and intact CnB and Cyp, as judged by SDS/PAGE. Mass spectrometry combined with enzymatic digestion confirmed that the truncated CnA contains residues 20–392; thus the N-terminal residues (including the His-tag), the calmodulin-binding domain, and the autoinhibitory helix were all removed by trypsin. The ternary complex was then purified by gel filtration chromatography (Superdex S-200, Amersham Pharmacia) and concentrated in 10 mM Tris⋅HCl/50 mM NaCl/1 mM CaCl2, pH 7.5.
Crystallization and Data Collection.
The ternary complex was crystallized by a microbatch method. A total of 30 μl of the ternary complex at 5 mg/ml were mixed with 30 μl of 150 mM Na3Citr/10 mM KH2PO4/24% PEG4000/30% glycerol, pH 4.6, centrifuged at 16,000 × g at 4°C for 10 min and sealed as 15 μl drops at 19°C. Crystals appeared in 2 days and continued to grow for another week to a maximum size of 0.1 × 0.2 × 0.5 mm3. Crystals were rapidly frozen in cryo-loops (Hampton Research, Riverside, CA) by direct dipping into liquid nitrogen. X-ray diffraction data were collected at 100 K at the Cornell High Energy Synchrotron Source (CHESS) F-1 beamline, using a Quantum-4 charge coupled device detector (Area Detector Systems, Poway, CA) and processed with HKL2000 (HKL Research, Charlottesville, VA) (12) and the CCP4 suite (13). The crystals belong to space group P212121, with unit cell dimensions a = 64.95 Å, b = 108.33 Å, and c = 112.84 Å. There is one ternary complex in the asymmetric unit, corresponding to a solvent content of 46%. Statistics are given in Table 1.
Table 1.
Summary of crystallographic data
| Resolution range | 15–3.1Å |
| Total no. of reflections | 14,195 |
| Completeness, % | 95.6 (81.7) |
| Rsym, % | 7.7 (37.7) |
| Average 〈I/σ〉 | 19.3 (3.3) |
| Mean redundancy | 5.3 |
| R factor, % | 25.5 (40.9) |
| Free R factor, % | 30.0 (45.0) |
| Number of non-H atoms per asymmetric unit | |
| CnA (residues 20-371) | 2,861 |
| CnB (residues 5-168) | 1,311 |
| CyP (residues 1-164) | 1,256 |
| CsA (residues 1-11) | 85 |
Solvent (1 PO , 4 Ca2+) , 4 Ca2+) |
9 |
| rmsd bond lengths, Å | 0.01 |
| rmsd bond angles, ° | 1.5 |
| Main-chain torsion angles, % | |
| Preferred | 82.2 |
| Allowed | 16.6 |
| Generously allowed | 0.7 |
| Disallowed | 0.5 |
The numbers in parentheses are for the outer shell (3.21-3.10 Å).
Rsym = ΣhΣi|I/(h) − I(h)i|/ΣhΣiI(h)iwhere I(h) is the measured mean intensity of the i observations of symmetry related reflections of h.
The free R factor of 1,052 randomly selected reflections (15.0-3.1 Å) after final round of refinement.
From cns; rmsd, rms deviation from ideal values.
Structure Determination and Refinement.
The structure of the ternary complex was determined by molecular replacement with cns (14), using human Cn (PDB ID ) and human Cyp (PDB ID ) as search models. Cn was first located by cross-rotation and translation searches. The orientation and position of Cyp were then found by fixing Cn and searching for Cyp. After rigid-body refinement, there was clear electron density for CsA in both 2Fo − Fc and Fo − Fc maps calculated with Cn and Cyp only. CsA was built into electron density when Rfree had dropped below 35% (Fig. 2A). The structure was improved by iterative rounds of manual rebuilding by using o (15) and refinement with energy-restrained conjugate gradient minimization in cns. Two rounds of grouped B-factor refinement were done at the end. A maximum likelihood amplitude target, with bulk solvent and anisotropic temperature factor corrections, was applied throughout the refinement, with Rfree as a monitor. Electron density maps were calculated in cns with solvent flipping and σA weighting. The R factor and Rfree for the final model are 25.5% and 30.0%, respectively. The final model contains CnA (residues 20–371), CnB (residues 5–168), Cyp (residues 1–164), CsA, 4 Ca2+, and one phosphate ion. Some residues have poor side-chain density, due either to their flexibility or to the relatively low resolution at which the structure was determined. The area of buried surface was calculated in cns with a probe radius of 1.4 Å. Refinement statistics are presented in Table 1. Figures were produced with o and molscript (17).
Results and Discussion
Contacts Between Cyp/CsA and Cn.
Cn, CsA and Cyp form a precisely configured 1:1:1 ternary complex. The Cyp/CsA binary complex, essentially unaltered from its conformation in various crystal structures (5, 6), lies at the base of the CnB-binding helix of CnA (Fig. 1A). It contacts both chains of Cn, in two distinct patches with approximately 1,870 Å2 of total buried surface.
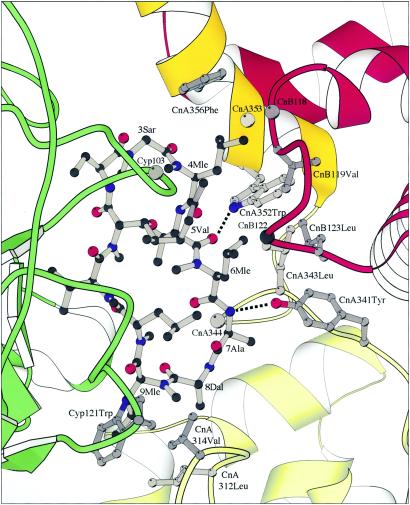
The primary interface includes residues 3–7 of CsA and a hydrophobic surface to which CnA343Leu, CnA344Pro, CnA352Trp, CnA353Ser, CnA356Phe, CnB118Met, CnB119Val, CnB122Asn, and CnB123Val are important contributors (Fig. 3). The side chain of CsA4Mle fits into a pocket defined by the aromatic rings of CnA352Trp and CnA356Phe on two of its sides, by the backbone and Cβ atoms of CnB118Met and CnB119Val on a third side, and by the backbone and Cβ atoms of CnA353Ser at its base; this same pocket receives the C21-allyl group of FK506 in the FKBP/FK506/Cn ternary complex (2). The side chain of CsA6Mle lies against the opposite face of the CnA352Trp indole ring from that of CsA4Mle, in contact with CnA341Tyr, CnA343Leu, and CnB123Leu. The side chains of CsA5Val and CsA7Ala and the Cα and carbonyl of CsA3Sar also contribute through van der Waals contacts with CnA344Pro and CnA356Phe, respectively. In the midst of this extended hydrophobic interface are two hydrogen bonds—one from the indole NH of CnA352Trp to the carbonyl of CsA5Val and the other from the amide NH of CsA7Ala to the phenolic OH of CnA341Tyr. The dependence of these polar interactions on distance and directionality probably adds to the specificity and precision of the fit, reducing the opportunities for other small adjustments in response to modifications of CsA or mutations in Cn. The only interactions between Cyp and Cn in the primary interface are a hydrogen bond from CnA359Glu, presumably protonated at the pH of crystallization, to the carbonyl of Cyp73Thr and a potential hydrogen bond, rather long in the current coordinate set, from Nδ of CnB122Asn to the carbonyl of Cyp103Ala. The asparagine side chain, which extends over one face of CsA (the proximal face in Figs. 2–3), also contributes a hydrophobic contact for CsA6Mle, and it forms, together with the methyl leucine, a nonpolar pocket for the 2-butenyl substituent on CsA1Bmt.
Fig 3.
Details of the interaction between CsA and Cn. CsA, ball-and-stick; CnA, yellow; CnB, red; Cyp, green. Some Cn and Cyp residues involved in CsA binding are represented as ball-and-sticks. The backbone positions of other residues mentioned in the text (CnA344Pro, CnA353Ser, CnB118Met, CnB122Asn, Cyp103Ala) are shown as gray spheres. There are two hydrophobic surfaces for CsA binding, one formed by residues from CnA and CnB (upper right) and the other formed jointly by CnA and Cyp (lower left). The two hydrogen bonds between CsA and CnA are indicated as broken lines.
The secondary interface includes Cyp121Trp and residues 8 and 9 of CsA on the surface of the composite inhibitor and CnA312Leu, and CnA314Val on Cn (Fig. 3). There is an open, presumably water-filled cavity between the primary and secondary interfaces, where the CsA rings “turns the corner” between residues 7 and 8. This cavity is large enough to accommodate the heterocyclic moiety introduced into CsA by Alberg and Schreiber to form a tricyclic variant (TCsA) (18).
Conformational Changes in Cn and in Cyp/CsA.
Comparison of the structure of Cn in our complex with those of free Cn and Cn bound with FKBP/FK506 shows that the only significant conformational change is in the hinge angle between the catalytic domain and the CnB-binding helix. The interdomain angle closes by about 10°, through flexion of a hinge region (residues CnA341 to CnA348). A slightly smaller domain closure is seen in the ternary complex with FKBP/FK506 (Fig. 1B) (2). Side-chain conformations in our structure are essentially the same as those in uncomplexed Cn, even in the contact regions and in the catalytic cleft. A phosphate ion lies close to CnA122Arg, just as in FKBP/FK506/Cn. There is no electron density for Zn2+ or Fe3+ at the active center of CnA. We do not believe that binding of Cyp/CsA has expelled these ions, but rather that the low pH (4.6) of crystallization has led to their loss through displacement by protons. The ions are present in crystals of free Cn, purified by the same protocol but crystallized at pH 7.5 (data not shown).
The only significant differences between CsA in binary complexes with Cyp and in the ternary complex described here are in the conformations of the 4Mle and 6Mle side-chains (Fig. 2B). These residues project from the effector edge of the macrocycle. Unconstrained by Cyp in binary complexes, they form the core of the primary interface with Cn.
Mechanism of Cn Inhibition.
The position of Cyp/CsA requires that it displace the autoinhibitory helix and calmodulin-binding segment of Cn. Cyp/CsA does not, however, substitute for the autoinhibitory helix by contacting the Cn active site directly. Instead, it lies over the active site in such a way as to block access to catalytic residues by a protein substrate, just as for inhibition by FKBP/FK506 (Fig. 1). Kinetic studies indeed show that the inhibitory mechanism is noncompetitive (19).
Structure-Activity Relationships.
The structure is consistent with the effects of various mutations in Cn on its susceptibility to inhibition by Cyp/CsA and with relevant structure-activity relationships for the drug.
Substitutions in the loop of CnB that contains residues 118 to 125 (the so-called “latch region”) confer varying degrees of resistance to CsA and FK506, when assayed by the capacity of these drugs (with their cognate immunophilin) to inhibit phosphatase activity in vitro and to suppress transcriptional activation in culture T cells (20). This loop is an important part of the primary interface in the Cyp/CsA/Cn complex (Fig. 3), as well as a critical element in the corresponding ternary complex with FKBP and FK506 (2). Two mutations in CnA, one in the primary interface (Tyr341Phe) and the other in the secondary interface (Val314Arg) also confer resistance to CsA (but not to FK506) (21). These effects are fully consistent with observed contacts (Fig. 3). Conferral of resistance by Tyr341Phe demonstrates that the buried hydrogen bond between the phenolic oxygen of CnA341Tyr and the main-chain amide group of CsA7Ala is indeed an energetically significant contact (Fig. 3).
Extensive studies with modified CsA point to the importance of N-methyl leucines 4 and 6 in the interaction with Cn (4, 22). Changing these residues to N-methyl valine decreases immunosuppressive activity, whereas changing 4Mle to Me[4(s)-Me]norLeu increases it, presumably by affecting affinity for Cn. The negative effects of a β-branched side chain and the positive effect of changing one of the 4Mle methyl groups to ethyl are easily reconciled with the geometry of the primary interface and the properties of the 4Mle pocket.
Conclusion
CsA and FK506 have famously resisted substantial efforts to improve on them as drugs. The structure of the Cyp/CsA/Cn complex, although it brings no major surprises, does suggest potential strategies for renewing such a quest. In particular, alteration of the butenyl substituent of CsA1Bmt might improve its fit against the hydrophobic primary-contact surface of Cn, and modification of TCsA (18) to include a hydrogen-bond donor for the carbonyl of CnA342Trp or that of CnA312Leu might enhance affinity for Cn while retaining the desired rigidification of CsA. Comparison with the FKBP/FK506/Cn complex demonstrates a remarkable similarity in the way two unrelated molecules recruit two rather different small proteins (although both are prolyl isomerases) to create a bulky intracellular inhibitor out of a much smaller, freely diffusible drug.
Acknowledgments
We thank Dr. Jun O. Liu (The Johns Hopkins University, Baltimore) for the full-length human calcineurin α expression plasmid and Dr. Wesley I. Sundquist (University of Utah, Salt Lake City) for the human cyclophilin expression plasmid. Mass spectrometric analysis was performed by using the Taplin Mass Spectrometry Facility at Harvard Medical School. X-ray diffraction data were collected at the Cornell High Energy Synchrotron Source (CHESS), using the Macromolecular Diffraction at CHESS (MacCHESS) facility. We are grateful to Drs. Christopher T. Walsh and Anjana Rao for careful reading of the manuscript. This work is supported by a National Institute of General Medical Sciences Program Project grant (to S.C.H.). S.C.H. is an investigator in the Howard Hughes Medical Institute.
Abbreviations
Cn, calcineurin
CnA, Cn A subunit
CnB, Cn B subunit
CsA, cyclosporin A
Cyp, cyclophilin
FKBP, FK506-binding protein
NFAT, nuclear factor of activated T cells
Mle, N-methyl leucine
Data deposition: Coordinates and structure factor amplitudes of the Cyp/CsA/Cn ternary complex crystal structure have been deposited in the Protein Data Bank, www.rcsb.org (PDB ID ).
References
- 1.Aramburu J., Rao, A. & Klee, C. B. (2000) Curr. Top. Cell. Regul. 36, 237-295. [DOI] [PubMed] [Google Scholar]
- 2.Griffith J. P., Kim, J. L., Kim, E. E., Sintchak, M. D., Thomson, J. A., Fitzgibbon, M. J., Fleming, M. A., Caron, P. R., Hsiao, K. & Navia, M. A. (1995) Cell 82, 507-522. [DOI] [PubMed] [Google Scholar]
- 3.Kissinger C. R., Parge, H. E., Knighton, D. R., Lewis, C. T., Pelletier, L. A., Tempczyk, A., Kalish, V. J., Tucker, K. D., Showalter, R. E., Moomaw, E. W., et al. (1995) Nature 378, 641-644. [DOI] [PubMed] [Google Scholar]
- 4.Fliri H., Baumann, G., Enz, A., Kallen, J., Luyten, M., Mikol, V., Movva, R., Quesniaux, V., Schreier, M., Walkinshaw, M., et al. (1993) Ann. N.Y. Acad. Sci. 696, 47-53. [PubMed] [Google Scholar]
- 5.Mikol V., Kallen, J., Pflugl, G. & Walkinshaw, M. D. (1993) J. Mol. Biol. 234, 1119-1130. [DOI] [PubMed] [Google Scholar]
- 6.Pflugl G., Kallen, J., Schirmer, T., Jansonius, J. N., Zurini, M. G. & Walkinshaw, M. D. (1993) Nature 361, 91-94. [DOI] [PubMed] [Google Scholar]
- 7.Theriault Y., Logan, T. M., Meadows, R., Yu, L., Olejniczak, E. T., Holzman, T. F., Simmer, R. L. & Fesik, S. W. (1993) Nature 361, 88-91. [DOI] [PubMed] [Google Scholar]
- 8.Zydowsky L. D., Etzkorn, F. A., Chang, H. Y., Ferguson, S. B., Stolz, L. A., Ho, S. I. & Walsh, C. T. (1992) Protein Sci. 1, 1092-1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mondragon A., Griffith, E. C., Sun, L., Xiong, F., Armstrong, C. & Liu, J. O. (1997) Biochemistry 36, 4934-4942. [DOI] [PubMed] [Google Scholar]
- 10.Liu J., Albers, M. W., Chen, C. M., Schreiber, S. L. & Walsh, C. T. (1990) Proc. Natl. Acad. Sci. USA 87, 2304-2308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Swanson S. K., Born, T., Zydowsky, L. D., Cho, H., Chang, H. Y., Walsh, C. T. & Rusnak, F. (1992) Proc. Natl. Acad. Sci. USA 89, 3741-3745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Otwinowski Z. & Minor, W. (1997) Methods Enzymol. 276, 307-326. [DOI] [PubMed] [Google Scholar]
- 13.Collaborative Computational Project Number 4 (1994) Acta Crystallogr. D 50, 760-763.15299374 [Google Scholar]
- 14.Brünger A. T., Adams, P. D., Clore, G. M., DeLano, W. L., Gros, P., Grosse-Kunstleve, R. W., Jiang, J. S., Kuszewski, J., Nilges, M., Pannu, N. S., et al. (1998) Acta Crystallogr. D 54, 905-921. [DOI] [PubMed] [Google Scholar]
- 15.Jones T. A., Zou, J. Y., Cowan, S. W. & Kjeldgaard, M. (1991) Acta Crystallogr. A 47, 110-119. [DOI] [PubMed] [Google Scholar]
- 16.Laskowski R. A., MacArthur, M. W., Moss, D. S. & Thornton, J. M. (1993) J. Appl. Crystallogr. 26, 283-291. [Google Scholar]
- 17.Kraulis P. J. (1991) J. Appl. Crystallogr. 24, 946-950. [Google Scholar]
- 18.Alberg D. G. & Schreiber, S. L. (1993) Science 262, 248-250. [DOI] [PubMed] [Google Scholar]
- 19.Etzkorn F. A., Chang, Z. Y., Stolz, L. A. & Walsh, C. T. (1994) Biochemistry 33, 2380-2388. [DOI] [PubMed] [Google Scholar]
- 20.Milan D., Griffith, J., Su, M., Price, E. R. & McKeon, F. (1994) Cell 79, 437-447. [DOI] [PubMed] [Google Scholar]
- 21.Zhu D., Cardenas, M. E. & Heitman, J. (1996) Mol. Pharmacol. 50, 506-511. [PubMed] [Google Scholar]
- 22.Papageorgiou C., Florineth, A. & Mikol, V. (1994) J. Med. Chem. 37, 3674-3676. [DOI] [PubMed] [Google Scholar]