Abstract
Intermolecular nuclear Overhauser effects (NOEs) between the integral outer membrane protein OmpX from Escherichia coli and dihexanoylphosphatidylcholine (DHPC) provided a detailed description of protein–detergent interactions. The NOEs were measured in 3D 15N- and 13C-resolved [1H,1H]-NOESY spectra recorded with selectively methyl-protonated and otherwise uniformly 2H,13C,15N-labeled OmpX in micelles of DHPC at natural isotope abundance. In these mixed micelles the NMR structure of OmpX consists of an eight-stranded antiparallel β-barrel. The OmpX surface area covered with intermolecular NOEs to the DHPC hydrophobic tails forms a continuous cylinder jacket of approximately 28 Å in height, which is centered about the middle of the long axis through the β-barrel. In addition, some intermolecular NOEs with methyl groups of the DHPC polar head were identified along both boundaries of this cylinder jacket. The experimental data suggest that the hydrophobic surface areas of OmpX are covered with a monolayer of DHPC molecules, which appears to mimic quite faithfully the embedding of the β-barrel in a double-layer lipid membrane.
Keywords: NOE‖detergents‖solvation of membrane proteins‖intermolecular NOEs
Membrane proteins constitute about one-third of all proteins encoded by the genomes of living organisms. However, they are strongly underrepresented in the database of 3D protein structures, which reflects the big challenge presented by this class of proteins to structural biologists. Apart from difficulties related to high-yield expression, purification, and refolding, considerable additional effort is usually required for finding either suitable crystallization conditions or solution conditions for NMR measurements, whereby detergent micelles, bicelles, lipid bilayers, or lipid vesicles are commonly used as a replacement of the natural membrane environment (1–4). For solution NMR studies, the overall size of the mixed protein/detergent/lipid supramolecular structure is an important factor, in addition to the preservation of the natural structure and function of the protein, and this combined demand has most promisingly been met with protein–detergent micelles. The use of transverse relaxation-optimized spectroscopy (TROSY) (5–9) and advanced isotope labeling strategies (10) has now actually opened avenues for NMR structure determination of integral membrane proteins reconstituted in detergent micelles (1, 11–13). In this context, the nature of the protein–detergent interactions in mixed micelles is of keen interest.
Various different schemes for the interaction between detergent molecules and membrane proteins have been suggested (14, 15). Based on lipid binding quantification by chromatographic methods and model calculations on sarcoplasmic reticulum Ca2+-ATPase, Møller and le Maire (14) proposed that formation of a monolayer rather than a bilayer type of interaction is the basis for solubilization of membrane proteins by detergents. With reference to unfolding studies of the Escherichia coli outer membrane protein OmpA in micelles formed by detergent molecules with different chain lengths, Kleinschmidt et al. (15) advanced the idea that a monolayer or a prolate ellipsoid arrangement of detergent molecules on the hydrophobic protein surface prevails in the mixed micelles. In this paper, we use solution NMR spectroscopy for further experimental studies of membrane protein–detergent interactions.
The potentialities of high-resolution NMR spectroscopy for studies of the architecture of mixed polypeptide–detergent micelles have long been recognized (16–19), and the technique also has recently been applied with membrane protein fragments (20–22). Here, we present a study of the intact integral outer membrane protein OmpX from E. coli in mixed micelles with dihexanoylphosphatidylcholine (DHPC) (11, 12, 23). Results on the solvation of OmpX by DHPC were obtained using 3D 15N- and 13C-resolved [1H,1H]-NOESY spectra recorded with selectively methyl-protonated and otherwise uniformly 2H,13C,15N-labeled OmpX, [u-2H,13C,15N/L,V,Iδ1-13CH3]-OmpX, in mixed micelles with DHPC at natural isotope abundance.
Materials and Methods
Production of OmpX and NMR Sample Preparation.
OmpX selectively methyl-protonated at Val, Leu, and Ile-δ1, and otherwise uniformly 2H,13C,15N-labeled, [u-2H,13C,15N/L,V,Iδ1-13CH3]-OmpX, was prepared as reported in ref. 23. Protein isolation, purification, refolding, and reconstitution into detergent micelles was performed as described (11). The NMR sample contained 2 mM OmpX, 20 mM phosphate at pH 6.5, 100 mM NaCl, 0.05% NaN3, and 200 mM DHPC at natural isotope abundance.
NMR Spectroscopy.
All spectra were measured at 30°C on a Bruker Avance DRX-800 spectrometer equipped with four radio-frequency channels and a 1H-{13C,15N}-triple resonance probe head with an actively shielded z-gradient coil. The following parameters were used. Three-dimensional 15N-resolved [1H,1H]-NOESY spectrum: time domain data size 150(t1) × 40(t2) × 1,024(t3) complex points; maximal acquisition times t1max(1H) = 14.4 ms, t2max(15N) = 16.4 ms, and t3max(1HN) = 98.3 ms; τm = 200 ms. Three-dimensional 13C-resolved [1H,1H]-NOESY spectrum: time domain data size 175(t1) × 40(t2) × 1,024(t3) complex points; t1max(1H) = 18.2 ms, t2max(13C) = 9.9 ms, and t3max(1H) = 106.5 ms; τm = 200 ms. The spectra were transformed with the program prosa (24) and analyzed with the program xeasy (25). Chemical shifts were referenced to 2,2-dimethyl-2-silapentane-5-sulfonate sodium salt (DSS) (26).
Results
The NMR spectral analysis presented here was based on the previous sequence-specific resonance assignments of the OmpX amide groups (11) and the Val, Leu, and Ile-δ1 methyl groups (23), and on the availability of the NMR structure of OmpX in DHPC micelles (11, 12). Three-dimensional 15N- and 13C-resolved [1H,1H]-NOESY spectra were recorded with [u-2H,13C,15N/L,V,Iδ1-13CH3]-OmpX/DHPC in micelles with DHPC at natural isotope abundance. In both experiments, a mixing time of τm = 200 ms was selected, with the primary purpose of obtaining additional nuclear Overhauser effect (NOE) distance constraints for the refinement of the OmpX structure in the micelles (data not shown). The high extent of deuteration of OmpX enabled the observation of intramolecular NOEs between protons separated by up to ∼7 Å in the 3D protein structure (11, 12). Correspondingly, at a NOESY mixing time of 200 ms, spin-diffusion within the protein was not a limiting factor either for the structure determination or for the present solvation study. The NOE data were analyzed along similar lines as in the NMR studies of protein hydration (27). All DHPC molecules were considered to form a continuous hydrophobic phase surrounding the protein, where individual DHPC molecules cannot be distinguished, and the NOEs were assigned by reference to the sequence-specific assignments of the OmpX resonances (11, 23). The NOEs then provide information about the distribution of DHPC on the protein surface, similar to the result of a hydration study by NMR (27).
The orientation of the detergent molecules relative to the protein surface can also be determined with the same experiments. This additional information is based on the resonance assignments for DHPC, which were obtained with [1H,1H]- and [1H,13C]-correlation experiments. The 1H NMR spectrum of DHPC contains the resonance of the lipophilic −CH3 groups at the chemical shift of δ = 0.78 ppm, the penultimate and antepenultimate −CH2− groups are overlapped at δ = 1.22 ppm, the remaining hexanoyl −CH2− groups are at δ = 1.50 ppm and 2.21 ppm, the choline methyl resonance is at δ = 3.16 ppm (Fig. 1 c and d), and all other methylene proton resonances are at lower field than 3.5 ppm (not shown). Because of the large size of the mixed OmpX/DHPC micelles, efficient spin-diffusion links the methyl and methylene protons of the hydrophobic tails of DHPC. It is readily apparent in the spectra of Fig. 1 that NOEs involving the hexanoyl methyl groups are accompanied throughout by NOEs to some or all of the methylene protons in the lipophilic tails. In contrast, there is no spin-diffusion between the hydrophobic end of DHPC and the choline methyl groups. As a result, NOEs to the two chain ends (Fig. 1d) can unambiguously be distinguished, so that the orientation of the DHPC molecules relative to the protein surface can be determined.
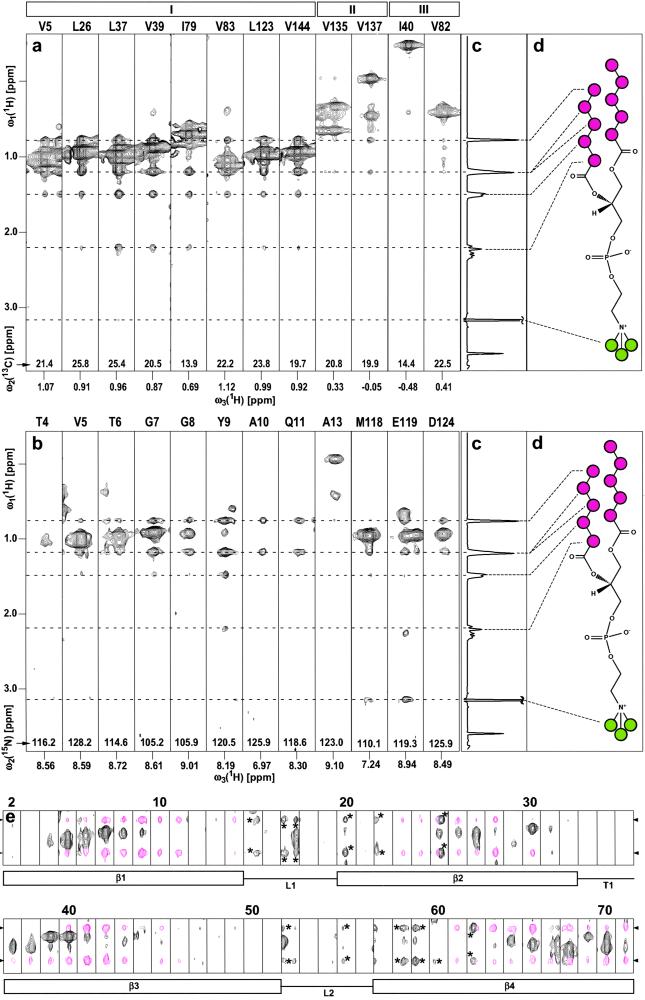
Figure 1.
(a) Selection of ω1(1H)/ω3(1H) strips from an 800-MHz 3D 13C-resolved [1H,1H]-NOESY spectrum measured with a sample of [u-2H,13C,15N/L,V,Iδ1-13CH3]-OmpX (23) in protonated DHPC micelles at 30°C (see Materials and Methods for details of the experimental set-up). The strips were taken at the 13C chemical shifts of the methyl groups for the residues indicated at the top, and they are centered about the methyl proton chemical shifts. The roman numbers I–III at the top denote different locations of the selectively protonated Val, Leu, and Ile(δ1) methyl groups relative to the surface of the NMR structure of OmpX/DHPC (see text). (b) Selection of ω1(1H)/ω3(1H) strips from an 800-MHz 3D 15N-resolved [1H,1H]-NOESY spectrum measured at 30°C with the same sample as in a. The strips were taken at the 15N chemical shifts of the residues indicated at the top and are centered about the respective amide proton chemical shifts. (c) One-dimensional 1H NMR spectrum of DHPC, measured with the same sample and the same experimental conditions as the spectra in a and b. (d) Chemical structure of DHPC. The CHn moieties of interest in this study are color-coded, with magenta circles indicating the CHn groups of the hydrophobic tails, and green circles identifying the polar head methyls of DHPC. The positions of the signals arising from the hydrophobic end (−CH3, δ = 0.78 ppm; penultimate and antepenultimate −CH2−, δ = 1.22 ppm; remaining −CH2− at δ = 1.50 and 2.21 ppm) and the choline N+-bound methyls (δ = 3.16 ppm) are marked with broken lines. (e) ω1(1H)/ω3(1H) strips for the polypeptide segment of residues 2–71 (same data set as in b), showing NOE cross peaks between backbone amide protons of the protein and the hydrophobic end groups of the DHPC. The chemical shifts of the DHPC protons shown here are indicated with arrowheads on the left and right. The NOE cross peaks of interest are colored magenta, and peaks labeled with asterisks represent tails of signals assigned in neighboring planes along ω2(15N). Below the strips, the secondary structure elements in this polypeptide segment are indicated, with β, L, and T standing for β-strand, loop, and turn, respectively.
Intermolecular NOEs Between OmpX Methyl Protons and the Hydrophobic Tails of DHPC.
Using the previously determined sequence-specific assignment of the protonated methyl groups (23), DHPC–methyl intermolecular NOEs could be assigned in a 3D 13C-resolved [1H,1H]-NOESY spectrum (Fig. 1a). From the total of 19 Val, Leu, and Ile residues in the 3D structure of OmpX (11, 28), 13 residues have their methyl groups located on the hydrophobic surface of the β-barrel, and they all show strong NOEs to the hydrophobic ends of DHPC (Fig. 1a, residue group I). The three residues Ile-132, Val-135, and Val-137 are located in the extracellular loop L4 (11, 28), which presumably lies on the surface of the membrane in the natural OmpX environment. In the DHPC micelles, these methyl groups are apparently also somewhat separated from the lipid phase, because they display only weak NOEs to the hydrophobic ends of DHPC (II in Fig. 1a). The residues Ile-40, Val-82, and Ile-141 have the methyl groups pointing toward the center of the β-barrel, and these core residues show no, or at most very weak, NOEs to DHPC (III in Fig. 1a).
Intermolecular NOEs Between OmpX Amide Protons and the Hydrophobic Tails of DHPC.
Similar to the data in Fig. 1a, intermolecular NOEs between amide protons of OmpX and the hydrophobic ends of DHPC were assigned in a 3D 15N-resolved [1H,1H]-NOESY spectrum (Fig. 1b). The intensities of these intermolecular NOEs are largest for residues located centrally on the barrel surface and are decreased toward the edges of the barrel surface. No amide proton–detergent NOEs were detected in the polypeptide segments corresponding to the loops and turns (11, 28). This finding is documented in Fig. 1e with the amide proton–detergent NOE data for the polypeptide segment 2–71, which comprises four β-strands (β1–β4), two loops (L1 and L2), and one tight turn (T1). Note that all of the apparent peaks in the strips attributed to loops or turns are “tails” from peaks located in neighboring planes along the ω2(15N)-dimension (these peaks have been marked with asterisks in Fig. 1e).
Intermolecular NOEs Between OmpX Amide Protons and the Polar Head Methyl Groups of DHPC.
In the 3D 15N-resolved [1H,1H]-NOESY spectrum, intermolecular NOEs were identified from the N-bound methyls of the DHPC choline group at δ = 3.16 ppm (Fig. 1 c and d) to the amide backbone protons of Met-118 and Glu-119 (Fig. 1b), and to the indole protons of Trp-76 and Trp-140 (data not shown).
Discussion
Protein Surface Structure and Detergent Interactions.
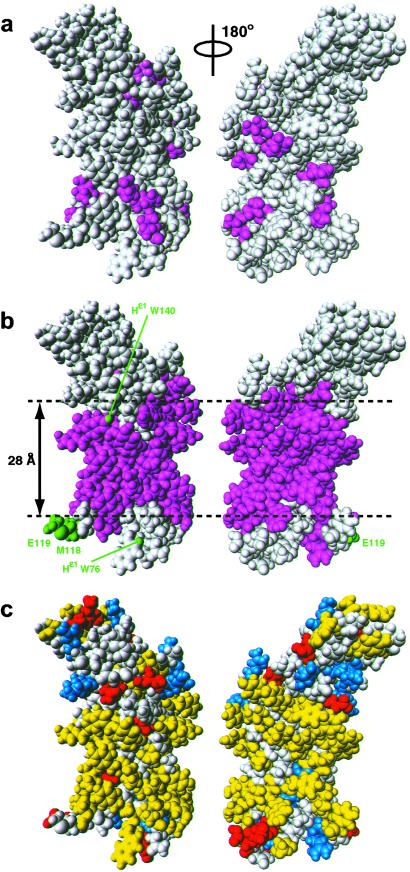
As a platform for rationalizing the experimental data of Fig. 1, Fig. 2 compares the distribution of intermolecular NOEs with DHPC on the surface of the NMR solution structure of OmpX (refs. 11 and 12; Fig. 2 a and b) with the surface charge distribution (Fig. 2c). The key data come from the 3D 15N-resolved [1H,1H]-NOESY spectrum, which shows that the intermolecular NOEs between the DHPC hydrophobic tails and the amide protons cover the surface of OmpX over a range of approximately 28 Å centered about the middle of the β-barrel (magenta area in Fig. 2b). This surface region coincides closely with the hydrophobic surface area of OmpX (Fig. 2c). The observation that close contacts with the hydrophobic end of DHPC molecules are confined to the central surface of the OmpX β-barrel is nicely confirmed by the methyl–DHPC NOEs in the 3D 13C-resolved [1H,1H]-NOESY spectrum (Fig. 1a), because all of the protonated surface methyl groups in [u-2H,13C,15N/L,V,Iδ1-13CH3]-OmpX are located within the confines of or immediately adjacent to the cylindrical shell defined by the DHPC–amide proton NOEs (Fig. 2 a and b).
Figure 2.
Space-filling all-atom presentations affording views of the surface of the NMR structure of OmpX in DHPC micelles (11, 12). (a) Val, Leu, and Ile residues that showed NOEs in a 3D 13C-resolved [1H,1H]-NOESY spectrum between their methyl protons and the hydrophobic tails of DHPC are colored magenta (groups I and II in Fig. 1a). (b) The residues that showed NOEs between the backbone amide proton and the hydrophobic tails of DHPC in a 3D 15N-resolved [1H,1H]-NOESY spectrum are colored magenta. Residues that had NOEs between the backbone amide proton and the polar head methyls of DHPC, and Trp indole protons with NOEs are colored green and labeled with green lettering. Gray residues in a and b did not show intermolecular NOEs to DHPC. The horizontal broken lines indicate the boundaries between the central hydrophobic surface and the peripheral hydrophilic surface areas of OmpX, and the approximate height of the hydrophobic cylinder jacket is indicated. (c) Distribution of hydrophobic and charged residues on the surface of OmpX in DHPC micelles. Hydrophobic residues are yellow, positively charged and negatively charged residues are blue and red, respectively, and polar residues are gray. The figure has been prepared with the program molmol (36).
The intermolecular NOEs of the −N+(CH3)3 moieties of DHPC with the amide protons of Met-118 and Glu-119 from OmpX could be rationalized by electrostatic interactions between the positively charged nitrogen atom in the choline group and the negatively charged Glu side chain. Along similar lines, the NOEs between the polar head methyls of DHPC and the indole protons of Trp-76 and Trp-140 may reflect close approach of these groups due to polar interactions between the choline head group and the indole rings, which may also include hydrogen bonding interactions. Hydrogen bonds involving tryptophan indole groups have previously been proposed to play an important role in aligning membrane proteins with respect to the bilayer plane of biological membranes for optimal functional activity (29, 30). The present observation of a small number of highly selective NOEs between OmpX and DHPC polar head moieties contrasts with the uniform, continuous area of lipophilic interactions (Fig. 2b). These amino acid type-specific interactions might also contribute to the stability of the protein–detergent micelles, and their presence offers a rationale for the observation that membrane proteins may behave widely differently in solutions with different detergents (2, 4).
Modeling DHPC–OmpX Interactions in Micelles.
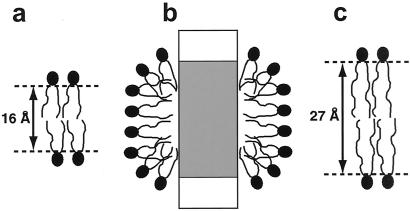
The length of the hydrophobic chains of DHPC in the extended conformation is about 8 Å, which would give rise to a bilayer with a hydrophobic core diameter of about 16 Å (Fig. 3a). Formation of a torus-like bilayer around the protein therefore seems improbable, because there would be a discrepancy of about 10 Å between the bilayer thickness (Fig. 3a) and the height of the experimentally determined hydrophobic contact area on the OmpX surface (Fig. 2b). Therefore, an arrangement of DHPC molecules on the hydrophobic protein surface as a distorted monolayer, with the polar head groups forming the surface of a prolate ellipsoid (Fig. 3b), seems more probable. In this arrangement, the detergent molecules are oriented perpendicular to the protein surface, forming a cylindrical belt around the hydrophobic surface (Fig. 2c). With this interpretation, the experimental results presented in this paper would be in line with previous theoretical calculations and experimental observations on the embedding of other membrane proteins in detergent micelles (14, 15).
Figure 3.
Three schematic drawings of lipid bilayers and a mixed protein–lipid micelle. (a) Hypothetical DHPC bilayer with indication of the thickness of the lipophilic phase. (b) Model of a mixed micelle of DHPC and OmpX, inspired by the data in this paper. The protein is represented as a rectangle, with its hydrophobic surface region depicted in gray. (c) Lipid bilayer in the outer membrane of E. coli (37); there is a close match between the thickness of the lipophilic phase of this bilayer and the height of the hydrophobic surface in OmpX (Fig. 2b).
In addition to the data on the protein–detergent contacts (Fig. 2 a and b), the approximate molecular mass of the OmpX/DHPC micelles is known to be in the range 50–70 kDa. Nitrogen-15 NMR relaxation data indicated a correlation time of τc = 21 ns for the rotational molecular tumbling of OmpX/DHPC micelles (11, 23), corresponding to an apparent particle size of about 60 kDa. Analytical ultracentrifugation and protein–detergent titration yielded a size estimate of 50–70 kDa. Clearly, an adequate structural model for rationalizing the observed protein–detergent contacts must also be compatible with the measured size of the mixed micelles. In this context, we estimated the molecular weight for a dense packing arrangement of DHPC molecules on the OmpX surface.
For the calculation, we assumed that the surface area of OmpX occupied by DHPC is a cylinder jacket with a height of 28 Å (Fig. 2b). To account also for the volume occupied by the amino acid side chains, the cylinder diameter was taken to be 25 Å. Each of the two hydrophobic tails of DHPC was considered to be a cylinder with a diameter of 4 Å. Assuming that the DHPC molecules cover the hydrophobic surface of OmpX with the tightly packed tips of the hydrophobic tails, it was found that about 80 DHPC molecules can thus contact OmpX. The molecular masses of DHPC and OmpX are 453 Da and 16,384 Da, respectively, so that the total micelle size is about 52 kDa. This is at the lower end of the experimentally observed size range for OmpX/DHPC-micelles, and therefore the same calculation was repeated for a micelle containing an OmpX dimer, with the two β-barrels arranged side by side and parallel to each other. With the accessible hydrophobic surface of the dimer covered by DHPC molecules, a mass of 90 kDa was obtained. Although in these models the hydrophobic tails of the DHPC molecules are tightly packed against each other in the contact area with the protein, the polar head groups will be more loosely arranged and hydrated on the micelle surface (Fig. 3b). Because the hydration water has not been considered, the molecular masses estimated with this calculation represent a lower limit of the particle size that would actually be manifested in the hydrodynamic properties of the mixed micelles.
Mimicking Biological Membranes with Mixed Protein–Detergent Micelles.
Considering that membrane proteins need to be extracted from their natural environment and reconstituted in artificial milieus for 3D structure determination (1, 12, 31, 32), comparative studies of the protein interactions with the different environments are of special interest. The present results have led to a quite precise definition of the stoichiometry of OmpX/DHPC micelles in aqueous solution, and a structural characterization of the interaction between the two components of the mixed micelles. The observation of a continuous distribution of detergent molecules all around the β-barrel (Fig. 2b) and comparison of the molecular weights estimated for DHPC micelles with monomeric or dimeric OmpX indicate that there is only one OmpX molecule contained per micelle. It is seen that the DHPC interactions cover the best-defined regions of the protein NMR structure (11, 12), indicating that these interactions may have a crucial role in stabilizing the protein fold (33–35). Furthermore, the protein surface area covered by the lipophilic moieties of DHPC in the micelles corresponds closely to the area that is assumed to be lipid-exposed in a biological lipid bilayer membrane (Figs. 2b and 3c). Because the orientation of the lipid molecules relative to the protein surface is distinctly different in the two structures (Fig. 3 b and c), it is quite intriguing that a similar hydrophobic coating bounded by polar groups appears to be achieved with the two different arrangements of the lipid molecules. The experimental methods used here should also be applicable with other membrane proteins and different detergents or lipids, and it will be of interest to see how the data obtained for OmpX compare with the results of similar studies with different systems.
Complementary Information from NMR Spectroscopy and X-Ray Single-Crystal Diffraction.
Vogt and Schulz (28) reported that two glycerol molecules and one detergent molecule could be located in the crystal structure of OmpX. The information from the present studies is thus clearly complementary to the crystallographic data. In addition to the static view of the protein solvation (Fig. 2 a and b), the fact that the OmpX–DHPC intermolecular NOEs have negative signs shows that the lifetime of the individual DHPC molecules on the protein surface is longer than about 0.3 ns (27). Furthermore, the observation of a single set of DHPC chemical shifts would be compatible with the assumption of submillisecond lateral mobility of the individual DHPC molecules in the detergent layer covering the surface of OmpX (Fig. 3b).
Acknowledgments
This work was supported by the National Center of Competence in Research (NCCR) Structural Biology, the Kommission für Technologie und Innovation (KTI; Project 3392.1), and the Schweizerischer Nationalfonds (Project 31-66427.01).
Abbreviations
- NOE
nuclear Overhauser effect
- DHPC
dihexanoylphosphatidylcholine (1,2-dihexanoyl-sn-glycero-3-phosphocholine)
References
- 1.Arora A, Tamm L K. Curr Opin Struct Biol. 2001;11:540–547. doi: 10.1016/s0959-440x(00)00246-3. [DOI] [PubMed] [Google Scholar]
- 2.Sanders C R, Oxenoid K. Biochim Biophys Acta. 2000;1508:129–145. doi: 10.1016/s0005-2736(00)00308-4. [DOI] [PubMed] [Google Scholar]
- 3.Marassi F M, Opella S J. Curr Opin Struct Biol. 1998;8:640–648. doi: 10.1016/s0959-440x(98)80157-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vinogradova O, Sönnichsen F, Sanders C R. J Biomol NMR. 1998;11:381–386. doi: 10.1023/a:1008289624496. [DOI] [PubMed] [Google Scholar]
- 5.Pervushin K, Riek R, Wider G, Wüthrich K. Proc Natl Acad Sci USA. 1997;94:12366–12371. doi: 10.1073/pnas.94.23.12366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wüthrich K. Nat Struct Biol. 1998;5:492–495. doi: 10.1038/728. [DOI] [PubMed] [Google Scholar]
- 7.Wider G, Wüthrich K. Curr Opin Struct Biol. 1999;9:594–601. doi: 10.1016/s0959-440x(99)00011-1. [DOI] [PubMed] [Google Scholar]
- 8.Riek R, Pervushin K, Wüthrich K. Trends Biochem Sci. 2000;25:462–468. doi: 10.1016/s0968-0004(00)01665-0. [DOI] [PubMed] [Google Scholar]
- 9.Pervushin K. Q Rev Biophys. 2000;33:161–197. doi: 10.1017/s0033583500003619. [DOI] [PubMed] [Google Scholar]
- 10.Gardner K H, Kay L E. Annu Rev Biophys Biomol Struct. 1998;27:357–406. doi: 10.1146/annurev.biophys.27.1.357. [DOI] [PubMed] [Google Scholar]
- 11.Fernández C, Adeishvili K, Wüthrich K. Proc Natl Acad Sci USA. 2001;98:2358–2363. doi: 10.1073/pnas.051629298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fernández C, Hilty C, Bonjour S, Adeishvili K, Pervushin K, Wüthrich K. FEBS Lett. 2001;504:173–178. doi: 10.1016/s0014-5793(01)02742-9. [DOI] [PubMed] [Google Scholar]
- 13.Arora A, Abildgaard F, Bushweller J H, Tamm L K. Nat Struct Biol. 2001;8:334–338. doi: 10.1038/86214. [DOI] [PubMed] [Google Scholar]
- 14.Møller J V, le Maire M. J Biol Chem. 1993;268:18659–18672. [PubMed] [Google Scholar]
- 15.Kleinschmidt J H, Wiener M C, Tamm L K. Protein Sci. 1999;8:2065–2071. doi: 10.1110/ps.8.10.2065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lauterwein J, Bösch C, Brown L R, Wüthrich K. Biochim Biophys Acta. 1979;556:244–264. doi: 10.1016/0005-2736(79)90046-4. [DOI] [PubMed] [Google Scholar]
- 17.Bösch C, Brown L R, Wüthrich K. Biochim Biophys Acta. 1980;603:298–312. doi: 10.1016/0005-2736(80)90376-4. [DOI] [PubMed] [Google Scholar]
- 18.Brown L R, Bösch C, Wüthrich K. Biochim Biophys Acta. 1981;642:296–312. doi: 10.1016/0005-2736(81)90447-8. [DOI] [PubMed] [Google Scholar]
- 19.Braun W, Wider G, Lee K H, Wüthrich K. J Mol Biol. 1983;169:921–948. doi: 10.1016/s0022-2836(83)80143-0. [DOI] [PubMed] [Google Scholar]
- 20.Seigneuret M, Le Guernevé C. J Biomol NMR. 1999;13:31–41. doi: 10.1023/A:1008357824038. [DOI] [PubMed] [Google Scholar]
- 21.Williams K A, Farrow N A, Deber C M, Kay L E. Biochemistry. 1996;35:5145–5157. doi: 10.1021/bi952897w. [DOI] [PubMed] [Google Scholar]
- 22.Papavoine C H M, Konings R N H, Hilbers C W, Vandeven F J M. Biochemistry. 1994;33:12990–12997. doi: 10.1021/bi00248a007. [DOI] [PubMed] [Google Scholar]
- 23. Hilty, C., Fernández, C., Wider, G. & Wüthrich, K. (2002) J. Biomol. NMR, in press. [DOI] [PubMed]
- 24.Güntert P, Dötsch V, Wider G, Wüthrich K. J Biomol NMR. 1992;2:619–629. [Google Scholar]
- 25.Bartels C, Xia T H, Billeter M, Güntert P, Wüthrich K. J Biomol NMR. 1995;6:1–10. doi: 10.1007/BF00417486. [DOI] [PubMed] [Google Scholar]
- 26.Markley J L, Bax A, Arata Y, Hilbers C W, Kaptein R, Sykes B D, Wright P E, Wüthrich K. J Biomol NMR. 1998;12:1–23. doi: 10.1023/a:1008290618449. [DOI] [PubMed] [Google Scholar]
- 27.Otting G, Liepinsh E, Wüthrich K. Science. 1991;254:974–980. doi: 10.1126/science.1948083. [DOI] [PubMed] [Google Scholar]
- 28.Vogt J, Schulz G E. Struct Fold Des. 1999;7:1301–1309. doi: 10.1016/s0969-2126(00)80063-5. [DOI] [PubMed] [Google Scholar]
- 29.Hu W, Cross T A. Biochemistry. 1995;34:14147–14155. doi: 10.1021/bi00043a020. [DOI] [PubMed] [Google Scholar]
- 30.Schiffer M, Chang C H, Stevens F J. Protein Eng. 1992;5:213–214. doi: 10.1093/protein/5.3.213. [DOI] [PubMed] [Google Scholar]
- 31.Byrne B, Iwata S. Curr Opin Struct Biol. 2002;12:239–243. doi: 10.1016/s0959-440x(02)00316-0. [DOI] [PubMed] [Google Scholar]
- 32.Wiener M C. Curr Opin Coll Int Sci. 2001;6:412–419. [Google Scholar]
- 33.Gouaux E, White S H. Curr Opin Struct Biol. 2001;11:393–396. doi: 10.1016/s0959-440x(00)00222-0. [DOI] [PubMed] [Google Scholar]
- 34.Booth P J, Templer R H, Meijberg W, Allen S J, Curran A R, Lorch M. Crit Rev Biochem Mol Biol. 2001;36:501–603. doi: 10.1080/20014091074246. [DOI] [PubMed] [Google Scholar]
- 35.White S H, Wimley W C. Annu Rev Biophys Biomol Struct. 1999;28:319–365. doi: 10.1146/annurev.biophys.28.1.319. [DOI] [PubMed] [Google Scholar]
- 36.Koradi R, Billeter M, Wüthrich K. J Mol Graphics. 1996;14:51–55. doi: 10.1016/0263-7855(96)00009-4. [DOI] [PubMed] [Google Scholar]
- 37.Wimley W C. Protein Sci. 2002;11:301–312. doi: 10.1110/ps.29402. [DOI] [PMC free article] [PubMed] [Google Scholar]