Abstract
By using single-molecule measurements, we demonstrate that the elongation kinetics of individual Escherichia coli RNA polymerase molecules are remarkably homogeneous. We find no evidence of distinct elongation states among RNA polymerases. Instead, the observed heterogeneity in transcription rates results from statistical variation in the frequency and duration of pausing. When transcribing a gene without strong pause sites, RNA polymerase molecules display transient pauses that are distributed randomly in both time and distance. Transitions between the active elongation mode and the paused state are instantaneous within the resolution of our measurements (<1 s). This elongation behavior is compared with that of a mutant RNA polymerase that pauses more frequently and elongates more slowly than wild type.
Transcription elongation is a processive but discontinuous process, with active synthesis of mRNA punctuated by transient pauses (1–4). Both bacteria and eukaryotes control gene expression in vivo by regulating pausing at specific sites on the DNA template (5, 6). Several regulatory pauses lead to long-lived isomerizations of the elongation complex, allowing transcription factors to bind the paused RNA polymerase (RNAP) and modify subsequent elongation (6–8). However, not all hesitation by the RNAP is thought to be regulatory; transcription of any naturally occurring template reveals numerous pauses with short half-lives, reflecting a distinct type of pausing that is inherent to the RNAP enzyme mechanism. At present, little is known about the RNAP configurations during these brief pauses or what governs the transition from active elongation to a paused state in the absence of specific regulatory signals.
These dynamic aspects of elongation are obscured when studying a population of RNAP molecules in which the asynchronous behavior of individual RNAPs is smeared into an ensemble-average. Analyzing the motion of single RNAP molecules in real time eliminates the complication of population dynamics, revealing the kinetic interplay between active elongation and nonproductive states (9–12). Furthermore, continuous tracking of individual RNAPs exposes any variation in the behavior of single RNAP molecules as well as the differences that exist between RNAPs.
There is more asynchrony during transcription elongation than would be generated by an enzymatic reaction with a single rate-limiting step, but the mechanisms leading to the additional asynchrony have yet to be clearly defined (13). Transcriptional pausing, where a fraction of RNAP stop at discrete sites for a variable duration, is known to induce dispersion of the population. However, whether or not the stochastic behavior of a structurally homogeneous population is sufficient to generate the observed levels of asynchrony, or if one must also invoke alternate stable RNAP conformations is a subject of debate. A recent study of single RNAP molecules suggests that an elongating RNAP population is composed of RNAPs in distinct states that elongate at different intrinsic rates and are more or less likely to pause (12). However, the average elongation rates reported in the previous work were significantly slower than solution rates, which complicates an interpretation of the fast and slow elongation modes observed (12). In this report, we have characterized elongation and transient pauses (1–30 s) by single RNAP molecules on a template that lacks known regulatory pause sites. The average elongation rates in these experiments were identical to those obtained from bulk solution assays of transcription, indicating that the RNAP molecules studied were fully active. We show that individual RNAPs exhibit homogeneous elongation dynamics, with differences among RNAPs arising from random switching between a single active elongation mode and the paused state.
Materials and Methods
Cloning and Purification of WT and RpoB8 RNAPs.
Plasmid p706a encodes an N-terminal 6-His-tagged version of rpoA (the Escherichia coli α-subunit) under control of a lac-repressor-regulated trc promoter. This plasmid was subjected to PCR-directed mutagenesis to remove the stop codon following the α coding sequence. The resulting plasmid was digested with the appropriate restriction enzymes, and annealed oligonucleotides encoding the 9-aa hemagglutinin (HA) epitope tag and stop codon, flanked by the appropriate sticky ends, were ligated into the vector, creating plasmid pKA1. The resulting N-terminal His-6 and C-terminal HA-tagged α-subunit was expressed at low levels in wild type (WT, DH5α) and rpoB8 polymerase mutant E. coli strains to yield WT and RpoB8 mutant RNAPs with a single HA-tag. RNA polymerases were purified to homogeneity by using a modification of the method of Burgess and Jendrisak (14) to include chromatography on nickel agarose. In vitro transcription assays confirmed that neither the HA-tag nor the anti-HA antibody (Covance) altered the rate or efficiency of transcription. rpoB8 is a point mutation in the β-subunit, which substitutes Q513 with P. The homologous residue in the yeast Rpb2 subunit (Q481) contacts the nascent mRNA between positions −5 and −6 (15).
Transcription Assays.
Ternary complexes formed in transcription buffer [25 mM Tris⋅Cl, pH 7.9/100 mM KCl/4 mM MgCl2/1 mM DTT/3% (vol/vol) glycerol] with 25 nM HA-tagged RNAP and 5 nM template (pRL574; containing the T7A1 promoter and rpoB gene; ref. 9) were supplied with ApU (200 μM), GTP, CTP, and ATP (10 μM) and incubated for 20 min at 37°C to allow formation of a 20-nt transcript. Heparin was added to 0.2 μg/μl, and the stalled elongation complexes were transferred to ice. For bulk transcription assays, stalled ternary complexes were radiolabeled by using [α-32P]CTP [5 μCi (1 Ci = 37 GBq) at 3,000 Ci/mmol]. Transcription was restarted with unlabeled NTPs at 1 mM, and aliquots were stopped at 5-s time intervals. Transcripts were separated on polyacrylamide sequencing gels and analyzed by using a PhosphorImager (Molecular Dynamics) to determine elongation rates and pausing patterns under these conditions. The level of pausing at a given position was estimated by quantitating the percentage of total RNA with a defined length, which was consistently <15% at any position at all time points. To improve the resolution of pausing patterns and elongation rates on sequences >1 kb from the promoter on standard sequencing gels, restriction fragments of increasing sizes were excised from within rpoB, bringing more distant template sequences closer to the promoter.
Experimental Configuration.
Stalled ternary complexes and disposable flow cells were prepared much as described (9–11), except that the anti-HA antibody was adsorbed nonspecifically to the coverslip surface and the remaining surface was blocked with 4% (wt/vol) milk protein before introducing stalled RNAP complexes, which were diluted 1:200 into transcription buffer plus heparin (0.2 μg/μl). After the initial DNA tether length (4,103 bp) was confirmed, elongation was restarted by the addition of 1 mM NTPs. Under these conditions, both WT RNAP and B8 RNAP resumed elongation with an efficiency of >80%. Data acquisition began after the flow of NTPs had stopped and the RNAP had begun elongation, which led to some variation in template position at the beginning of each trace. A digital feedback loop held the trapping force constant at 4 pN by moving a piezo-controlled microscope stage toward the trap as elongation proceeded. Although a trapping force of 4 pN was used for all of the data presented herein, the use of forces within the range of 2–8 pN yields indistinguishable elongation rates and pausing patterns on this template, both for WT RNAP and the B8 mutant RNAP (data not shown; refs. 11 and 12).
Tether length was determined from the position of the piezo stage and the displacement of the microsphere from the trap center based on published methods (11, 16). Analog signals were low-pass filtered through an 8-pole butterworth filter to remove frequency components above 5 kHz, sampled at 13 kHz with a 16-bit analog-to-digital converter, and digitally averaged to 130 Hz. Conversion of distance to LDNA was performed as described (11). The uncertainty in the absolute position of the RNAP on the DNA template is ≈±100 bp over an ≈4,000-bp long tether. However, for a given molecule, the resolution in the relative position is significantly better: the uncertainty is ±10 bp over an ≈200-bp movement. WT RNAP data sets averaged 1,504 nt and 126 s. The average WT elongation rate was 12.0 ± 2.1 nt/s (mean ± SD). Mutant B8 RNAP data sets averaged 607 nt and 176 s. The average elongation rate was 3.4 ± 1.3 nt/s. Increasing the NTP concentration above 1 mM (up to 5 mM) did not detectably increase the rate of elongation for either B8 RNAP or WT RNAP.
Data Analysis.
The instantaneous velocity at time t was determined by fitting a 3-s window of data centered at t (sampled at 130 Hz) to a straight line. The data were fit by using a Gaussian weight function with SD of 1 s, thus effectively imposing a 1-Hz Gaussian low-pass filter. Curves for instantaneous velocity and position are obtained from the slope and position of the linear fit by incrementing t by 30 ms per point (32 samples per s).
The combined velocity distributions were fit with a normalized function composed of two Gaussian functions, with their means, SDs, and ratio of their areas as fitting parameters. When fitting the data from B8 RNAPs, the center of the first component was fixed at 0.9 nt/s to allow for direct comparison with the WT RNAP. We also generated combined distributions of velocity in which equal statistical weight is assigned to each position on the template that is traversed, rather than to each time interval. This method gives no statistical weight to pauses, because the change in template position as a function of time is, by definition, zero during a pause. As a result, the distribution of velocities around zero is removed, and the distributions for active elongation by each RNAP were fit by a single Gaussian function (WT center = 14.27 nt/s, SD = 4.5 nt/s; B8 center = 5.0 nt/s; SD = 3.0 nt/s).
A computer algorithm was developed to detect pauses from a plot of dwell time vs. filtered transcript position. An RNAP was considered to have paused when the time spent at a single nucleotide position exceeded the cutoff duration. Integration of the area under the peak, using the most likely dwell time as baseline, yielded pause duration. The cutoff duration was determined by examining histograms of the dwell times for all WT or B8 RNAPS. The histograms had skewed Gaussian shapes with long tails stretching out to extended dwell times. The cutoff duration for both RNAPs was chosen to be 3.5× the most likely dwell time, which is 59 ms for WT and 137 ms for B8 RNAP, to define only the tail portion of these curves as the paused state. Data analysis included only pauses that lasted for <30 s, because measurements of RNAPs that have paused for longer periods are subject to increasing effects of instrument drift. Although this cutoff improves the position resolution of the resulting data, it precludes measurement of the frequency of transcriptional arrest. A total of 128 WT pauses and 421 B8 pauses were analyzed. Calculation of the time elapsed and distance traveled between consecutive pauses was determined by using 99 pairs of pauses by WT RNAP and 391 pairs for B8 RNAP. Pause distributions were fit to a single exponential decay model, where the deviation of the fit from each point is normalized by the estimated experimental uncertainty. The P values for determining the quality of these fits were calculated from the χ2 value and the number of degrees of freedom.
Results and Discussion
Elongation Kinetics of Single RNAP Molecules Are Homogeneous.
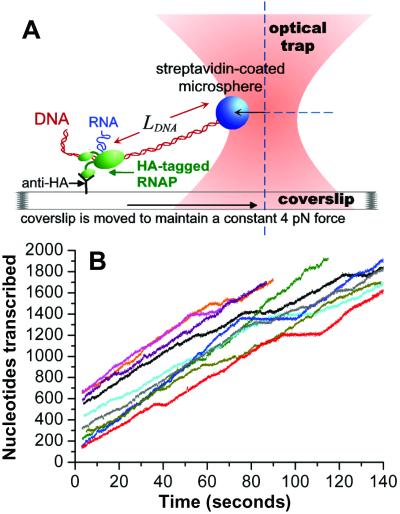
Relevant single-molecule analysis of RNAP elongation requires that the collection of single molecules studied resembles an RNAP population in bulk solution. To achieve this goal, we developed a method for specific, flexible immobilization of an HA epitope-tagged RNAP on a coverslip surface coated with antibody against this tag. The HA tag was fused to the α-subunit C terminus, which has a radius of motion of 70 Å with respect to the central core of the RNAP (17). RNAP elongation complexes immobilized in this way (Fig. 1A) are highly active; their behavior on the surface is nearly identical to that observed in bulk solution (see Materials and Methods). Transcript elongation by 10 single RNAP molecules is shown in Fig. 1B (also, see Fig. 5, which is published as supporting information on the PNAS web site, www.pnas.org, for additional traces). The average elongation rate for the 30 individual WT RNAPs studied in this work was 12.0 nt/s (nucleotides transcribed/time interval). In vitro transcription assays performed under identical conditions [22°C and 1 mM NTPs] on this template, which bears the E. coli rpoB gene, yielded elongation rates of 12–13 nt/s and revealed no sites at which a large proportion of RNAPs paused (data not shown; ref. 9). The transient pauses observed occurred with low probability throughout the gene, allowing us to perform our initial characterization of RNAP elongation in the absence of dominant sequence effects.
Figure 1.
Experimental setup and raw data. (A) A stalled ternary elongation complex composed of E. coli RNAP bearing an HA-epitope tag, template DNA, and a short RNA was specifically immobilized on a coverslip surface through interaction with a nonspecifically adsorbed anti-HA antibody. A streptavidin-coated microsphere was attached to a biotin located on the downstream end of the DNA. The microsphere was held at a fixed position relative to the optical trap throughout elongation. During transcription, the template DNA was pulled through the RNAP, leading to a decrease in length of downstream DNA (LDNA). Feedback control moved the coverslip toward the optical trap so that a constant force of 4 pN was maintained on the RNAP. Force of this magnitude has been shown to have no detectable effect on elongation rate or pausing (11, 12). (B) Elongation profiles of 10 single RNAP molecules are plotted as nucleotides transcribed vs. time.
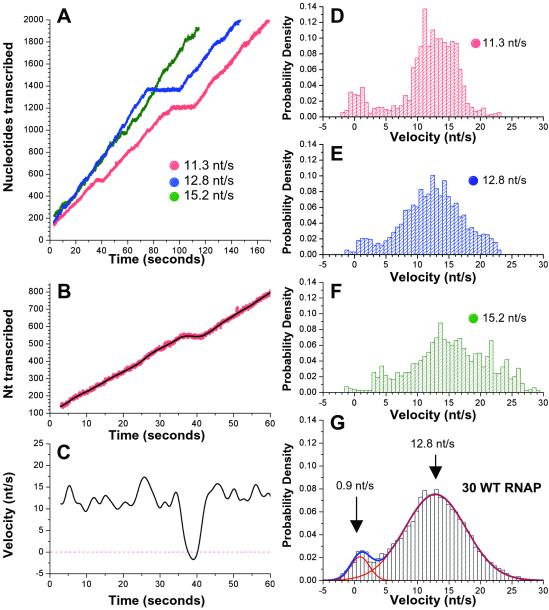
Data obtained from three representative RNAP molecules are plotted in Fig. 2A, showing that the pausing pattern is highly variable among RNAPs. However, during periods of relatively uninterrupted movement, the three traces have comparable slopes, indicating that individual RNAPs display similar rates of active elongation. Although long pauses are apparent at this level of resolution, numerous shorter-lived pauses and minor rate fluctuations are less evident. To examine elongation dynamics in greater detail, we calculated the instantaneous velocity throughout the time interval with an algorithm that effectively applies a Gaussian low-pass filter to the trace of nucleotides transcribed vs. time (see Materials and Methods). A close-up of transcription is shown for one RNAP molecule (as in Fig. 2A) with the unfiltered trace shown in red and the smoothed curve that results from filtering in black (Fig. 2B). The instantaneous velocity derived for this RNAP molecule (Fig. 2C) varies within the range of 8–18 nt/s throughout the majority of this time interval. Because these variations in instantaneous velocity are significantly larger than the standard deviation (SD) of our measurement uncertainty (1.5 nt/s), they reveal continual fluctuations in elongation velocity, consistent with reports that the rate of nucleotide addition varies appreciably as a function of template position (13, 18). Pausing is reflected by a sharp drop in velocity that is followed by an equally sharp transition back to the actively elongating state.
Figure 2.
Analysis of elongation velocity. (A) Elongation by three RNAP molecules, expressed as nucleotides transcribed vs. time. Overall transcription rates are designated for each RNAP. (B and C) Magnified views of the first 60 s of elongation by one RNAP (red, as in A). The black curves are the nucleotide position (B) and the instantaneous velocity derived from our filtering procedure (C). (D–F) Normalized distributions of instantaneous velocity (s/nt) for the RNAPs shown in A. (G) The combined normalized distribution for 30 WT RNAPs is fit by two Gaussian functions (shown in red, overall fit in blue). The component that represents the paused state is centered at 0.9 nt/s (SD = 1.5 nt/s; area = 7.8%) and the component reflecting active elongation is centered at 12.8 nt/s (SD = 4.9 nt/s; area = 92.2%).
The instantaneous velocity data are summarized by normalized distributions that indicate the fraction of time that an RNAP moved at a given velocity. Fig. 2 D–F demonstrates these distributions for the three RNAP molecules shown in Fig. 2A. The velocity distributions are characterized by two distinct components: one that corresponds to pausing centered near 0 nt/s, and a second that represents active elongation, centered around 13 nt/s. Strikingly, the most probable velocity is essentially the same for all WT RNAPs observed, despite their differing average rates, with run-to-run variations attributable to statistical fluctuations. The major difference among RNAP molecules is reflected in the area under the distribution function near 0 nt/s, with slower-than-average RNAP spending a larger fraction of time in the paused state. For a given molecule, there is a single peak corresponding to active elongation, which argues against the existence of the long-lived fast and slow elongating forms of RNAP proposed in a recent single-molecule study of RNAP elongation (12). The combined normalized distribution for 30 WT RNAP (Fig. 2G) reveals that all RNAPs display essentially the same dynamics, because the data are fit extremely well by two Gaussian functions. The first component, centered at 0.9 nt/s, represents the paused state. The small positive value for this distribution arises from slight smearing of the pauses by our finite averaging window. The second distribution describing active elongation is fit by a single Gaussian function centered at 12.8 nt/s, indicating that a single elongation mode is sufficient to account for the data. The SD in instantaneous velocity of the active elongation mode is 4.9 nt/s. After subtraction of the estimated fluctuations attributable to the Brownian motion of the microsphere in the trap, the corrected value is 4.7 nt/s. This value is larger than the 2.1 nt/s that would be predicted if each nucleotide addition were a single-rate kinetic process (meaning that each nucleotide addition only involves a single rate-limiting step), consistent with a previous solution analysis of RNA chain elongation (13).
Elongation Behavior of the RNAP Mutant RpoB8.
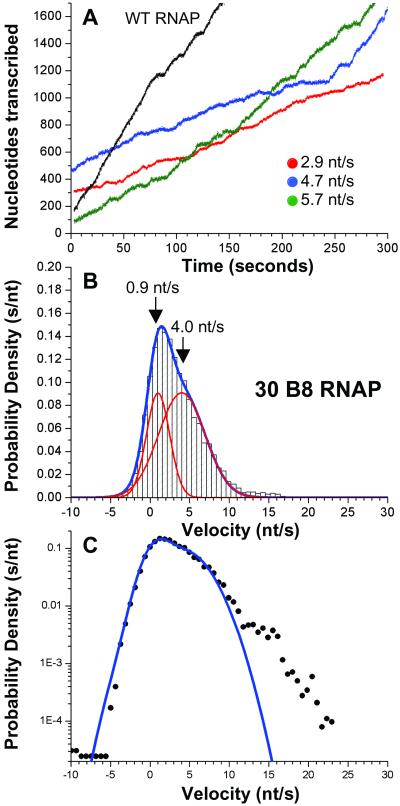
A point mutation in RNAP, rpoB8, leads to enhanced pausing and termination and has been suggested to decrease elongation rate (19–22). The mutation in RpoB8 (B8) RNAP likely removes a specific contact with the nascent mRNA within the RNA-DNA hybrid more than 20 Å away from the active site (15), making it improbable that the mutation directly alters the catalytic rate. To distinguish unambiguously the effects of this mutation on active elongation velocity and pausing we investigated elongation by single B8 RNAP molecules. B8 RNAPs transcribe significantly more slowly than WT, with an average overall elongation rate of 3.4 nt/s (Fig. 3A and Fig. 6, which is published as supporting information on the PNAS web site). Elongation by individual B8 RNAPs proceeds at highly variable rates, with significant changes in slope and bursts of nearly WT velocity noted in many traces (Fig. 3A, see blue trace). However, the most probable instantaneous velocities for B8 RNAPs were markedly slower than for WT. The combined normalized distribution for 30 B8 RNAP molecules (Fig. 3B) demonstrates that the component representing active elongation velocity is centered around 4.0 nt/s. The greater-than-threefold decrease in elongation velocity compared with WT is coupled with considerable effects on pausing. The area under the curve that represents pausing encompasses 33.3% of total elongation time for B8 RNAP, compared with 7.8% for WT RNAP. Interestingly, it is known that that other conditions that slow the rate of transcription, such as decreased temperature or NTP concentration, also increase pausing (18). Inspection of the combined distribution reveals higher velocity movement that falls outside the realm of the dual Gaussian fit (≈2% of total elongation time). This observation is emphasized by plotting the combined dual-Gaussian function with the data on a semilog graph (Fig. 3C), revealing that the residual is largely composed of velocities between 12 and 24 nt/s. Remarkably, the occurrence of these anomalously large velocities shows that the B8 RNAP retains the ability to elongate at WT rates.
Figure 3.
Elongation by the RpoB8 mutant RNAP. (A) Elongation by three representative B8 RNAP molecules shown in color, with one WT trace shown in black for comparison. (B) The combined normalized distribution of velocity from 30 B8 RNAP is fit by two Gaussian functions (each shown in red, overall fit in blue). The distribution that represents pausing, centered at 0.9 nt/s (SD = 1.5 nt/s; area = 33.3%) significantly overlaps the component that corresponds to active elongation, centered around 4.0 nt/s (SD = 5.9 nt/s; area = 66.7%). (C) The fit to two Gaussian functions (blue line) and the normalized distribution (filled circles) plotted vs. velocity on a semilog graph.
Investigation of Pausing by WT and B8 RNAPs.
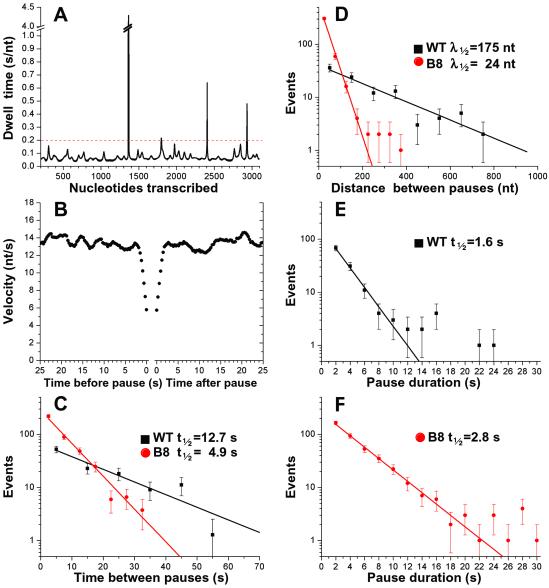
To determine whether the increase in pausing by B8 RNAPs reflects enhanced pause frequency, duration, or both, we characterized pauses by both WT and B8 RNAP. As shown in Fig. 4A, increased dwell time at a given nucleotide position leads to a peak in this graph; peaks are defined as pauses when they exceed the cutoff duration, shown as a dotted line (see Materials and Methods). Given the tendency of slower-moving RNAPs to pause, we investigated whether the WT RNAP molecules show any signs of slowing down before pausing by compiling the velocity of all WT RNAP molecules as a function of time before and after each pause. These data show no evidence of reduced elongation rate before or after a pause, within the resolution limit (1 s) imposed by the Gaussian filter (Fig. 4B), indicating that any conformational change associated with entry into or exit from a pause must be so short-lived that it is undetectable by these methods. Thus, these data indicate that pausing induces no long-term changes in the RNAP that affect elongation behavior after the RNAP has resumed transcription; i.e., the RNAP retains no “memory” of having paused.
Figure 4.
Pause frequency and duration. (A) Dwell time vs. nucleotide position for one WT RNAP molecule (shown in Fig. 2A, blue). (B) The average of the instantaneous velocity as a function of the time before the beginning of a pause (Left) or since the end of a pause (Right) for all pauses by WT RNAPs (0.3-s intervals). (C) The number of pauses that occurred within a given time interval after the previous pause (10-s bins for WT RNAP, 5-s bins for B8 RNAP) is plotted vs. time on a semilog graph. The lines are the fits to exponential decay, (WT RNAP t1/2 = 12.7 s, P = 0.37; B8 RNAP t1/2 = 4.9 s; P = 0.29). (D) The distribution of distances between consecutive pauses (100-nt bins for WT RNAP, 50-nt bins for B8 RNAP) is plotted vs. distance on a semilog graph. Single exponential fits of the data yield the half-distances between pauses (WT RNAP λ1/2 = 175 nt; P = 0.29; B8 RNAP fit between 0–200 nt λ1/2 = 24 nt; P = 0.18). The distribution of pause durations (2-s bins) is shown for WT RNAP (E) or B8 RNAP (F). Shown are exponential fits to the data representing pauses that last ≤10 s for WT RNAP (t1/2 = 1.6 s; P = 0.81) and ≤16 s for B8 RNAP (t1/2 = 2.8 s; P = 0.32).
To investigate entry into the paused state, we calculated the time between the end of one pause and the beginning of the next pause for all sets of consecutive pauses by WT and B8 RNAPs. The distributions of times between consecutive pauses for each RNAP population are fit by a single exponential decay (Fig. 4C), suggesting that pauses are stochastic, uncorrelated events with respect to time. The stochastic nature of pausing likely reflects the fact that this gene contains only low probability pause sites that are located throughout the template. This simple distribution of times between pauses permits a direct comparison of the pause frequencies for the WT vs. B8 RNAPs. Consistent with the decreased rate of forward synthesis, B8 RNAPs pause significantly more frequently than WT (Fig. 4C). If active elongation velocity is similar among RNAPs and the distribution of pauses is stochastic with respect to time, pauses also should be distributed randomly with respect to distance. Although this holds true for the WT RNAPs (Fig. 4D), the distribution of pauses by B8 RNAPs shows that whereas the majority of pauses are fit by an exponential decay, longer distances deviate from the fit. These outlying points are derived from B8 RNAPs that have transiently adopted the fast elongating state, yielding distances between pauses that are more like WT RNAP. Examining the duration of pauses by WT RNAP demonstrates that escape from the majority of pauses on this sequence can be described by a single-rate constant (Fig. 4E). Pauses that deviate from these kinetics resume elongation on a much longer time scale, suggesting that these RNAP have adopted altered configurations. The extended pause duration is largely sequence dependent, because 8 of the 10 long (>10 s) pauses observed occurred within experimental uncertainty of template positions 1,360 or 1,710 (4 pauses each), which are two of the longest-lived pause sites noted in solution assays on this template (data not shown). B8 RNAPs exhibit a modest increase in pause half-life (Fig. 4F); yet, approximately the same percentage of B8 pauses deviate from a single exponential decay (6%, compared with 3% for WT). Thus, although B8 RNAPs pause significantly more frequently than WT RNAPs, escape of B8 RNAPs from the paused state is less dramatically affected by the mutation.
Conclusions.
By ensuring that the single RNAP molecules investigated behaved like RNAP in bulk solution, we have determined the fundamental kinetic features of transcription elongation under conditions that allow direct comparison to population studies. Our examination of single WT RNAP molecules reveals that the asynchrony of transcription elongation does not result from distinct modes of RNAP movement. Instead, the observed heterogeneity in elongation rates on the rpoB gene results from statistical variations caused by the stochastic switching of RNAPs from a single active elongation mode to a transiently paused state.
This conclusion contrasts with the results of a recent single-molecule study by Davenport et al. (12), who reported that a transcribing RNAP population is heterogeneous, with distinct fast and slow elongation modes. Although we cannot explain these discrepancies, there are several differences in the experimental approaches that may have contributed to them. Some differences may be caused by the distinct DNA templates used and/or activity levels of the immobilized RNAPs, but these possibilities are difficult to assess because the previous study did not provide a comparative solution analysis of elongation under their conditions. It is worth noting that Davenport et al. (12) reported a slower average overall elongation rate: 8.0 ± 3 nt/s compared with 12.0 ± 2.1 nt/s from our studies, under similar conditions. It is also possible that the discrepancies arise from the differing methods of calculation of instantaneous velocity combined with the use of a “peak” velocity distribution in the prior work (12). To calculate an instantaneous velocity, Davenport et al. averaged data over 15 s, which is significantly longer than our averaging window. Because we measure that the half-life of active elongation between pauses for WT RNAPs is 12.7 s, and the pause half-life is 1.6 s, the instantaneous velocities based on a 15-s filter were likely complicated by inclusion of pauses.
Our investigation of the B8 RNAP mutant demonstrates a reduced active elongation velocity concomitant with enhanced pause frequency. However, our studies also indicate that the B8 RNAP maintains the capacity to elongate at WT velocities, suggesting that B8 RNAPs may occasionally assume a conformation that resembles WT RNAP. Interestingly, the B8 RNAP can maintain this fast conformation through a pause (for example, see blue trace, Fig. 3A and supporting information).
In summary, by defining the elongation dynamics of WT and a mutant RNAP on a gene without strong pause sites, these data provide a foundation for future single-molecule measurements of transcription elongation and investigation of the mechanisms by which regulatory pause sites and transcription factors alter the elongation process.
Supplementary Material
Acknowledgments
We thank Steve Koch, Richard Yeh, and Alla Shundrovskaya for construction of the optical trapping instrument, and Brent Brower-Toland, Lee Kraus, and Lu Bai for helpful discussions. This work was supported by National Institutes of Health grants (to J.T.L., J.W.R., and M.D.W.), a Beckman Young Investigator Award, and the Keck Foundation's Distinguished Young Scholar Award (to M.D.W.).
Abbreviations
- RNAP
RNA polymerase
- HA
hemagglutinin
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Guajardo R, Sousa R. J Mol Biol. 1997;265:8–19. doi: 10.1006/jmbi.1996.0707. [DOI] [PubMed] [Google Scholar]
- 2.von Hippel P H. Science. 1998;281:660–665. doi: 10.1126/science.281.5377.660. [DOI] [PubMed] [Google Scholar]
- 3.Nudler E. J Mol Biol. 1999;288:1–12. doi: 10.1006/jmbi.1999.2641. [DOI] [PubMed] [Google Scholar]
- 4.Korzheva N, Mustaev A. Curr Opin Microbiol. 2001;4:119–125. doi: 10.1016/s1369-5274(00)00176-4. [DOI] [PubMed] [Google Scholar]
- 5.Lis J T. Cold Spring Harbor Symp Quant Biol. 1998;63:347–356. doi: 10.1101/sqb.1998.63.347. [DOI] [PubMed] [Google Scholar]
- 6.Roberts J W, Yarnell W, Bartlett E, Guo J, Marr M, Ko D C, Sun H, Roberts C W. Cold Spring Harbor Symp Quant Biol. 1998;63:319–325. doi: 10.1101/sqb.1998.63.319. [DOI] [PubMed] [Google Scholar]
- 7.Komissarova N, Kashlev M. J Biol Chem. 1997;272:15329–15338. doi: 10.1074/jbc.272.24.15329. [DOI] [PubMed] [Google Scholar]
- 8.Artsimovitch I, Landick R. Cell. 2002;109:193–203. doi: 10.1016/s0092-8674(02)00724-9. [DOI] [PubMed] [Google Scholar]
- 9.Schafer D A, Gelles J, Sheetz M P, Landick R. Nature. 1991;352:444–448. doi: 10.1038/352444a0. [DOI] [PubMed] [Google Scholar]
- 10.Yin H, Wang M D, Svoboda K, Landick R, Block S M, Gelles J. Science. 1995;270:1653–1657. doi: 10.1126/science.270.5242.1653. [DOI] [PubMed] [Google Scholar]
- 11.Wang M D, Schnitzer M J, Yin H, Landick R, Gelles J, Block S M. Science. 1998;282:902–907. doi: 10.1126/science.282.5390.902. [DOI] [PubMed] [Google Scholar]
- 12.Davenport R J, Wuite G J L, Landick R, Bustamante C. Science. 2000;287:2497–2500. doi: 10.1126/science.287.5462.2497. [DOI] [PubMed] [Google Scholar]
- 13.Matsuzaki H, Kassevetis G A, Geiduschek E P. J Mol Biol. 1994;235:1173–1192. doi: 10.1006/jmbi.1994.1072. [DOI] [PubMed] [Google Scholar]
- 14.Burgess R R, Jendrisak J J. Biochemistry. 1975;14:4634–4638. doi: 10.1021/bi00692a011. [DOI] [PubMed] [Google Scholar]
- 15.Gnatt A L, Cramer P, Fu J, Bushnell D A, Kornberg R D. Science. 2001;292:1876–1882. doi: 10.1126/science.1059495. [DOI] [PubMed] [Google Scholar]
- 16.Wang M D, Yin H, Landick R, Gelles J, Block S M. Biophys J. 1997;72:1335–1346. doi: 10.1016/S0006-3495(97)78780-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Naryshkin N, Revyakin A, Kim Y, Mekler V, Ebright R H. Cell. 2000;101:601–611. doi: 10.1016/s0092-8674(00)80872-7. [DOI] [PubMed] [Google Scholar]
- 18.Richardson J P, Greenblatt J. In: Escherichia coli and Salmonella: Cellular and Molecular Biology. 2nd Ed. Neidhardt F C, Curtiss R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger M E, editors. Washington, DC: Am. Soc. Microbiol.; 1996. pp. 822–848. [Google Scholar]
- 19.Jin D J, Gross C. J Mol Biol. 1988;202:45–58. doi: 10.1016/0022-2836(88)90517-7. [DOI] [PubMed] [Google Scholar]
- 20.Jin D J, Walter W A, Gross C. J Mol Biol. 1988;202:245–253. doi: 10.1016/0022-2836(88)90455-x. [DOI] [PubMed] [Google Scholar]
- 21.Jin D J, Gross C. J Biol Chem. 1991;266:14478–14485. [PubMed] [Google Scholar]
- 22.McDowell J C, Roberts J W, Jin D J, Gross C. Science. 1994;266:822–825. doi: 10.1126/science.7526463. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.