Abstract
The bacterial outer membrane enzyme PagP transfers a palmitate chain from a phospholipid to lipid A. In a number of pathogenic Gram-negative bacteria, PagP confers resistance to certain cationic antimicrobial peptides produced during the host innate immune response. The global fold of Escherichia coli PagP was determined in both dodecylphosphocholine and n-octyl-β-d-glucoside detergent micelles using solution NMR spectroscopy. PagP consists of an eight-stranded anti-parallel β-barrel preceded by an amphipathic α helix. The β-barrel is well defined, whereas NMR relaxation measurements reveal considerable mobility in the loops connecting individual β-strands. Three amino acid residues critical for enzymatic activity localize to extracellular loops near the membrane interface, positioning them optimally to interact with the polar headgroups of lipid A. Hence, the active site of PagP is situated on the outer surface of the outer membrane. Because the phospholipids that donate palmitate in the enzymatic reaction are normally found only in the inner leaflet of the outer membrane, PagP activity may depend on the aberrant migration of phospholipids into the outer leaflet. This finding is consistent with an emerging paradigm for outer membrane enzymes in providing an adaptive response toward disturbances in the outer membrane.
Certain essential molecular components that are uniquely associated with infectious agents, including the lipid A or endotoxin component of lipopolysaccharide from Gram-negative bacteria (1), can provide molecular signals that alert the host innate immune system to infection (2). Subtle variations in the structure of lipid A can greatly impact the nature and efficacy of the immune response. During severe infections, lipid A can elicit a cascade of cytokine production that may lead to massive inflammation and septic shock as the lethal consequence. On the other hand, certain lipid A analogs can act as excellent antagonists of endotoxin signaling (3).
Several regulated covalent modifications of lipid A have been implicated in the virulence of pathogenic Gram-negative bacteria (4, 5). Lipid A modifications in Salmonella depend largely on the PhoPQ two-component signal transduction pathway, which responds to Mg2+-limited growth conditions encountered during infection (6). The PhoPQ regulon controls at least 40 genes, many of which directly promote infection and/or provide an adaptive response toward Mg2+ limitation. Deregulation of the Salmonella PhoPQ regulon is known to attenuate virulence and intracellular survival within macrophages (7).
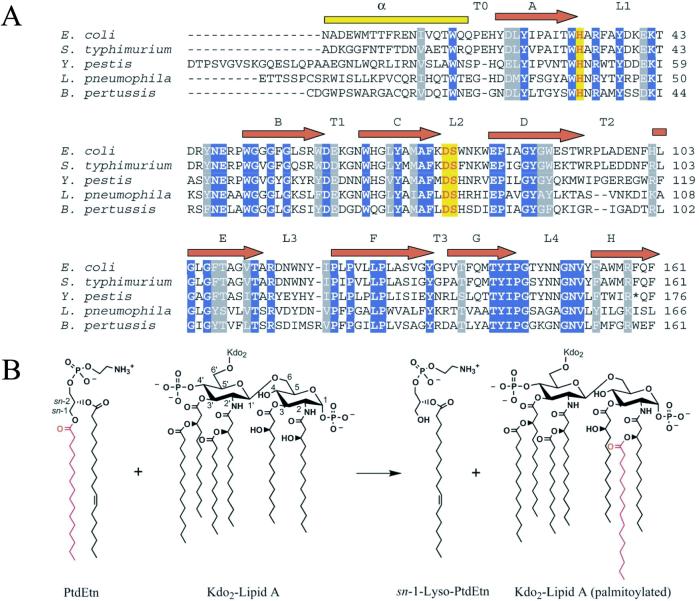
Palmitoylation of lipid A can both convert lipid A into an antagonist of endotoxin signaling (8) and provide bacterial resistance against vertebrate cationic antimicrobial peptides (9). The reaction is governed by a novel PhoPQ-activated outer membrane enzyme known as PagP (10). Homologues of PagP are implicated in virulence of Salmonella (9) and Legionella (11) species and are also present in Yersinia, Bordetella, and Escherichia coli genomes (Fig. 1A). Design of potent inhibitors of PagP would be facilitated by a knowledge of its structure and catalytic mechanism.
Figure 1.
(A) Alignment of PagP from five different pathogenic Gram-negative bacterial genera. Only representative sequences from each genus are shown when multiple species exist. The alignment was constructed after removal of signal peptides, either known (10) or predicted by signalp (44). The three residues shown to be essential for catalysis by site-directed mutagenesis are highlighted in red with yellow shading. Absolutely conserved residues are shaded in blue, and highly conserved residues in gray. The Yersinia pestis homologue appears to contain a premature stop codon at position 174. (B) PagP-catalyzed palmitoylation of lipid A. PagP can transfer a palmitate chain from the sn-1 position of a phospholipid, such as phosphatidylethanolamine (PtdEtn), to the free hydroxyl-group of the N-linked R-3-hydroxymyristate chain on the proximal glucosamine unit of lipid A or its precursors (10). The simplest lipid A acceptor for PagP in the outer membrane contains two units of 3-deoxy-d-manno-octulosonic acid (Kdo) and is known as Kdo2-lipid A (10).
PagP catalyzes the transfer of a palmitate chain from the sn-1 position of a phospholipid to the hydroxyl group of the N-linked R-3-hydroxymyristate chain on the proximal glucosamine unit of lipid A (Fig. 1B). The reaction bears some resemblance to lecithin-cholesterol acyltransferase (LCAT), which transfers a fatty acid from the sn-2 position of phosphatidylcholine to cholesterol (12). Like LCAT, PagP is also reported to slowly transfer palmitate to water in vitro, thus acting as a phospholipase.†† However, in its native environment, PagP functions as a transacylase, and its activity is absolutely essential for the formation of hepta-acylated lipid A (9, 10). Because lipid A is found exclusively in the outer leaflet of the outer membrane, one might also expect to find the PagP active site in that location.
Materials and Methods
PagP Mutagenesis, Expression, and Assay.
Site-directed mutations were generated by using the QuickChange protocol according to the manufacturer's instructions (Stratagene), using pETCrcAH (10) as the template. All oligonucleotide sequences used in this study can be obtained from the authors on request. PagP expression, membrane isolation, and protein analysis were performed as described previously (10) by using 15% SDS/PAGE without boiling the samples. Palmitoyl transferase reactions were performed as described previously (10) by using lipid IVA as the acyl acceptor.
Expression and Purification of 2H,15N,13C-Labeled PagP.
All isotopically labeled media and detergents were purchased from Cambridge Isotope Laboratories (Cambridge, MA). To express PagP in cytoplasmic inclusion bodies, the original pETCrcAH plasmid (10) was mutated by PCR so that the leader sequence was replaced by a single Met residue. E. coli BL21 (DE3) cells were transformed with the new pETCrcAHΔS vector and grown in 1 liter of 99.9% D2O M9 media containing 0.3% 2H,13C-glucose and 0.1% 15NH4Cl according to a previously published protocol (13). The cells were harvested and then suspended in 20 ml of chilled 50 mM Tris⋅HCl (pH 8.0) with 5.0 mM EDTA. After disrupting the cells by sonication, the lysate was spun down (35,000 × g). The pellet was resuspended in 20 ml of 50 mM Tris⋅HCl (pH 8.0) containing 2% Triton X-100 by vigorous stirring at room temperature for 1 h. After washing the pellet in the same buffer without Triton X-100, it was dissolved in 10 ml of 6 M Gdn-HCl, 50 mM Tris⋅HCl (pH 8.0), and purified by using standard nickel affinity chromatography procedures. Column fractions were assessed for purity by using SDS/PAGE. The purest fractions were pooled and then dialyzed twice (molecular mass cutoff of 3,500 Da) against 4 liters of 50 mM sodium phosphate (pH 6.0), yielding a white precipitate.
Refolding of PagP and NMR Sample Preparation.
For dodecylphosphocholine (DPC)-containing NMR samples, precipitated PagP was dissolved in 5 ml of 6 M Gdn-HCl, 50 mM sodium phosphate (pH 6.0). The final concentration was about 0.1 mM, determined by using an estimated extinction coefficient, ɛ280, of 82,630 M−1⋅cm−1 (14). The denatured solution was diluted dropwise into 35 ml of 50 mM sodium phosphate solution (pH 6.0), containing a final concentration of 0.2% d38-dodecylphosphocholine. Folding was quantitative, as monitored by SDS/PAGE. The sample was dialyzed twice in 4 liters of 50 mM sodium phosphate (pH 6.0) for 2 h and then concentrated down to 0.45 ml. Finally, 50 μl of D2O was added, yielding a 0.8 mM PagP, ≈16% DPC NMR sample.
For n-octyl-β-d-glucoside (β-OG)-containing NMR samples, precipitated PagP was dissolved in 1 ml of 5% perdeuterated SDS and dialyzed (molecular mass cutoff of 3,500 Da) for 5 days against 50 mM sodium phosphate (pH 6.0), to remove SDS. Thirty-two milligrams of perdeuterated β-OG was slowly dissolved, and then d5-EtOH was added to 1%. The sample was concentrated down to 450 μl and mixed with 50 μl of D2O, yielding a 1 mM PagP NMR sample in ≈200 mM β-OG. Both SDS and ethanol were required for 100% folding.
NMR Spectroscopy.
Backbone assignment experiments were carried out at 45°C on a Varian Inova 600-MHz spectrometer. Six different transverse relaxation-optimized spectroscopy (TROSY)-type (15) triple-resonance experiments (16) were recorded: HNCA, HN(CO)CA, HN(CA)CB, HN(COCA)CB, HNCO, and HN(CA)CO. Data were processed with NMRPipe (17) and analyzed with nmrview (18). The 4D 15N-edited NOESY experiments (19, 20) with 175-ms mixing times were recorded on a Varian Inova 800-MHz spectrometer; TROSY was not implemented because sensitivity gains were not realized in test experiments. All indirect dimensions were processed with linear prediction.
Relaxation and solvent exchange experiments were carried out on a Varian Inova 800-MHz spectrometer at 45°C. Relaxation rates were obtained by using standard experiments (21) modified to make use of the TROSY principle. For T1 experiments, peak volumes were fit against delay times ranging from 10 ms to 2 s. Delay times for T1ρ experiments ranged from 10 to 50 ms, during which the transverse 15N magnetization was subject to a 1.6-kHz spin-lock field. Peak volumes were fit to monoexponential decay curves to yield T1ρ time constants, which were subsequently converted to T2 (22). To measure heteronuclear 1H-15N nuclear Overhauser effects (NOEs), peak intensities in a spectrum obtained with a 7-s recycle delay followed by 5-s 1H presaturation were compared with a spectrum with no presaturation (12-s recycle delay). For solvent exchange studies, H2O NMR samples were lyophilized and then redissolved in D2O. 1H-15N TROSY-heteronuclear single quantum correlation (HSQC) spectra were recorded at various time intervals, ranging from 8 min to over 1 day.
Structure Calculations.
Backbone dihedral angles were calculated with talos (23) using chemical shift assignments. The chemical shifts were first corrected for deuterium isotope shifts. 1H-1H NOEs, obtained from 4D 15N-edited NOESY spectra, were separated into strong, medium, and weak, corresponding to distance restraints with upper limits of 3.5 Å, 5.0 Å, and 6.0 Å, respectively. Distances were calibrated by noting that cross-strand HN-HN distances in an antiparallel β-sheet are, on average, 3.3 Å. Slowly exchanging amide protons were identified through solvent exchange studies. In most cases, hydrogen bonding partners could be unambiguously assigned based on NOE patterns, and two distance restraints were used to simulate a hydrogen bond. NOE- and hydrogen bond-based distance restraints and backbone dihedral angle restraints were used as input for the simulated annealing protocol of cns v. 1.0 (24). Default settings were used, except that additional steps were necessary to ensure proper folding (2,000 high temperature steps, 6,000 torsion angle dynamics cooling steps, and 5,000 Cartesian coordinate dynamics cooling steps). Two hundred fifty structures were generated for PagP-DPC and for PagP-β-OG. Initial attempts at measuring dipolar couplings by using strain-induced alignment in a gel (ref. 25) have been hampered by poor signal-to-noise, due to an inability to obtain a sufficiently high protein concentration within the gel.
Results and Discussion
Mutagenesis Studies.
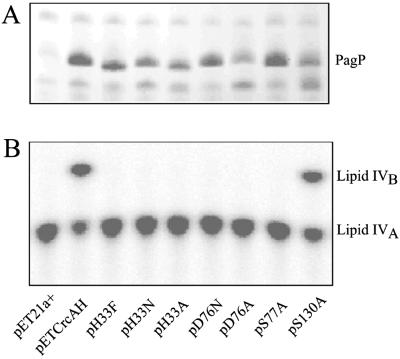
Many hydrolases and transferases possess an Asp-His-Ser catalytic triad (26), and PagP was initially suspected to use a similar arrangement in its catalytic mechanism. In PagP, His-33, Asp-76, and Ser-77 are absolutely conserved (Fig. 1A), and mutations at these loci drastically reduced enzymatic activity while leaving expression and membrane insertion intact (Fig. 2). Ser-130 was also mutated because it had been invariant before the discovery of the Legionella homologue (ref. 11; Fig. 1A). Not surprisingly, the S130A mutant was found to be fully functional (Fig. 2). Although these studies have identified three residues that are important for the enzymatic reaction in vitro, structural studies are needed to clarify their precise role in catalysis.
Figure 2.
Expression and activity of PagP-His6 and its site-directed mutant derivatives. (A) Membrane proteins were analyzed by SDS/PAGE and stained with Coomassie blue dye (10). The band corresponding to fully folded PagP is indicated. Note that unfolded PagP migrates more rapidly and did not accumulate for any of the mutants. (B) Palmitoyl transferase reactions were performed with induced membranes from the host strain BL21(DE3)pLysE at 100 ng/ml using 32P-lipid IVA as the acyl-acceptor, and the production of the palmitoylated metabolite lipid IVB was detected by TLC (10). The compounds on the plate were visualized by overnight exposure to a PhosphorImager (Molecular Dynamics) screen. pET21a+-transformed cells were used as a negative control, whereas pETCrcAH-transformed cells expressing wild-type PagP served as a positive control.
NMR Studies.
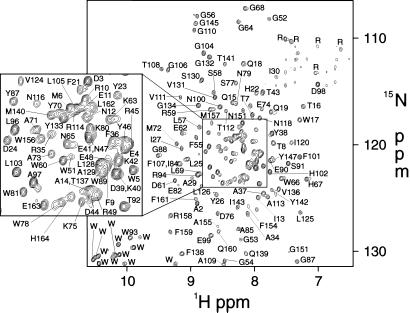
Recent developments in NMR, like TROSY (15), have allowed the successful global fold determinations of the outer membrane proteins OmpA (27) and OmpX (28), indicating that solution NMR would be a rapid and effective method for studying PagP structure. To examine the effects of various lipid environments, we studied PagP in both dodecylphosphocholine (PagP-DPC), a zwitterionic detergent, and n-octyl-β-d-glucoside (PagP-β-OG), a small nonionic detergent. To obtain sufficient quantities of protein for NMR analysis, PagP was overexpressed without its N-terminal signal peptide into cytoplasmic inclusion bodies. The protein was then denatured, purified, and refolded into detergent micelles (see Materials and Methods). The specific activity of refolded PagP was indistinguishable from that of native PagP purified from membranes (10). For PagP-DPC, chemical shift assignments (Fig. 3) were obtained for 153 of 162 residues (excluding Met-0, Asn-1, and the last six residues of the His-tag). All unassigned residues localized to the extracellular loops L1 [31–33], L3 [115, 117, and 119], and L4 [148, 149, 152, and 153], similar to NMR studies of OmpA, which were hampered by weak signals in the extracellular loop regions (27). For PagP-β-OG, the spectral quality for the extracellular loops was much improved, so that only L3 [115 and 119] could not be completely assigned. However, some periplasmic residues in T1 [60] and T2 [95 and 96] were broadened beyond detection. The different linewidths of these exposed residues illustrate the particular effects of different detergents on membrane protein structure.
Figure 3.
1H-15N TROSY-HSQC spectrum of 0.8 mM PagP-DPC, 45°C, at 800 MHz.
NMR Solution Folds.
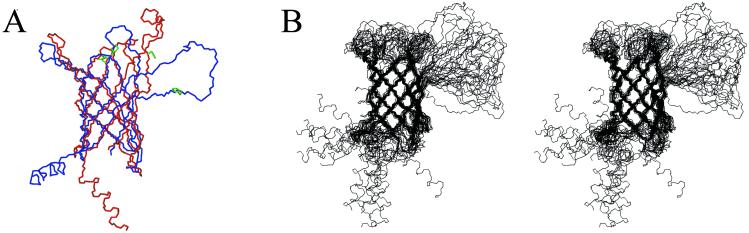
Global folds of PagP-DPC and PagP-β-OG were calculated based on HN-HN NOEs, hydrogen bond restraints, and chemical shift-derived backbone dihedral angle restraints (Table 1). The rms deviation (rmsd) between the average PagP-DPC and PagP-β-OG structures was 0.91 Å in the well defined region (see Fig. 4A), demonstrating that the protein can adopt its native fold in a variety of detergents and under different refolding conditions (see Materials and Methods). The main structural features of PagP are an eight-stranded antiparallel β-barrel formed by residues 22–161 preceded by an N-terminal α helix. The core of the barrel is well defined, whereas the solvent exposed surfaces are relatively disordered, particularly the large extracellular loop L1 (Fig. 4B). Although a precise positioning of the helix was not possible, it probably lies on the surface of the membrane, given its amphipathic nature. N-terminal α helices may be a feature common to other outer membrane proteins as well. In the electron crystallographic structure of the outer membrane phospholipase, OMPLA, an elongated domain was observed protruding from the β-barrel (29), most likely the first 18 residues at the N terminus (30). Unfortunately, this region was not visible in the high-resolution x-ray crystal structure of the protein (26).
Table 1.
Structural statistics for PagP-DPC and PagP-β-OG*
| PagP-DPC | PagP-β-OG | |
|---|---|---|
| Structural Information | ||
| HN-HN NOEs | 147 | 160 |
| Hydrogen bond restraints | 148 | 154 |
| Dihedral angle restraints | 234 | 248 |
| Backbone rmsd for well defined region, ņ | 0.90 ± 0.19 | 0.80 ± 0.13 |
| Backbone rmsd for all residues, ņ | 7.45 ± 1.00 | 6.44 ± 1.21 |
| Ramachandran map analysis‡ | ||
| Most favored regions | 77.6% | 74.8% |
| Additionally allowed regions | 16.8% | 19.6% |
| Generously allowed regions | 4.9% | 4.2% |
| Disallowed regions | 0.7% | 1.4% |
| Deviations from restraints and idealized geometry | ||
| Distance restraints, Å | 0.0021 ± 0.0005 | 0.0040 ± 0.0008 |
| Dihedral angle restraints, ° | 0.020 ± 0.006 | 0.039 ± 0.008 |
| Bonds, Å | 0.0007 ± 0.00001 | 0.0007 ± 0.00003 |
| Angles, ° | 0.2391 ± 0.0005 | 0.243 ± 0.002 |
| Impropers, ° | 0.0783 ± 0.0007 | 0.081 ± 0.002 |
The 20 lowest energy structures were included.
Shows rmsd to the average structure. Residues 22–27, 53–61, 65–75, 81–93, 102–113, 121–133, 136–143, and 153–162 were superimposed.
procheck-nmr (43) analysis was performed on the lowest energy conformer.
Figure 4.
(A) Superposition of the lowest energy PagP-DPC structure (blue) and the lowest energy PagP-β-OG structure (red). The side chains of the residues implicated in catalysis, His-33, Asp-76, and Ser-77, are shown in green. (B) Stereoview of the 20 lowest energy structures (residues 3–163) of PagP in DPC micelles.
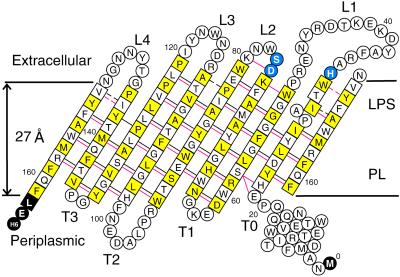
All outer membrane β-barrels seem to fold according to a set of rules (31). For example, the N and C termini of the protein are always found facing the periplasm. In PagP, the amphipathic N-terminal α helix is therefore on the periplasmic side of the membrane. The extracellular loops are generally longer than their periplasmic counterparts, a feature seen in the other β-barrel structures. The β-strands are tilted with respect to the bilayer normal, so that the shear number (32) of the β-barrel is the expected n + 2 = 10, where n is the number of β-strands in the barrel (eight in PagP). This configuration places His-33, Asp-76, and Ser-77 just beyond the outer edge of the 27-Å membrane bilayer (ref. 33; Fig. 5). These catalytically important residues are thus positioned at the depth required to interact with polar headgroups in the outer membrane outer leaflet. Surprisingly, however, Asp-76 and Ser-77 (L2) are well separated from His-33 (L1), suggesting that His-33 may not be part of a catalytic triad. [In the PagP-DPC ensemble of structures, the mean distance between Cɛ1 of His-33 and Oγ of Ser-77 is 23.4 Å; the closest distance is 15.1 Å. In contrast, the corresponding distance in the OMPLA crystal structure (26) is only 4.8 Å.] The absence of His-Ser contact is similar to what has been observed for human cytosolic phospholipase A2 (cPLA2; ref. 34). In cPLA2, the nucleophilic Ser-228 is flanked by Asp-549, but there is no histidine residue important to catalytic activity (35). Arg-200 is also important for activity, but it is located 9 Å from Ser-228 and believed to play a role in substrate binding. His-33 may play a similar role in PagP. (However, it is still possible that in vivo a dimer is formed that might juxtapose His-33 between Asp-76 and Ser-77 from another monomer.) As was suggested for cPLA2, the increase in activation energy by not having a catalytic triad could be compensated by improved stabilization of the tetrahedral transition state, consistent with the high selectivity for palmitate displayed by PagP (10, 36). There are a number of highly conserved polar residues near His-33, Asp-76, and Ser-77 that could also play a role in catalysis. Unfortunately, a precise picture of the active site could not be obtained because this region is not well ordered in the NMR structures, prompting a further investigation into the conformational dynamics of PagP.
Figure 5.
Topology model of PagP. Slowly exchanging backbone amides (exchange time constant >5 min) are shown with black lines, and HN-HN NOE connectivities are shown in red. Dashed lines indicate observations made for PagP-β-OG but not PagP-DPC, due to exchange broadening in the latter case. Residues 13–19, 64, 92, 96, 98, and 101 exhibit slow hydrogen exchange as well. Residues in squares are part of β-strands. Secondary structure is based on hydrogen bonding. Residues in yellow squares have side chains facing the membrane bilayer, whereas white squares indicate side chains lining the interior of the β-barrel. The presence of the β-bulge with the extension of strand A to Trp-32 is based on NOE data from PagP-β-OG; residues 31–33 and 152–153 are broadened beyond detection in PagP-DPC. The three residues important for catalytic activity as determined by site-directed mutagenesis, His-33, Asp-76, and Ser-77, are shown in blue.
PagP Dynamics.
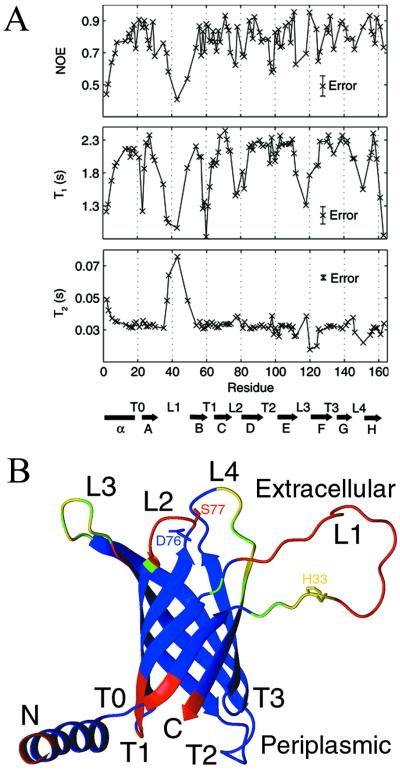
Backbone 15N T1, 15N T2 (from T1ρ), and 1H-15N NOE values were measured at 800 MHz and used to calculate a rotational diffusion tensor according to a previously published protocol (37). Anisotropic tumbling models were unable to account for the wide range of observed T1 values (see Fig. 6A), yielding only a very modest, and not statistically significant, improvement in fit. The overall isotropic rotational correlation time for PagP-DPC is 20 ns at 45°C. Based on the 18.6-ns correlation time for the 42-kDa maltose binding protein (MBP)-β-cyclodextrin complex at 37°C (37), the PagP-DPC micelle is estimated to have a molecular mass between 50 and 60 kDa, similar to what was estimated for the monomeric OmpA (27) and OmpX (28) protein-detergent micelles by NMR. Moreover, PagP migrates predominantly as a monomer in SDS/PAGE (10), which seems to preserve oligomeric states of β-barrel membrane proteins (38). Hence, under the conditions studied, PagP appears to exist as a monomer.
Figure 6.
(A) Heteronuclear 1H-15N NOE, 15N T1, and 15N T2 values for PagP-DPC measured at 800 MHz plotted against PagP sequence and secondary structure. The relaxation measurements performed on PagP-β-OG displayed similar trends. (B) Ribbon representation of PagP in DPC. Residues colored in red have backbone 15N T1s that are less than 80% of the value predicted for a 20-ns overall correlation time, S2 = 0.85, τe = 10 ps (2.3 s). Yellow residues have NMR signals too faint to be detected in 1H-15N HSQC spectra, and green residues have a weak but observable signal. All other residues are shown in blue.
T1 values are exquisitely sensitive to the nanosecond timescale internal dynamics exhibited by PagP because the molecular tumbling time is large (20 ns). T1 times drop considerably for most loops (Fig. 6A), with the exceptions of T2 and T3. (The long T2 loop is actually fairly structured, as evidenced by nonrandom coil chemical shifts, medium-range 1H-1H NOEs, and slowly exchanging backbone amides.) Nanosecond timescale motions tend to increase T2, but motions with characteristic times in the μs-ms range can reduce T2 through exchange broadening. In mobile regions of PagP (for example, L3), T2 values seem to fluctuate about the mean, indicating that areas with high mobility on the nanosecond timescale may also experience μs-ms motions.
As shown in Fig. 6B, there appear to be three highly mobile clusters in PagP. The long extracellular loop, L1, is particularly dynamic. Two proline residues, Pro-28 and Pro-50, further contribute to its mobility by limiting the number of hydrogen bonds that can form between strands A and B. Pro-28 is part of a β-bulge that is well defined in β-OG (see Fig. 5) but could not be well characterized in DPC due to exchange broadening, demonstrating how the stability and dynamics of interfacial regions are sensitive to the local micelle environment. In addition to Pro-28 and Pro-50, there are a number of conserved proline residues found in the β-strands of PagP, highly unusual given the distinct conformational preferences of proline (39). It is interesting to note the high degree of mobility in the extracellular region of PagP, which also contains the active site.
The positioning of its active site on the extracellular boundary of the outer membrane, comprised of a high concentration of lipid A, explains why PagP exclusively palmitoylates lipid A in vivo (9). What remains unresolved is how phospholipids, which are not normally found in the outer leaflet of the outer membrane, find their way to the active site of PagP. It may be that PagP is unnecessary under resting physiological conditions. However, during infection, PagP expression is up-regulated by the PhoPQ regulon in response to Mg2+ limitation. The lack of Mg2+ in lipopolysaccharide binding sites can also destabilize the outer membrane (40), permitting phospholipids to migrate to the outer leaflet where they can serve as acyl donors for PagP. Antimicrobial cationic peptides are also known to promote lipid flip-flop in a membrane bilayer (41). The ensuing disruption of lipid asymmetry can be “sensed” by PagP, which responds by acylating lipid A and conferring resistance to the bactericidal peptides. PagP may thus provide an adaptive response to stressors that challenge the integrity of the outer membrane, similar to what has been proposed for the outer membrane enzymes OMPLA (26) and OmpT (42).
Acknowledgments
We thank Dr. Ranjith Muhandiram for assistance in setting up some of the NMR experiments. P.M.H. acknowledges the Canadian Institutes of Health Research (CIHR) for a predoctoral award. This research was supported by operating grants from the CIHR (to G.G.P., J.D.F.-K., R.E.B., and L.E.K.) and by National Institutes of Health Grant GM-51310 (to C.R.H.R.). L.E.K. holds a Canada Research Chair in Biochemistry.
Abbreviations
- PagP
PhoPQ-activated gene P
- DPC
dodecylphosphocholine
- β-OG
n-octyl-β-d-glucoside
- TROSY
transverse relaxation-optimized spectroscopy
- NOE
nuclear Overhauser effect
- HSQC
heteronuclear single quantum correlation
- rmsd
rms deviation
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.rcsb.org (PDB ID codes 1MM4 and 1MM5).
Bishop, R. E. & Raetz, C. R. H., Fortieth Interscience Conference on Antimicrobial Agents and Chemotherapy, September 17–20, 2000, Toronto, ON, Canada, abstr. 2290.
References
- 1.Raetz C R H, Whitfield C. Annu Rev Biochem. 2002;71:635–700. doi: 10.1146/annurev.biochem.71.110601.135414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aderem A, Ulevitch R J. Nature. 2000;406:782–787. doi: 10.1038/35021228. [DOI] [PubMed] [Google Scholar]
- 3.Christ W J, Asano O, Robidoux A L, Perez M, Wang Y, Dubuc G R, Gavin W E, Hawkins L D, McGuinness P D, Mullarkey M A, et al. Science. 1995;268:80–83. doi: 10.1126/science.7701344. [DOI] [PubMed] [Google Scholar]
- 4.Guo L, Lim K B, Gunn J S, Bainbridge B, Darveau R P, Hackett M, Miller S L. Science. 1997;276:250–253. doi: 10.1126/science.276.5310.250. [DOI] [PubMed] [Google Scholar]
- 5.Ernst R K, Yi E C, Guo L, Lim K B, Burns J L, Hackett M, Miller S I. Science. 1999;286:1561–1565. doi: 10.1126/science.286.5444.1561. [DOI] [PubMed] [Google Scholar]
- 6.Garcia Vescovi E, Soncini F C, Groisman E A. Cell. 1996;84:165–174. doi: 10.1016/s0092-8674(00)81003-x. [DOI] [PubMed] [Google Scholar]
- 7.Groisman E A. J Bacteriol. 2001;183:1835–1842. doi: 10.1128/JB.183.6.1835-1842.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tanamoto K, Azumi S. J Immunol. 2000;164:3149–3156. doi: 10.4049/jimmunol.164.6.3149. [DOI] [PubMed] [Google Scholar]
- 9.Guo L, Lim K B, Poduje C M, Daniel M, Gunn J S, Hackett M, Miller S I. Cell. 1998;95:189–198. doi: 10.1016/s0092-8674(00)81750-x. [DOI] [PubMed] [Google Scholar]
- 10.Bishop R E, Gibbons H S, Guina T, Trent M S, Miller S I, Raetz C R. EMBO J. 2000;19:5071–5080. doi: 10.1093/emboj/cdd507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Robey M, O'Connell W, Cianciotto N P. Infect Immun. 2001;69:4276–4286. doi: 10.1128/IAI.69.7.4276-4286.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Aron L, Jones S, Fielding C J. J Biol Chem. 1978;253:7220–7226. [PubMed] [Google Scholar]
- 13.Gardner K H, Kay L E. Annu Rev Biophys Biomol Struct. 1998;27:357–406. doi: 10.1146/annurev.biophys.27.1.357. [DOI] [PubMed] [Google Scholar]
- 14.Gill S C, von Hippel P H. Anal Biochem. 1989;182:319–326. doi: 10.1016/0003-2697(89)90602-7. [DOI] [PubMed] [Google Scholar]
- 15.Pervushin K, Riek R, Wider G, Wüthrich K. Proc Natl Acad Sci USA. 1997;94:12366–12371. doi: 10.1073/pnas.94.23.12366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yang D, Kay L E. J Am Chem Soc. 1999;121:2571–2575. [Google Scholar]
- 17.Delaglio F, Grzesiek S, Vuister G W, Zhu G, Pfeifer J, Bax A. J Biomol NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 18.Johnson B A, Blevins R A. J Biomol NMR. 1994;4:603–614. doi: 10.1007/BF00404272. [DOI] [PubMed] [Google Scholar]
- 19.Grzesiek S, Wingfield P, Stahl S, Kaufman J, Bax A. J Am Chem Soc. 1995;117:9594–9595. [Google Scholar]
- 20.Venters R A, Metzler W J, Spicer L D, Mueller L, Farmer B T. J Am Chem Soc. 1995;117:9592–9593. [Google Scholar]
- 21.Farrow N A, Muhandiram R, Singer A U, Pascal S M, Kay C M, Gish G, Shoelson S E, Pawson T, Forman-Kay J D, Kay L E. Biochemistry. 1994;33:5984–6003. doi: 10.1021/bi00185a040. [DOI] [PubMed] [Google Scholar]
- 22.Peng J W, Wagner G. J Magn Reson. 1992;98:308–332. [Google Scholar]
- 23.Cornilescu G, Delaglio F, Bax A. J Biomol NMR. 1999;13:289–302. doi: 10.1023/a:1008392405740. [DOI] [PubMed] [Google Scholar]
- 24.Brunger A T, Adams P D, Clore G M, DeLano W L, Gros P, Grosse-Kunstleve R W, Jiang J S, Kuszewski J, Nilges M. Acta Crystallogr D Biol Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 25.Ishii Y, Markus M A, Tycko R. J Biomol NMR. 2001;21:141–151. doi: 10.1023/a:1012417721455. [DOI] [PubMed] [Google Scholar]
- 26.Snijder H J, Ubarretxena-Belandia I, Blaauw M, Kalk K H, Verheij H M, Egmond M R, Dekker N, Dijkstra B W. Nature (London) 1999;401:717–721. doi: 10.1038/44890. [DOI] [PubMed] [Google Scholar]
- 27.Arora A, Abildgaard F, Bushweller J H, Tamm L K. Nat Struct Biol. 2001;8:334–338. doi: 10.1038/86214. [DOI] [PubMed] [Google Scholar]
- 28.Fernandez C, Adeishvili K, Wüthrich K. Proc Natl Acad Sci USA. 2001;98:2358–2363. doi: 10.1073/pnas.051629298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Boekema E J, Stuart M, Koning R I, Keegstra W, Brisson A, Verheij H M, Dekker N. J Struct Biol. 1998;123:67–71. doi: 10.1006/jsbi.1998.4013. [DOI] [PubMed] [Google Scholar]
- 30.Dekker N. Mol Microbiol. 2000;35:711–717. doi: 10.1046/j.1365-2958.2000.01775.x. [DOI] [PubMed] [Google Scholar]
- 31.Schulz G E. Curr Opin Struct Biol. 2000;10:443–447. doi: 10.1016/s0959-440x(00)00120-2. [DOI] [PubMed] [Google Scholar]
- 32.Liu W M. J Mol Biol. 1998;275:541–545. doi: 10.1006/jmbi.1997.1501. [DOI] [PubMed] [Google Scholar]
- 33.Wimley W C. Protein Sci. 2002;11:301–312. doi: 10.1110/ps.29402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dessen A, Tang J, Schmidt H, Stahl M, Clark J D, Seehra J, Somers W S. Cell. 1999;97:349–360. doi: 10.1016/s0092-8674(00)80744-8. [DOI] [PubMed] [Google Scholar]
- 35.Pickard R T, Chiou X G, Strifler B A, DeFelippis M R, Hyslop P A, Tebbe A L, Yee Y K, Reynolds L J, Dennis E A, Kramer R M, Sharp J D. J Biol Chem. 1996;271:19225–19231. doi: 10.1074/jbc.271.32.19225. [DOI] [PubMed] [Google Scholar]
- 36.Brozek K A, Bulawa C E, Raetz C R H. J Biol Chem. 1987;262:5170–5179. [PubMed] [Google Scholar]
- 37.Hwang P M, Skrynnikov N R, Kay L E. J Biomol NMR. 2001;20:83–88. doi: 10.1023/a:1011226512421. [DOI] [PubMed] [Google Scholar]
- 38.Dekker N, Tommassen J, Lustig A, Rosenbusch J P, Verheij H M. J Biol Chem. 1997;272:3179–3184. doi: 10.1074/jbc.272.6.3179. [DOI] [PubMed] [Google Scholar]
- 39.Li S-C, Goto N K, Williams K A, Deber C M. Proc Natl Acad Sci USA. 1996;93:6676–6681. doi: 10.1073/pnas.93.13.6676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kotra L P, Golemi D, Amro N A, Liu G-Y, Mobashery S. J Am Chem Soc. 1999;121:8707–8711. [Google Scholar]
- 41.Matsuzaki K. Biochim Biophys Acta. 1998;1376:391–400. doi: 10.1016/s0304-4157(98)00014-8. [DOI] [PubMed] [Google Scholar]
- 42.Vandeputte-Rutten L, Kramer R A, Kroon J, Dekker N, Egmond M R, Gros P. EMBO J. 2001;20:5033–5039. doi: 10.1093/emboj/20.18.5033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Laskowski R A, Rullmannn J A, MacArthur M W, Kaptein R, Thornton J M. J Biomol NMR. 1996;8:477–486. doi: 10.1007/BF00228148. [DOI] [PubMed] [Google Scholar]
- 44.Nielsen H, Engelbrecht J, Brunak S, von Heijne G. Protein Eng. 1997;10:1–6. doi: 10.1093/protein/10.1.1. [DOI] [PubMed] [Google Scholar]