Abstract
The nuclei of eukaryotic cells consist of discrete substructures. These substructures include the nuclear bodies, which have been implicated in a number of biological processes such as transcription and splicing. However, for most nuclear bodies, the details of involvement in these processes in relation to their three-dimensional distributions in the nucleus are still unclear. Through the analysis of TDP, a protein functional in both transcriptional repression and alternative splicing, we have identified a new category of nuclear bodies within which the TDP molecules reside. Remarkably, the TDP bodies (TBs) colocalize or overlap with several different types of nuclear bodies previously suggested to function in transcription or splicing. Of these nuclear bodies, the Gemini of coiled bodies (GEM) seems to associate with TB through the interaction between survival motor neuron (SMN) protein and TDP. Furthermore, TB sometimes appears to be the bridge of two or more of these other nuclear bodies. Our data suggest the existence of a hierarchy and possibly functional arrangement of the nuclear bodies within the eukaryotic nuclei.
Keywords: nuclear structure‖splicing‖transcription
The eukaryotic nucleus is a cellular compartment in which a number of biological processes occur, such as DNA replication and repair, gene expression through transcription and splicing, protein modification and degradation, etc. However, the variety of biological materials in the nucleus is not distributed randomly (1–5). For instance, the interphase chromosomes each occupy a distinct territory, and this organization has been suggested to play a regulatory role in gene expression from the chromosomes (5). In addition, there exist the structurally and functionally well defined nucleoli and the different types of nuclear bodies identified through the use of cytological and immunohistochemical tools (1–4).
The nuclear bodies range in sizes from tenths of a micrometer to several micrometers, and each type could be easily identified immunologically with antibodies against specific “signature proteins” or other factors such as the small nuclear (sn)RNAs (1–4). Among the known nuclear bodies are the Cajal body or coiled body (CB; ref. 3 and references therein), the promyelocytic leukemia (PML) body or POD (ref. 6 and references therein), a class of splicing-related bodies including the SC35 speckles or interchromatin granule cluster (ref. 2 and references therein), the GEM body (7), and the more recently identified matrix-associated deacetylase body (8), HAP body (9), and nucleoli-associated paraspeckles (10). The integrity of a nuclear body could be disrupted after depletion of its normal component(s), as evidenced by the disruption of PODs in acute PML (reviewed in ref. 1). Because of the relatively limited materials for biochemical assays (11), the functions of most of these nuclear bodies are not well defined, although some of them have been suggested to be the storage sites of specific factors and/or stations for assembly of functional complexes to carry out specific biological processes (refs. 1–4; see Discussion). Except for the Cajal body and GEM, which are obviously colocalized (7), there appears to be not much association among the different nuclear bodies. Finally, unlike the interphase chromosomes, it is not known whether there exist specific patterns of interconnection and distributions of nuclear bodies in the nucleus, either among the individual ones of the same type or between the different categories.
TDP-43 was cloned initially as a human protein capable of binding to a polypyrimidine-rich motif in the HIV transactive response DNA (12). Presumably through this binding, TDP-43 represses transcription in vitro from the HIV long-terminal repeat promoter but not the major late adenovirus promoter. In addition, coexpression of TDP-43 resulted in repression of the HIV-1 proviral gene expression (12). TDP-43 was independently identified later as part of a 50-kDa complex that is involved in the splicing of the cystic fibrosis transmembrane conductance regulator gene, or CTFR (13). In particular, overexpression of TDP-43 could cause exon-skipping of the CTFR exon 9, and this skipping is through TDP-43 binding to a (UG) m(U)n motif near the 3′ splice site of the CTFR intron 8 (13). Indeed, TDP-43 contains the so-called RNA-recognition motifs (RRMs) and a glycine-rich sequence, both of which are functional modules commonly found in RNA-binding proteins (refs. 14 and 15 and references therein). Interestingly, at least the first RRMs of TDP-43 are required for TDP binding to DNA and RNA (12, 16), and by implication they bifunction to repress transcription and enhance exon skipping.
We present evidence below that the mammalian TDP proteins are also concentrated in discrete nuclear substructures. Significant overlapping and/or colocalization of this TDP-concentrated substructure with several previously identified nuclear bodies suggest that different types of nuclear bodies could be linked extensively to one another to carry out tightly coupled functions such as transcription and splicing.
Materials and Methods
Plasmid Constructs.
The molecular, cellular, and immunological methods used in this study followed the standard protocols (17, 18). The expression plasmids pEF-FLAG-mTDP-L, pFF-FLAG-mTDP-S, and pCMV-eGFP-mTDP-S contain the full-length mouse (m)TDP-L or mTDP-S coding sequence fused with either FLAG or enhanced GFP (eGFP). Both cDNAs of mTDP-L and mTDP-S were generated by standard RT-PCR of total RNA isolated from the mouse brain with Trizol (GIBCO). cDNA synthesis was carried out with Superscriptase II (GIBCO), and PCR was performed with specific primer pairs. The sequences of the primers are: forward, 5′-aagcttatgtctgaatatattcgggtaac-3′ (for cloning in the pEF-FLAG vector) and 5′-aagcttcgatgtctgaatatattcgggtaac-3′ (for the pCMV-eGFP vector); backward, 5′-gcggccgcattccccagccagaagacttag-3′, which is complementary to nucleotides 1,300–1,322 of human (h)TDP-43 cDNA (for cloning of pEF-FLAG-mTDP-L; Fig. 1), 5′-gcggccgctggaacacccaccgtcaaag-3′ (for pEF-FLAG-mTDP-S), and 5′-ggatcctggaacacccaccgtcaaag-3′ (for pCMV-eGFP-mTDP-S). The latter two primers are complementary to nucleotides 2,016–2,036 of the hTDP-43 3′-untranslated region. The PCR products were sequenced and cloned as a partial HindIII–NotI fragment into pEF-FLAG (a kind gift from Xin Chen, National Taiwan Hospital, Taiwan, China), and as a HindIII–BamHI fragment into pCMV-eGFP (CLONTECH). Further details are available upon request. The reporter plasmids pG0-α-Luc and pG0-TK-Luc have been used and described (19).
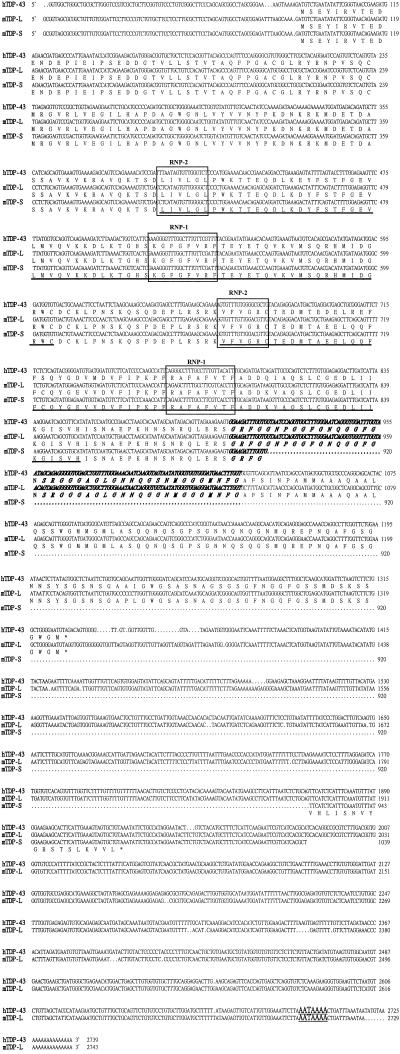
Figure 1.
Sequence alignment of hTDP-43, mTDP-L, and mTDP-S. The nucleotide and amino acid sequences of the cDNAs encoding hTDP-43, mTDP-L, and mTDP-S, respectively, are aligned for comparison. The mTDP-L sequence was put together through sequencing of the RT-PCR products and of a mouse EST clone (GenBank accession no. AA116656). The two RRM domains conserved in all three proteins are underlined, and the RNP-1 and RNP-2 motifs within the two domains are boxed. The glycine-rich region present in hTDP-43 and mTDP-L, but not in mTDP-S, are indicated with italic letters. The polyadenylation signal near the 3′ ends of the hTDP-L and mTDP-L cDNAs are indicated with large, underlined letters.
Antibodies.
The anti-PML rabbit antibody was purchased from Santa Cruz Biotechnology. The anti-SC35 and anti-FLAG(M2) mouse antibodies were from Sigma. The anti-survival motor neuron (SMN) monoclonal mouse antibody was from Transduction Laboratories (Lexington, KY). Anti-SMN and anticoilin rabbit antibodies were kind gifts from Hung Li and C.-S. Wu, respectively. The secondary antibodies labeled with Cy5 or Cy3 were from Santa Cruz Biotechnology.
Cell Culture and DNA Transfection.
Human 293T or 293 cells were maintained as monolayers in DMEM supplemented with 100 units/ml penicillin-streptomycin and 10% FCS (GIBCO). Cultures were maintained at 37°C and 5% CO2. One day before transfection, the cells were seeded at 50–70% confluence. The calcium phosphate precipitation method was used for transfection with 1 μg of the reporter plasmid and varying amounts of the expression plasmids. After 12 h of transfection incubation, the medium was changed, and the cells were incubated further for 24 h. The expression of the FLAG-tagged mTDP-L or mTDP-S protein then was determined by Western blot analysis of the extracts by using anti-FLAG(M2) as the hybridization probe. The luciferase activity from the reporter plasmid, pG0-α-Luc or pG0-TK-Luc, was assayed also. Because the mTDP proteins behave as a general transcription repressor, it was not feasible to use other reporter plasmids as the internal standard. Instead, the cell numbers were used for calibration of the luciferase activities.
Immunohistochemistry.
For the immunostaining experiments, cells were plated onto gelatin-coated coverslides and transfected with pEF-FLAG-mTDP-L, pEF-FLAG-mTDP-S, or pCMV-eGFP-mTDP-S. After 12–24 h of incubation with the transfection mixture, the cells were fixed with 3.7% formaldehyde in PBS at 4°C for 15 min and then permeabilized with 0.5% Triton X-100. Antigen localization was determined after incubation of the permeabilized cells with appropriate antibodies at 4°C overnight. Secondary antibodies conjugated to fluorescein, Cy5 or Cy3, were applied then for 2 h at room temperature. When DNA staining was needed, the cells were treated also with bis-benzimide (Hoechst 33258, Molecule Probes). Finally, the coverslides were mounted with Fluoromount G (Fisher Scientific), and the fluorescent images from eGFP, the secondary antibodies, and Hoechst were analyzed in a Zeiss laser confocal microscope (LSM510). Each confocal image is from a single optical section. 3D modeling was carried out by analysis of images from serial sections with the Zeiss 3D VISART program.
Cross-Immunoprecipitation (IP).
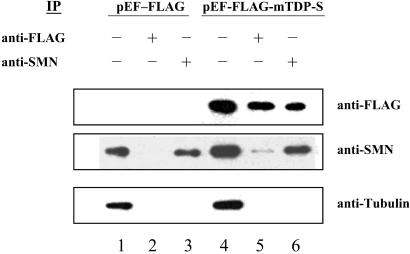
The interaction between SMN protein and FLAG-mTDP in vivo was studied by cross-IP analysis of extracts prepared from 293 cells transiently transfected with pEF-FLAG vector, pEF-FLAG-mTDP-S, or pEF-FLAG-mTDP-L. For IP by anti-FLAG(M2), the cells at 24 h posttransfection were lysed by three freeze-thaw cycles in isotonic buffer (10 mM Hepes/142.5 mM KCl/5 mM MgCl2/1 mM EGTA/0.2% Nonidet P-40/1 mM PMSF/0.2% protease inhibitor mixture). For IP by anti-SMN rabbit antibody, the cells were lysed in the modified RIPA buffer (50 mM Tris⋅HCl, pH 8.0/150 mM NaC1/10 mM MgCl2/0.4% Nonidet P-40/0.05% SDS/1 mM PMSF/0.2% protease inhibitor mixture). The cell lysates were precleaned with 50% protein A beads in the lysis buffer for 30 min at 4°C. They then were incubated with anti-FLAG(M2) or anti-SMN rabbit antibody at 4°C. The bound immunoprecipitates to protein-A beads were washed, boiled, and analyzed by Western blot using anti-FLAG(M2), anti-SMN mouse antibody, or antitubulin as the hybridization probe. The hybridizing bands were developed with the enhanced chemiluminescence Western blotting detection system (Perkin–Elmer).
Results
Cloning of the mTDP-43 Homolog and a Shorter Isoform.
hTDP-43 has an approximate molecular mass of 43 kDa on SDS/PAGE gel, and it is 414-aa long (12). Two partial cDNA sequences corresponding to hTDP-43 could be found in the mouse EST database (GenBank accession nos. BC012873 and AA116656, respectively). Expression of the mTDP-43 ortholog was confirmed by cDNA cloning using degenerate primers complementary to the 5′ end and 3′ region of the hTDP-43 cDNA (see Materials and Methods). Interestingly, a shorter isoform was also identified in this way. We termed the two mouse cDNAs mTDP-L and mTDP-S, respectively.
As shown in Fig. 1, mTDP-L polypeptide is identical in length to hTDP-43, and the two proteins share 96% in identity and 99% in similarity. The two RRM domains and the glycine-rich region previously identified in hTDP-43 are all conserved in mTDP-L. Sequence alignment of mTDP-L and mTDP-S cDNAs showed that they are identical in the 5′-untranslated region (88 bp) and the first 840 bp of the coding region. Thus, the two proteins most likely are generated by alternative splicing of an mTDP primary transcript. After amino acid 281, mTDP-L and mTDP-S become different in their amino acid sequences as well as the lengths (see also Discussion).
Transcriptional Repression by the mTDP Proteins.
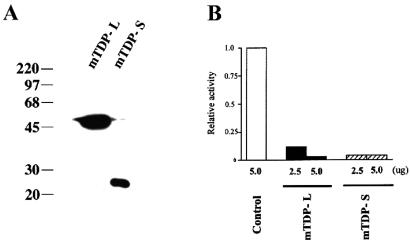
Because the RRM domains of hTDP-43 have been implicated in transcriptional repression and exon skipping, presumably through their abilities to bind to both DNA and RNA (12, 13, 16), we tested by DNA transfection assay whether the two mTDP proteins could also function in transcriptional regulation. As shown in Fig. 2A, both mTDP-L and mTDP-S could be expressed in transfected cell cultures to generate FLAG-tagged polypeptides with approximate molecular masses of 45 and 27 kDa, respectively, as expected from the cDNA sequence analysis in Fig. 1. Either protein could repress the activities of cotransfected human α-globin promoter (Fig. 2B) or mouse TK promoter (data not shown) in human 293 cells. Thus, as their human counterpart, mTDP-L and mTDP-S could function as transcription repressors.
Figure 2.
Transcriptional repression by mTDP-L and mTDP-S. 293 cells were cotransfected with 1 μg of the reporter plasmids (pG0-α-Luc) and increasing amounts (2.5 and 5 μg) of the expression plasmids (pEF-FLAG-mTDP-L or pEF-FLAG-mTDP-S) or the vector. At 24 h posttransfection, the cells were harvested for analysis of expression of the FLAG-tagged mTDP-L and mTDP-S by Western blotting (A) and for an assay of their effects on the expression of the cotransfected reporter plasmids (B).
Cellular Localization of mTDP-L and mTDP-S.
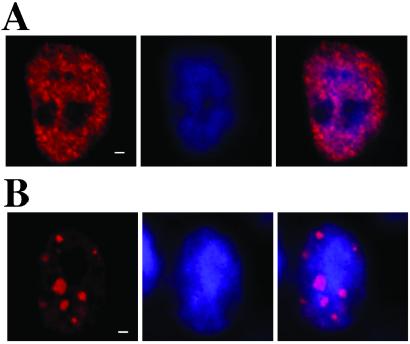
The cellular localizations of mTDP-L and mTDP-S were determined by transient expression of the two proteins tagged with either FLAG or eGFP followed by analysis of the fluorescent images under the confocal microscope. As exemplified for the FLAG-tagged proteins in transfected 293 cells, ≈90% of mTDP-L molecules are confined in the nucleus with a punctuated pattern (Fig. 3A). On the other hand, while also located in the nucleus, mTDP-S exhibited mainly a speckle-like pattern (Fig. 3B). The expression patterns of GFP fusions of the two mTDP proteins parallel those of the FLAG-tagged ones described above (see below). Limited study of a transfected mouse neuro2A cell line, either transiently or stably expressing the tagged mTDP-S, gave similar results (data not shown). Because of compatibility of available antibodies for the immunostaining experiments (see below), the human 293 cells, instead of the mouse cell lines, are used routinely for transfection.
Figure 3.
Cellular immunostaining patterns of FLAG-mTDP-L and FLAG-mTDP-S. 293 cells were transfected with pEF-FLAG-mTDP-L (A) or pEF-FLAG-mTDP-S (B) and analyzed by immunostaining with anti-FLAG antibody (red, A and B Left) or with the Hoechst DNA dye (blue, A and B Center). The merged images are shown in A and B Right. (Scale bar, 1 μm.)
Spatial Relationship Between the TDP Bodies (TBs) and Other Nuclear Bodies.
To facilitate the investigation of the structure and possible function of the speckles containing mTDP-S, which we now termed TBs, 293 cells were transiently transfected with plasmid pCMV-eGFP-mTDP-S, and the locations of the GFP-mTDP-S fusion molecules were examined in the confocal microscope at 12 h posttransfection. The distribution pattern of eGFP-mTDP-S is similar to the FLAG-tagged molecules, as exemplified by the cotransfection experiment with pCMV-eGFP-mTDP-S and pEF-FLAG-mTDP-S in Fig. 4A. The majority of the GFP-positive nuclei contain 1 to more than 12 TB speckles, the sizes of which in general vary in the range of 0.6–2.6 μm.
Figure 4.
Immunostaining patterns of eGFP-mTDP-S-expressing cells. 293 cells cotransfected with pCMV-eGFP-mTDP-S and pEF-FLAG-mTDP-S (A) or transfected with pCMV-eGFP-mTDP-S alone (B–D) were immunostained with antibodies against the FLAG epitope (A), coilin (B), SMN (C), or PML (D). The images of the green fluorescence from eGFP-mTDP-S (Aa–Da) and the red fluorescence from the secondary antibodies (Ab–Db) are merged in Ac–Dc. The overlapping/colocalized regions exhibit the yellow color, as shown in Ac and exemplified with the arrows in Bc–Dc. (Scale bar, 1 μm.)
To determine the spatial relationship between TB and the other nuclear substructures identified previously by others, 293 cells transfected with pCMV-eGFP-mTDP-S were immunostained with antibodies against the signature proteins of these other nuclear substructures (Fig. 4 B–D).
CB.
The CB, also known as the Cajal body or sn ribonucleoprotein (RNP) foci, contains high concentrations of snRNAs and snRNPs (refs. 3 and 20 and references therein). There are usually only a few CBs per nucleus, and they together with the GEM bodies are likely the “stations” for recycling of the snRNPs after mRNA splicing (21). It has been suggested also to be temporarily associated with chromatin containing actively transcribing genes. One of the signature proteins for CB is coilin (22). As exemplified in Fig. 4B, the CBs significantly overlap with TBs. Almost every TB-positive nucleus has at least one TB overlapping with a CB. Occasionally, a CB could colocalize with TB. Overall, 40% of the TBs examined overlap/colocalize with a CB. It should be noted that the average number of CBs per cell is ≈3 for either eGFP-mTDP-S-positive or eGFP-mTDP-S-negative 293 cells, indicating that there is little induced formation of CBs by the overexpressed eGFP-mTDP-S molecules.
GEM Body.
Previously, immunostaining with the use of antibodies against the Survival of Motor Neuron protein, or SMN, has defined a unique nuclear substructure named GEM (7). Reduction in the amount or mutation of SMN could lead to the development of spinal muscular atrophy, or SMA (23). It has been shown that SMN is involved in pre-mRNA splicing (21), and it forms complexes through the association with several snRNP proteins such as the Sm (ref. 24 and references therein). Interestingly, each of the GEMs colocalizes with a CB, with the CB appearing contained within the GEM (7). This association most likely results from direct interaction between coilin and SMN (25), and the two bodies have been proposed to function together in recycling and assembly of the snRNPs (21). In view of the associations of TB with CB (Fig. 4B) and of CB with GEM described above, we carried out immunostaining of eGFP-mTDP-S-expressing cells with anti-SMN mouse antibody. Indeed, more than 60% of the GEM bodies overlap with one TB, as shown in Fig. 4C. Occasionally, a TB could be found to associate with two GEM bodies.
The association between TB and GEM seems to be mediated through the interaction of TDP with SMN. As shown by cross-IP experiments, SMN interacts with FLAG-mTDP-S (Fig. 5) and FLAG-mTDP-L (data not shown) in transfected 293 cells.
Figure 5.
Cross-IP of SMN and TDP. Extracts from 293 cells transfected with pEF-FLAG vector (lanes 1–3) or pEF-FLAG-mTDP-S (lanes 4–6) were analyzed by Western blotting using anti-FLAG, anti-SMN, or antitubulin after IP with anti-FLAG (lanes 2 and 5) or anti-SMN (lane 3 and 6). Extract without the IP step was used as the control in lanes 1 and 4. The detections of the SMN band in lane 5 by anti-SMN and of the FLAG-mTDP-S band in lane 6 by anti-FLAG indicate the interaction between SMN and FLAG-mTDP-S in vivo.
POD.
The PML gene product oncogene domains, or PODs, are one class of nuclear bodies containing the PML and several transcription factors including CBP (26) and p53 (27). The PODs are involved in protein metabolism as well as transcriptional regulation (refs. 1 and 3 and references therein).
Antibodies against either PML, the signature protein of PODs, or CBP were used to immunostain pCMV-eGFP-mTDP-S-transfected 293 cells. As exemplified in Fig. 4D, significant associations between TBs and PODs were observed also. Approximately 50% of the TBs overlap with a POD. Again, the average numbers of PODs seen in eGFP-positive and eGFP-negative nuclei are approximately the same, indicating that there is no induced formation of PODs by the exogenous mTDP-S.
SC35 Speckles.
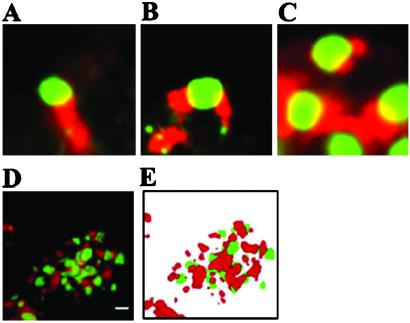
The SC35 speckles were first identified immnocytologically in the nuclei with an antibody against spliceosomal protein SC35 (28). It was subsequently found that many, but not all, of the SC35 speckles also contain another spliceosome protein, Sm (20). A subpopulation of the RNA polymerase II molecules also colocalizes with the SC35 speckles (29, 30). Interestingly, costaining experiments showed that more than 90% of the TBs overlap with at least one SC35 speckle (Fig. 6A). Often one TB could be found to overlap with two or more of the SC35 speckles as shown in Fig. 6B. Sometimes multiple TB and SC35 bodies are linked consecutively (Fig. 6C). Overall, these two types of nuclear bodies are interconnected significantly in the nuclei, as exemplified by 3D reconstruction of a nucleus transfected with pCMV-eGFP-mTDP-S and immunostained with anti-SC35 antibody (Fig. 6 D and E).
Figure 6.
Interconnection of TB and SC35 speckles. 293 cells transfected with pCMV-eGFP-mTDP-S were analyzed by immunostaining with anti-SC35. Examples of merged images of TB (green) and SC35 (red) are shown in A–D. The image in D is from one section of a nucleus with highly reticulated interconnection of TB and SC35 speckles. The 3D reconstruction of this nucleus is shown in E. (Scale bar in D, 2 μm.)
Links Among Different Nuclear Bodies: TB/GEM/CB, TB/GEM/POD, TB/SC35/CB, and TB/SC35/POD.
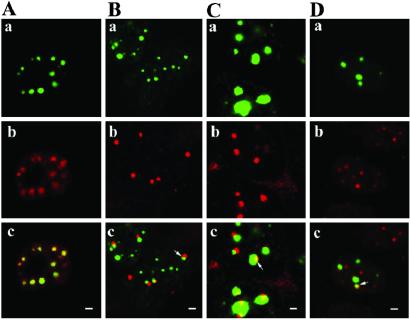
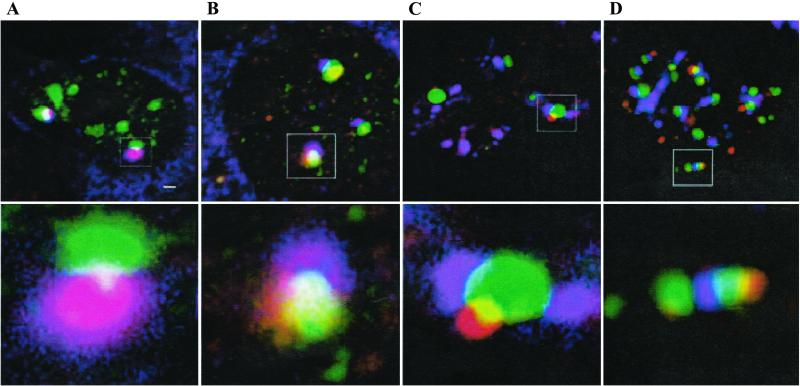
The associations of TBs with different nuclear bodies including CB, GEM, POD, and SC35 have prompted us to examine whether a TB could associate at the same time with more than one type of these other nuclear bodies. Indeed, double-immunostaining of pCMV-eGFP-mTDP-S-transfected 293 cells have demonstrated the existence of nuclear substructures containing TB/GEM/CB (Fig. 7A), TB/GEM/POD (Fig. 7B), TB/SC35/CB (Fig. 7C), or TB/SC35/POD (Fig. 7D). In fact, the same domain sometimes is shared by three different nuclear bodies (white area, Fig. 7 A and B).
Figure 7.
Interconnection among three different nuclear bodies. 293 cells transfected with pCMV-eGFP-mTDP-S were double-stained with different pairs of antibodies: (A) anti-coilin (red) and anti-SMN (blue); (B) anti-SMN (blue) and anti-PML (red); (C) anti-coilin (red) and anti-SC35 (blue); and (D) anti-PML (red) and anti-SC35 (blue). Note that the merged color is yellow for green plus red, purple for red plus blue, sky blue for green plus blue, and white for green plus red plus blue. The boxed regions in A–D Upper are magnified in A–D Lower. (Scale bar in A Upper, 1 μm.)
Lack of Association Between TB and Other Nuclear Substructures.
Immunohistochemistry was used also to examine whether TB associates with nuclear substructure other than the ones described above. We found little, if any, overlap between TB and the nucleolus, and TB does not associate with nuclear spots lighted by fluorescently labeled antibodies against MBD2b or HDAC5 (data not shown).
Discussion
In this study, we characterized mTDP-L and in more detail mTDP-S. As indicated by sequence alignment of the two cDNAs with the corresponding mouse genomic region, the two proteins are the products of alternative splicing with mTDP-S generated via an additional splicing event within the most 3′ exon of mTDP-L (data not down). In fact, alternative splicing of the TDP primary transcript seems to be a general phenomenon, because we have also detected, by RT-PCR, the expression of different TDP isoforms including TDP-S in human cells (H.-Y. Wang and B. Krishman, unpublished results). Furthermore, similar to hTDP-43 (12), both of the mTDP isoforms are expressed ubiquitously in a variety of tissues including the heart, lung, liver, spleen, muscle, brain, and kidney as well as in cell lines such as NIH 3T3 and neuro2A (I-F.W., unpublished data). As demonstrated in Fig. 2B, mTDP-L and mTDP-S both could repress transcription. Because they each contains the two RRM domains, very likely they also could function in exon skipping, as demonstrated previously for hTDP-43 (13, 16). The TDP gene is conserved from Caenorhabditis elegans (GenBank accession no. Nm063520) to Drosophila (31) and mammals. Together with the repression and exon-skipping data described above, the conservation and diversity of expression of the TDP gene across the animal kingdom indicate its essential regulatory roles in multiple transcription and splicing pathways.
We have used GFP/FLAG tagging and DNA transfection to examine the subcellular locations of the TDP proteins. A similar approach has been used previously to identify and/or analyze other nuclear bodies such as the matrix-associated deacetylase body (8), PODs (26), CB (32), and PEB-associated splicing factor (PSF; ref. 33). Among several advantages, the possible colocalization of GFP-tagged proteins with other molecules could be analyzed by costaining experiments with the use of antibodies from mouse or rabbit, as shown in this study. It should be cautioned, though, that staining patterns of transfected cells might not completely reflect those of the endogenous proteins. Bearing this in mind, our immunohistochemistry analysis showed that a major portion of the mTDP-S molecules are concentrated in discrete nuclear bodies, which we termed TBs (Figs. 3B and 4A). Interestingly, the cellular localizations of several other RRM domain-containing proteins including paraspeckle protein 1 (10), the hnRNP protein HAP (9), and the PSF (33) also exhibited speckle-like patterns in the nucleus. The RRM domain likely is required for the speckle localization of these proteins, as demonstrated for PSF (33). The cellular characteristics of the nuclear speckles containing these other RRM proteins suggest that they are distinct from TB. The difference of the nuclear staining patterns between mTDP-S and mTDP-L may be caused by the presence of the glycine-rich domain in mTDP-L; however, this awaits further examination.
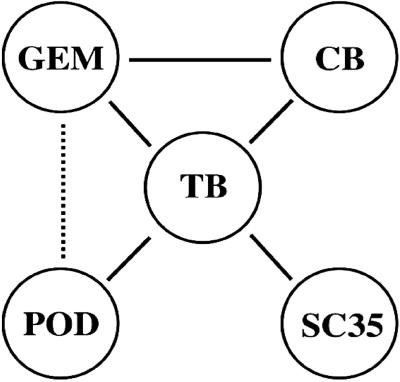
Remarkably though, many TBs are associated physically with several previously identified nuclear substructures including CB, GEM, POD, and SC35. (Figs. 4–7). Because a TB could associate with more than one of these other nuclear bodies of either the same (Fig. 6) or different types (Fig. 7), TBs seem to serve as a bridge to link different types of nuclear bodies (Fig. 8). In fact, we have noticed that data of several previous reports have already indicated the close proximity but not colocalization of different nuclear bodies, e.g. SC35 and the small snRNP foci/CB in HeLa cells (20). It is likely that in those cases the different bodies/speckles are interconnected physically through TB, as suggested by this study. We note here that the observed association of TB with these other nuclear bodies is not coincidental. First, the percentage of TBs associated with POD or SC35 is high. Second, almost every CB is associated with a TB. Third, several other types of nuclear bodies such as the HDAC5 and the nucleoli exhibited little or no association with TB.
Figure 8.
Association of TB with other nuclear bodies. The associations among five different nuclear bodies (TB, GEM, CB, SC35, and POD) are presented schematically. The association between GEM and CB has been described also in ref. 21. The less frequently observed association between POD and GEM is indicated by the dotted line.
Previous analyses have agreed in general that the eukaryotic transcription processes (initiation, elongation, splicing, 3′ cleavage, etc.) are coupled tightly, and they occur together at or near discrete sites in the nucleoplasm (1–5). Initially, these sites seem to include mainly the relatively larger nuclear substructures such as the SC35 speckles (34–36). However, later studies investigating the sites of UTP incorporation and locations of RNA polymerase II suggested the following scenario. That is, although active or abundant genes are transcribed at the speckled nuclear bodies, the bulk of the transcription process occurs throughout the nucleoplasm in a meshwork of punctuated spots (29, 30). The speckled nuclear bodies mainly serve as the stations for assembly of spliceosomal snRNPs (21) and as storage sites of these complexes and RNA polymerase II (29, 30). Also, CB has been found to associate with the so-called cleavage body, which contains factors required for 3′-end processing of mRNAs (37). Following the above, what are the functional implications of our demonstration that TB serves to connect different types of nuclear bodies/speckles? Most importantly, the networking through TB provides a way for effective trafficking and sorting of different nuclear factors among a wider range of categories than previously thought. For example, it has been shown that splicing factors such as SC35 could diffuse out of the speckles into transcriptionally active regions such as the perichromatin fibrils, where the factors participate in the splicing of pre-mRNAs (ref. 34 and references therein). In a separate set of analyses, it was suggested that the overlapping CBs and GEM bodies serve as the stations for recycling of the snRNPs required for assembly of the spliceosomes (21). However, most of the SC35 speckles do not contain several specific snRNAs or snRNPs (20). The CB and GEM, on the other hand, do not contain SC35 (21). Our data indicate that a more complete set of splicing components could be readily assembled together because of their close proximity within the physically linked TB, SC35, CB, and GEM bodies. The addition of POD to the picture and the bifunctionality of the TDP proteins further suggest that the multisubunit, nuclear body complexes play an important role in the tight coupling of the processes of splicing and regulated transcription. Finally, the finding of consecutive linking of TB and SC35 (Fig. 6D) suggests that some of the multisubunit, nuclear body structures may form a network in the nucleoplasm.
In summary, our data have suggested the existence of a higher order of arrangements of the eukaryotic nuclear bodies. We showed that nuclear bodies of either the same or different types could be linked to one another spatially within the eukaryotic nuclei. The linking, in this case through TB, leads to the formation of a lattice network, within which the coupled processes of transcription and splicing of genes could occur. Intriguing questions remain with the model. For instance, at least the TB and GEM seem to be interconnected through interaction between TDP and SMN (Fig. 5). What are the binding forces between TB and other nuclear bodies? Are there as-yet-identified nuclear bodies serving a similar linking role as TB ? Whether and how the higher order arrangement of the nuclear bodies differ among different cell types and along the cell cycle also awaits examination.
Acknowledgments
We thank the lab colleagues and Dr. Hung Li of the Institute of Molecular Biology, Dr. W. Y. Tarn of the Institute of Biomedical Sciences, and Dr. N.-H. Yeh of the National Yang-Ming University for insightful discussions. Dr. Hung-Yi Wang and Bose Krishnan's efforts in helping to prepare the graphs and sharing unpublished data are greatly appreciated. We also thank Dr. C.-S. Wu at the National Taiwan Hospital and Dr. Hung Li for providing the anticoilin and anti-SMN antibodies. Dr. Ann-Shyn Chiang of National Tsing Hua University helped us in the initial use of the Zeiss 3D visart software program. This research was supported by grants from the National Science Council, Foundation of Biomedical Sciences, the National Health Research Institute, and the Academia Sinica (Taiwan, Republic of China).
Abbreviations
- sn
small nuclear
- SMN
survival motor neuron
- CB
coiled body
- PML
promyelocytic leukemia
- Gem
Gemini of coiled bodies
- POD
PML body
- RRM
RNA-recognition motif
- eGFP
enhanced GFP
- m
mouse
- h
human
- IP
immunoprecipitation
- TB
TDP body
- RNP
ribonucleoprotein
References
- 1.Lamond A I, Earnshaw W C. Science. 1998;280:547–553. doi: 10.1126/science.280.5363.547. [DOI] [PubMed] [Google Scholar]
- 2.Misteli T, Spector D L. Curr Opin Cell Biol. 1998;10:323–331. doi: 10.1016/s0955-0674(98)80007-0. [DOI] [PubMed] [Google Scholar]
- 3.Matera A G. Trends Cell Biol. 1999;9:302–309. doi: 10.1016/s0962-8924(99)01606-2. [DOI] [PubMed] [Google Scholar]
- 4.Misteli T. Science. 2001;291:843–847. doi: 10.1126/science.291.5505.843. [DOI] [PubMed] [Google Scholar]
- 5.Cremer T, Cremer C. Nat Rev. 2001;2:292–301. doi: 10.1038/35066075. [DOI] [PubMed] [Google Scholar]
- 6.Salomoni P, Pandolfi P P. Cell. 2002;108:165–170. doi: 10.1016/s0092-8674(02)00626-8. [DOI] [PubMed] [Google Scholar]
- 7.Liu Q, Dreyfuss G. EMBO J. 2000;15:3555–3565. [PMC free article] [PubMed] [Google Scholar]
- 8.Downes M, Ordentlich P, Kao H Y, Alvarez-Jacqueline G A, Evans R M. Proc Natl Acad Sci USA. 2000;97:10330–10335. doi: 10.1073/pnas.97.19.10330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chiodi I, Biggiogera M, Denegri M, Corioni M, Weighardt F, Cobianchi F, Riva S, Biamonti G. J Cell Sci. 2000;113:4043–4053. doi: 10.1242/jcs.113.22.4043. [DOI] [PubMed] [Google Scholar]
- 10.Fox A H, Lam Y W, Leung-Anthony K L, Lyon C E, Andersen J, Mann M, Lamond A I. Curr Biol. 2002;12:13–25. doi: 10.1016/s0960-9822(01)00632-7. [DOI] [PubMed] [Google Scholar]
- 11.Mintz P J, Patterson S D, Neuwald A F, Spahr C S, Spector D L. EMBO J. 1999;18:4308–4320. doi: 10.1093/emboj/18.15.4308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ignatius S H, Wu F, Harrih D, Leon F, Martinez G, Gaynor R B. J Virol. 1995;69:3584–3596. doi: 10.1128/jvi.69.6.3584-3596.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Buratti E, Dork T, Zuccato E, Pagani F, Romano M, Baralle F E. EMBO J. 2001;20:1774–1784. doi: 10.1093/emboj/20.7.1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dreyfuss G, Matunis M J, Pinol-Roma S. Annu Rev Biochem. 1993;62:289–321. doi: 10.1146/annurev.bi.62.070193.001445. [DOI] [PubMed] [Google Scholar]
- 15.Wang J, Dong Z, Bell L R. J Biol Chem. 1997;272:22227–22235. doi: 10.1074/jbc.272.35.22227. [DOI] [PubMed] [Google Scholar]
- 16.Buratti E, Baralle F E. J Biol Chem. 2001;276:36337–34343. doi: 10.1074/jbc.M104236200. [DOI] [PubMed] [Google Scholar]
- 17.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd Ed. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 18.Harlow E, Lane D. Antibodies. Plainview, NY: Cold Spring Harbor Lab. Press; 1998. [Google Scholar]
- 19.Roder K, Hung M S, Lee T L, Lin T Y, Xiao H, Isobe K I, Juahg J L, Juang J L, Shen C-K J. Mol Cell Biol. 2000;20:7401–7409. doi: 10.1128/mcb.20.19.7401-7409.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Carmo-Fonseca M, Pepperkok R, Sproat B S, Ansorge W, Swanson M S, Lamond A I. EMBO J. 1991;10:1863–1873. doi: 10.1002/j.1460-2075.1991.tb07712.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pellizzoni L, Kataoka N, Charroux B, Dreyfuss G. Cell. 1998;95:615–624. doi: 10.1016/s0092-8674(00)81632-3. [DOI] [PubMed] [Google Scholar]
- 22.Andrade L E C, Chan E K L, Raska I, Peebled C L, Roos G, Tan E M. J Exp Med. 1991;173:1407–1419. doi: 10.1084/jem.173.6.1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lefebvre S, Burglen L, Reboullet S, Clermont O, Burlet P, Viollet L, Benichou B, Cruaud C, Millasseau P, Zeviani M, et al. Cell. 1995;80:155–165. doi: 10.1016/0092-8674(95)90460-3. [DOI] [PubMed] [Google Scholar]
- 24.Mister G, Buhler D, Laggerbauer B, Zobawa M, Lottspeich F, Fischer U. Hum Mol Genet. 2000;9:1977–1986. doi: 10.1093/hmg/9.13.1977. [DOI] [PubMed] [Google Scholar]
- 25.Hebert M D, Szymczyk P W, Shpargel K B, Matera A G. Genes Dev. 2001;15:2710–2729. doi: 10.1101/gad.908401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lamorte V J, Dyck J A, Ochs R L, Evans R M. Proc Natl Acad Sci USA. 1998;95:4991–4996. doi: 10.1073/pnas.95.9.4991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fogal V, Gostissa M, Sandy P, Zacchi P, Sternsdorf T, Jensen K, Pandolfi P P, Will H, Schneider C, Del Sal G. EMBO J. 2000;19:6185–6195. doi: 10.1093/emboj/19.22.6185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fu X D, Maniatis T. Nature. 1990;343:437–441. doi: 10.1038/343437a0. [DOI] [PubMed] [Google Scholar]
- 29.Bregman D B, Lu L, Van der zee S, Wassen S L. J Cell Biol. 1995;129:287–298. doi: 10.1083/jcb.129.2.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zeng C, Kim E, Warren S, Berget S. EMBO J. 1997;16:1401–1412. doi: 10.1093/emboj/16.6.1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lukacsovich T, Asztalos Z, Juni N, Awano W, Yamamoto D. Genomics. 1999;57:43–56. doi: 10.1006/geno.1999.5746. [DOI] [PubMed] [Google Scholar]
- 32.Platani M, Goldberg I, Swedlow J R, Lamond A I. J Cell Biol. 2000;151:1561–1574. doi: 10.1083/jcb.151.7.1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dye B T, Patton J G. Exp Cell Res. 2001;263:131–144. doi: 10.1006/excr.2000.5097. [DOI] [PubMed] [Google Scholar]
- 34.Spector D L. Annu Rev Cell Biol. 1993;9:265–315. doi: 10.1146/annurev.cb.09.110193.001405. [DOI] [PubMed] [Google Scholar]
- 35.Xing Y, Johnson C V, Dobner P, Lawsence J B. Science. 1993;259:1326–1330. doi: 10.1126/science.8446901. [DOI] [PubMed] [Google Scholar]
- 36.Van Driel R, Wansink D G, Van Steensel B, Grande M A, Schul W, de Jong L. Int Rev Cytol. 1995;162:151–189. doi: 10.1016/s0074-7696(08)61231-0. [DOI] [PubMed] [Google Scholar]
- 37.Schul W, Groenhout B, Koberna K, Takagaki Y, Jenny A, Manders E M, Raska I, vanDriel R, de Jong L. EMBO J. 1996;15:2883–2892. [PMC free article] [PubMed] [Google Scholar]