Abstract
Recent research on rainforest speciation has highlighted the importance of habitat variation in generating population diversification but lacks evidence of an associated reduction in gene flow. This paper describes a study in which molecular markers were used to examine the effects of allopatric divergence and habitat on levels of gene flow in the Caribbean lizard, Anolis roquet. Three study transects were constructed to compare variation in microsatellite allele frequencies and morphology across phylogenetic and habitat boundaries in northern Martinique. Results showed reductions in gene flow to be concordant with divergent selection for habitat type. No evidence could be found for divergence in allopatry influencing current gene flow. Morphological data match these findings, with multivariate analysis showing correlation with habitat type but no grouping by phylogenetic lineage. The results support the ecological speciation model of evolutionary divergence, indicating the importance of habitats in biodiversity generation.
Keywords: Anolis roquet‖gene flow‖Martinique‖microsatellite‖rainforest
Understanding speciation processes in rainforests is key to predicting changes in species number and planning conservation strategy (1). Ecological speciation due to divergent natural selection has emerged as an alternative theory to speciation in geographic isolation. Recent studies in support of an ecological gradient model of speciation in rainforests have shown morphological differences between habitats but have not tested for a reduction in gene flow (2, 3) or have not reported such a reduction where it has been tested (3, 4). Morphological variation along ecological gradients may indicate diversification, but speciation is not an inevitable consequence of population differentiation (5), and molecular evidence of reduced gene flow is needed to strengthen support for the theory of ecological speciation (6, 7). Here we use microsatellite markers to estimate gene flow and compare patterns of interbreeding among populations of a Caribbean lizard within a rainforest habitat and between habitats on the island of Martinique.
Martinique is typical of many mountainous islands in the Lesser Antilles arc, with a volcanic geological history producing faunal distributions representing past vicariance and a diverse microclimate resulting in pronounced habitat zonation. The endemic tree lizard, Anolis roquet, is found throughout the island and demonstrates marked morphological differentiation among geographic regions (8, 9). The north of Martinique is dominated by montane rainforest that is bisected north–south by a boundary between mitochondrial haplotype clades (Fig. 1, ref. 9). mtDNA sequence data give an estimated divergence time of 7.6 million years between the eastern and western lineages of A. roquet (maximum uncorrected sequence divergence of 10.6%, assuming 1.4% divergence per million years). The timing of divergence is consistent with geological evidence for the rise of corresponding precursor islands that were recently joined to form present-day Martinique, suggesting allopatric divergence between lineages (9, 10). The western lineage contains a steep ecological transition from central montane rainforest to xeric woodland along the Caribbean coast that occurs over a distance of 10 km [Fig. 1 (11)]. Such a system presents an opportunity to compare the effects of allopatric divergence and habitat on the evolution of A. roquet in this region.
Figure 1.
Northern Martinique, showing the position of the three study transects with sampling localities (symbols), the phylogenetic lineage boundary (black line), and habitat structure for the region under study (shading).
Methods
Sampling Regime.
We established three study transects, each consisting of between seven and ten localities in northern Martinique (Fig. 1). Localities were selected to cover regions of potential change while remaining separated by distances well in excess of single-generation migration distance for A. roquet. The transects were designed to allow a comparison between the effects of historical separation and habitat type on morphology and levels of gene flow in A. roquet. The “Lineage Transect” runs within a montane rainforest environment but crosses the deep north–south lineage boundary. The “Habitat Transect” runs within a single lineage but follows a sharp ecological gradient eastwards from xeric Caribbean coastal woodland to central montane rainforest. The “Control Transect” runs within a single lineage and a relatively constant habitat along the east coast. Sampling was undertaken from April to July 2001 with between 15 and 20 lizards caught per locality.
Gene Flow.
Gene flow was measured by using eight polymorphic microsatellite loci (12) to screen lizards from each locality along the three transects. The resulting allele frequencies were tested for linkage disequilibrium, null alleles, and departure from Hardy–Weinberg equilibrium (13, 14). Selection at individual loci was further tested for by examining the change in FIS with increased locality pooling (13, 15) before gene flow along each transect was estimated by using FST (16). In addition, we investigated population structure by using analysis of molecular variance (14), where localities were grouped into hypothetical populations based on mtDNA lineage (Lineage Transect), habitat type (Habitat Transect), or an arbitrary north–south split (Control Transect). These groupings were then tested to see whether the microsatellite allele frequencies supported the proposed population structure.
Morphology.
Sixteen morphological characters were recorded for each of five adult males per locality, as follows: snout-vent length (L), jaw L, head L, head width, head depth, upper leg L, lower leg L, tail depth, ventral scale count (sc), dorsal sc, postmental sc, between interparietal and supraorbital semicircles sc, light patch count (LPC) on head, LPC anterior body, LPC posterior body, and dark dorsal chevron count (see ref. 17 for illustrated definitions). These data were subjected to canonical variate analysis (18) for each transect, grouping by locality, to allow identification of change in multivariate morphology with geographic transect distance.
Results and Discussion
Before examining population structure using the FST metric, the implicit assumption that populations are at genetic equilibrium was tested by screening allele frequencies for linkage disequilibrium and departures from Hardy–Weinberg equilibrium (Table 1). There were no consistent departures from Hardy–Weinberg equilibrium or sign of linkage disequilibrium and no evidence of null alleles from heterozygote deficiency. These results support the assumption of migration-drift equilibrium and indicate that the microsatellite loci are not affected by selection. FIS was found to increase uniformly across all loci with locality pooling, further demonstrating that no individual locus was being selected. Consequently, pairwise FST was used to identify reductions in gene flow between adjacent localities.
Table 1.
Observed (HO) and expected (HE) heterozygosity and allele range for microsatellite loci (L031, L068, L062, L035, L120, L014, L065, L126) and transect localities (T1_1–T3_7)
| Locality | 031 | 068 | 062 | 035 | 120 | 014 | 065 | 126 | All loci | |
|---|---|---|---|---|---|---|---|---|---|---|
| T_1_1 | HO | 0.600 | 0.400 | 0.667 | 0.571 | 0.733 | 0.667 | 0.733 | 0.714 | 0.655 |
| HE | 0.508 | 0.405 | 0.706 | 0.651 | 0.690 | 0.687 | 0.864 | 0.765 | 0.663 | |
| T_1_2 | HO | 0.400 | 0.333 | 0.769 | 0.733 | 0.667 | 0.733 | 0.933 | 0.867 | 0.679 |
| HE | 0.561 | 0.370 | 0.643 | 0.715 | 0.759 | 0.579 | 0.903 | 0.839 | 0.671 | |
| T_1_3 | HO | 0.533 | 0.133 | 0.467 | 0.800 | 0.733 | 0.667 | 0.800 | 0.800 | 0.617 |
| HE | 0.522 | 0.239 | 0.501 | 0.802 | 0.763 | 0.756 | 0.784 | 0.862 | 0.654 | |
| T_1_4 | HO | 0.333 | 0.600 | 0.600 | 0.714 | 0.429* | 0.800 | 0.867 | 0.867 | 0.651 |
| HE | 0.561 | 0.480 | 0.625 | 0.714 | 0.751 | 0.784 | 0.818 | 0.818 | 0.694 | |
| T_1_5 | HO | 0.500 | 0.214 | 0.357 | 0.833 | 0.857 | 0.643 | 0.571 | 0.786 | 0.595 |
| HE | 0.495 | 0.198 | 0.542 | 0.826 | 0.810 | 0.640 | 0.688 | 0.751 | 0.619 | |
| T_1_6 | HO | 0.533 | 0.333 | 0.600 | 0.929 | 0.733 | 0.533 | 0.867 | 0.867 | 0.674 |
| HE | 0.522 | 0.384 | 0.609 | 0.892 | 0.818 | 0.660 | 0.894 | 0.837 | 0.702 | |
| T_1_7 | HO | 0.400 | 0.200 | 0.733 | 0.667 | 1.000 | 0.400 | 0.800 | 0.467 | 0.583 |
| HE | 0.405 | 0.370 | 0.685 | 0.630 | 0.839 | 0.641 | 0.763 | 0.692 | 0.628 | |
| T_2_1 | HO | 0.500 | 0.450 | 0.650 | 0.750 | 0.750 | 0.737 | 0.842 | 0.750 | 0.679 |
| HE | 0.559 | 0.481 | 0.674 | 0.796 | 0.762 | 0.721 | 0.892 | 0.835 | 0.715 | |
| T_2_2 | HO | 0.500 | 0.350 | 0.600 | 0.789 | 0.750 | 0.600 | 0.750 | 0.700 | 0.630 |
| HE | 0.497 | 0.368 | 0.583 | 0.866 | 0.804 | 0.691 | 0.886 | 0.740 | 0.679 | |
| T_2_3 | HO | 0.450 | 0.350 | 0.650 | 0.750 | 0.850 | 0.450 | 0.800 | 0.650 | 0.619 |
| HE | 0.409 | 0.368 | 0.677 | 0.683 | 0.821 | 0.654 | 0.812 | 0.779 | 0.650 | |
| T_2_4 | HO | 0.400 | 0.450 | 0.500 | 0.500 | 0.850 | 0.650 | 0.900 | 0.600 | 0.606 |
| HE | 0.467 | 0.409 | 0.492 | 0.511 | 0.797 | 0.700 | 0.913 | 0.687 | 0.622 | |
| T_2_5 | HO | 0.400 | 0.450 | 0.600 | 0.600 | 0.750 | 0.611 | 0.850 | 0.550 | 0.601 |
| HE | 0.345 | 0.499 | 0.555 | 0.629 | 0.763 | 0.611 | 0.864 | 0.495 | 0.595 | |
| T_2_6 | HO | 0.500 | 0.400 | 0.550 | 0.700 | 0.700 | 0.684 | 0.750 | 0.400 | 0.586 |
| HE | 0.492 | 0.328 | 0.569 | 0.751 | 0.751 | 0.670 | 0.809 | 0.435 | 0.601 | |
| T_2_7 | HO | 0.450 | 0.158 | 0.650 | 0.700 | 0.842 | 0.650 | 0.800 | 0.550 | 0.600 |
| HE | 0.465 | 0.152 | 0.606 | 0.677 | 0.772 | 0.701 | 0.856 | 0.642 | 0.609 | |
| T_2_8 | HO | 0.450 | 0.500 | 0.526 | 0.579 | 0.850 | 0.737 | 0.895 | 0.789 | 0.666 |
| HE | 0.465 | 0.512 | 0.502 | 0.529 | 0.823 | 0.710 | 0.876 | 0.774 | 0.649 | |
| T_2_9 | HO | 0.400 | 0.450 | 0.450 | 0.556 | 0.737 | 0.789 | 0.900 | 0.600 | 0.610 |
| HE | 0.431 | 0.409 | 0.479 | 0.648 | 0.775 | 0.667 | 0.874 | 0.665 | 0.619 | |
| T_2_10 | HO | 0.400 | 0.450 | 0.300 | 0.700 | 0.700 | 0.789 | 0.850 | 0.600 | 0.599 |
| HE | 0.396 | 0.478 | 0.276 | 0.695 | 0.759 | 0.698 | 0.844 | 0.659 | 0.601 | |
| T_3_1 | HO | 0.429 | 0.357 | 0.571 | 0.643 | 0.786 | 0.643 | 0.857 | 0.714 | 0.631 |
| HE | 0.349 | 0.548 | 0.531 | 0.653 | 0.730 | 0.659 | 0.884 | 0.712 | 0.639 | |
| T_3_2 | HO | 0.600 | 0.533 | 0.800 | 0.846 | 0.786 | 0.600 | 0.867 | 0.857 | 0.736 |
| HE | 0.545 | 0.515 | 0.724 | 0.757 | 0.722 | 0.683 | 0.899 | 0.881 | 0.716 | |
| T_3_3 | HO | 0.467 | 0.467 | 0.600 | 0.400* | 0.714 | 0.733 | 0.867 | 0.800 | 0.631 |
| HE | 0.476 | 0.480 | 0.667 | 0.722 | 0.791 | 0.715 | 0.823 | 0.786 | 0.682 | |
| T_3_4 | HO | 0.286 | 0.333 | 0.667 | 0.714 | 0.929 | 0.600 | 0.933 | 0.857 | 0.665 |
| HE | 0.323 | 0.434 | 0.653 | 0.735 | 0.892 | 0.749 | 0.906 | 0.836 | 0.691 | |
| T_3_5 | HO | 0.400 | 0.333 | 0.533 | 0.786 | 0.533 | 0.667 | 0.800 | 0.867 | 0.615 |
| HE | 0.460 | 0.287 | 0.531 | 0.762 | 0.559 | 0.697 | 0.892 | 0.880 | 0.633 | |
| T_3_6 | HO | 0.400 | 0.400 | 0.400 | 0.800 | 0.600 | 0.714 | 0.867 | 0.643 | 0.603 |
| HE | 0.405 | 0.460 | 0.349 | 0.793 | 0.793 | 0.672 | 0.825 | 0.794 | 0.636 | |
| T_3_7 | HO | 0.467 | 0.467 | 0.467 | 0.615 | 0.643 | 0.733 | 0.867a | 0.867a | 0.641 |
| HE | 0.513 | 0.480 | 0.598 | 0.606 | 0.817 | 0.763 | 0.908 | 0.855 | 0.693 | |
| Total alleles | 8 | 4 | 11 | 15 | 10 | 8 | 16 | 16 | ||
Asterisk (*) indicates a significant departure from Hardy–Weinberg equilibrium following sequential Bonferroni correction (α = 0.01, k = 24). Shared alphabetic superscript indicates pairwise linkage disequilibrium between those loci at that locality when departure from Hardy–Weinberg equilibrium is not involved.
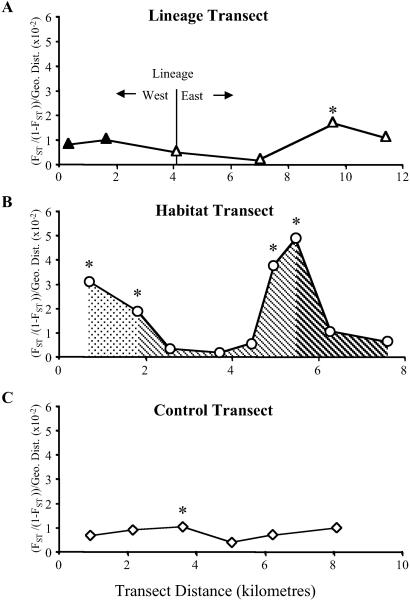
No reduction in gene flow between localities spanning the lineage boundary on the Lineage Transect was found (Fig. 2A). Only a single FST pairwise estimate along the entire transect was significant (P < 0.05), and when adjusted for between-locality distance, it can be seen to have minimal effect on the pattern of gene flow. Furthermore, there was no support for a lineage-based population structure in the microsatellite data under analysis of molecular variance (AMOVA) (P > 0.05, Table 2). In contrast, the Habitat Transect displayed marked reductions in gene flow in two regions (Fig. 2B). At the changes from xeric coastal woodland to transitional woodland and from transitional woodland to montane rainforest, pairwise FST values are both significant and show sharp reductions in gene flow per unit distance indicating reduced interbreeding between habitats. Population structuring by habitat was also supported by AMOVA (P < 0.01, Table 2). Estimates of gene flow for the Control Transect were similar to those on the Lineage Transect, with no sharp reductions in gene flow and no support for the arbitrary north–south grouping under AMOVA (P > 0.05, Table 2) (Fig. 2C). Taken together, the microsatellite results for the three transects strongly suggest that gene flow restriction is predominantly ecologically driven and not related to historical population boundaries.
Figure 2.
Pairwise FST/(1 − FST) divided by pairwise geographic distance plotted against midpair distance for adjacent localities. Asterisk (*) indicates significant FST values (P < 0.05). (A) Lineage Transect shows no reduction in nuclear gene flow at lineage boundary. (B), Habitat Transect shows reduced gene flow at transitions from xeric coastal woodland to lower transitional woodland and from upper transitional woodland to montane rainforest. (C) Control Transect shows no sharp reduction in gene flow with geographic distance. Symbols and shading follow Fig. 1.
Table 2.
Distribution of microsatellite allele covariance within and between hypothetical analysis of molecular variance (AMOVA) groupings
| % Covariance | Lineage transect | Habitat transect | Control transect |
|---|---|---|---|
| Among localities, within a grouping | 2.23* | 1.42* | 2.06* |
| Between groupings | 1.13NS | 3.20* | −0.02NS |
Significant between-group covariance on the Habitat Transect supports population structure by habitat type. AMOVA locality groupings: Lineage Transect, three western vs. 4 eastern lineages; Habitat Transect, three montane vs. five transitional vs. two xeric; Control Transect, four northern vs. three southern. Asterisk (*) signifies significance (P < 0.01). NS, nonsignificance.
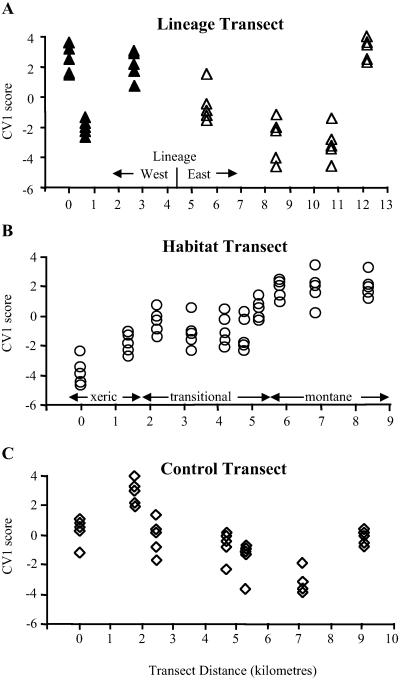
Morphological variation based on the canonical variate analysis results agrees with the pattern of population differentiation shown by the microsatellite data. Multivariate analysis shows that morphology is not linked to lineage on the Lineage Transect with first canonical variate score (CV1) scores showing no grouping congruent with the historical phylogenetic structure (Fig. 3A). In contrast, morphological variation parallels FST estimates of gene flow on the Habitat Transect indicating an association between morphology and gene flow. The change in CV1 score with transect distance shows a stepped clinal structure at the rainforest boundary with rapid transition in morphology correlating with the sharp change in habitat type as opposed to graduated geographic change (Fig. 3B). Morphological data for the Control Transect (Fig. 3C) are similar to those of the Lineage Transect with no indication of a clear categorical pattern of geographic variation. These results support the finding (9) that primarily selection, not phylogeny, is controlling morphology in A. roquet.
Figure 3.
Plots of first canonical variate score (CV1) against transect distance showing variation in multivariate morphology among localities on each transect. (A), Lineage Transect and (C) Control Transect show no pattern in morphology with transect distance. (B) Habitat Transect shows a stepwise change in morphology at the rainforest boundary, significantly correlated with change in habitat, not transect distance (partial correlation, P < 0.05). Symbols follow Fig. 1.
Rapid morphological differentiation and adaptation to local environments is a well-documented phenomenon (19) that has been previously demonstrated in a number of Anolis lizard species (20, 21). Selection on limb length for survival in different vegetation types has shown the potential pressure for a reduction in interbreeding between habitats (21), whereby adaptation to a particular niche could reduce migration between populations. Predation risk has also been suggested as a potential selective force on lizard morphology between habitats (2). Although changes in vegetation type are very rapid along the Habitat Transect, A. roquet is distributed continuously along its length, requiring an intrinsic barrier to gene flow to explain our results. Microniche variation, temporal display variation, and intersexual selection through female choice are currently being investigated as possible sources of behavioral isolation. Male intrasexual selection has previously been demonstrated in Anolis lizards (22, 23), and such behavior in A. roquet combined with differential adaptation by males to specific habitat type may account for reduced gene flow between habitats.
Until now, there has been little evidence that ecological selection has limited interbreeding between populations of tropical forest animals, and allopatric speciation models remain popular (24). However, the ecological hypothesis of speciation is gathering support (25), and examples of reductions in gene flow have been found in other ecosystems. In Cameroon, adaptation to either benthic or limnetic lifestyles within small lakes has led to reproductive isolation between habitats in cichlid fish (26), and in the Atlantic Canary Islands, reduced gene flow across a latitudinal ecotone has been observed in lacertid lizards (27). Such work typifies a recent shift in focus of evolutionary studies away from speciation models based solely on allopatry. Our findings here further this research by presenting one of the first examples of ecologically driven gene flow reduction across a habitat boundary.
Acknowledgments
We thank the Direction de l'Environnement de la Martinique for giving permission to undertake fieldwork; C. Gliddon, A. Malhotra, A. G. Stenson, and W. Wüster for helpful discussion; and three anonymous referees and the Member Editor for their comments. This research was supported by Biotechnology and Biological Sciences Research Council (Studentship 99/A2/G/5481 to R.O.), Flora and Fauna International and Herpetofauna Consultants International, Limited, for Biotechnology and Biological Sciences Research Council CASE sponsorship (to R.O.), the Linnean Society (to R.S.T.), and the Wellcome Trust (057257/z/99/z, 060384/z/00/z to R.S.T.).
Footnotes
References
- 1.Moritz C, Patton J L, Schneider C J, Smith T B. Annu Rev Ecol Syst. 2000;31:533–563. [Google Scholar]
- 2.Schneider C J, Smith T B, Larison B, Moritz C. Proc Natl Acad Sci USA. 1999;96:13869–13873. doi: 10.1073/pnas.96.24.13869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Smith T B, Schneider C J, Holder K. Genetica. 2001;112:383–398. doi: 10.1023/a:1013312510860. [DOI] [PubMed] [Google Scholar]
- 4.Smith T B, Wayne R K, Girman D J, Bruford M W. Science. 1997;276:1855–1857. [Google Scholar]
- 5.Magurran A E. Philos Trans R Soc London B. 1998;353:275–286. [Google Scholar]
- 6.Orr M R, Smith T B. Trends Ecol Evol. 1998;13:502–506. doi: 10.1016/s0169-5347(98)01511-0. [DOI] [PubMed] [Google Scholar]
- 7.Schluter D. In: Endless Forms–Species and Speciation. Howard D J, Berlocher S H, editors. New York: Oxford Univ. Press; 1998. p. 470. [Google Scholar]
- 8.Lazell J D. Bull Mus Comp Zool. 1972;131:359–401. [Google Scholar]
- 9. Thorpe, R. S. & Stenson, A. G. (2002) Mol. Ecol., in press.
- 10.Maury R C, Westbrook G K, Baker P E, Bouysse P, Westercamp D. In: The Caribbean Region. Dengo G, Case J E, editors. H. Boulder, CO: Geol. Soc. Am.; 1990. pp. 141–166. [Google Scholar]
- 11.Lassere G. Atlas des Department Francais d'Outre Mer. II. Paris: Centre National de la Recherche Scientifique–Institut Géographique National; 1977. [Google Scholar]
- 12.Ogden R, Griffiths T J, Thorpe R S. Conserv Genet. 2002;3:345–346. [Google Scholar]
- 13.Raymond M, Rousset F. GENEPOP. Montpellier, France: Institut des Sciences de l'Evolution, Univ. Montpellier; 1995. , Ver. 3.1. [Google Scholar]
- 14.Schneider S, Kueffer J-M, Roessli D, Excoffier L. arlequin (Genetics and Biometry Laboratory, Univ. Geneva, Geneva, Switzerland), Ver. 1.1. 1997. [Google Scholar]
- 15.Goudet J, de Meeüs T, Day A J, Gliddon C J. In: Genetics and Evolution of Aquatic Organisms. Beaumont A R, editor. London: Chapman & Hall; 1994. pp. 81–95. [Google Scholar]
- 16.Slatkin M. Annu Rev Ecol Syst. 1985;16:393–430. [Google Scholar]
- 17.Giannasi N C. Ph.D. thesis. Bangor, U.K.: Univ. Wales; 1997. [Google Scholar]
- 18.Dixon W J. BMDP Statistical Software. Los Angeles: BMDP; 1991. [Google Scholar]
- 19.Thompson J N. Trends Ecol Evol. 1998;13:81–87. doi: 10.1016/s0169-5347(98)01378-0. [DOI] [PubMed] [Google Scholar]
- 20.Malhotra A, Thorpe R S. Nature. 1991;353:347–348. [Google Scholar]
- 21.Losos J B, Warhelt K I, Schoener T W. Nature. 1997;387:70–73. [Google Scholar]
- 22.Jenssen T A, Nunez S C. Behaviour. 1998;135:981–1003. [Google Scholar]
- 23.Orrell K S, Jenssen T A. Anim Behav. 2002;63:1091–1102. [Google Scholar]
- 24.Haffer J. Biodivers Conserv. 1997;6:451–477. [Google Scholar]
- 25.Schluter D. Trends Ecol Evol. 2001;16:372–380. doi: 10.1016/s0169-5347(01)02198-x. [DOI] [PubMed] [Google Scholar]
- 26.Schliewen U, Rassmann K, Markmann M, Markert J, Kocher T, Tautz D. Mol Ecol. 2001;10:1471–1488. doi: 10.1046/j.1365-294x.2001.01276.x. [DOI] [PubMed] [Google Scholar]
- 27.Thorpe R S, Richard M. Proc Natl Acad Sci USA. 2001;98:3929–3934. doi: 10.1073/pnas.071576798. [DOI] [PMC free article] [PubMed] [Google Scholar]