Abstract
Population genetic theory shows that the efficacy of natural selection is limited by linkage—selection at one site interferes with selection at linked sites. Such interference slows adaptation in asexual genomes and may explain the evolutionary advantage of sex. Here, we test for two signatures of constraint caused by linkage in a sexual genome, by using sequence data from 255 Drosophila melanogaster and Drosophila simulans loci. We find that (i) the rate of protein adaptation is reduced in regions of low recombination, and (ii) evolution at strongly selected amino acid sites interferes with optimal codon usage at weakly selected, tightly linked synonymous sites. Together these findings suggest that linkage limits the rate and degree of adaptation even in recombining genomes.
Natural selection is imperfect. To become fixed, beneficial mutations must overcome both stochastic loss and interference from selection at linked loci. In asexual genomes, where linkage is complete, two kinds of interference compromise adaptation. The first, “ruby-in-the-rubbish” interference, occurs because beneficial mutations often appear on genetic backgrounds loaded with segregating deleterious mutations. Since deleterious mutations are, on average, probably more strongly selected than favorable ones, adaptation is mostly limited to those lucky few beneficial mutations that arise on unloaded backgrounds (1–4). The second form of interference, “clonal” interference, is caused by competition among multiple segregating beneficial mutations (1, 5–7). Because only one asexual genome can be fixed at a time, adaptive substitutions are forced to be nearly sequential. Both kinds of interference limit the rate of adaptation in asexuals (8–10).
The effects of both kinds of interference can be thought of as a reduction in the effective population size (Ne) caused by selection at linked loci (11–13). Recombination, by alleviating interference between linked sites, alleviates this reduction in Ne. Consequently, genomic regions that differ in recombination rate also differ in effective size—indeed, this is the basis of the well-known correlation between recombination rate and levels of neutral polymorphism (4Ne times the neutral mutation rate) seen in the genomes of Drosophila, humans, and others (14–18). That variation in linkage affects levels of neutral polymorphism suggests that it may also affect rates of nonneutral substitution. In particular, adaptive evolution may be limited in regions of low recombination (i.e., where Ne is reduced) or in situations of extreme linkage (e.g., among sites within the same gene).
Here we ask whether linkage systematically constrains adaptation in the Drosophila genome. We use divergence estimates from 255 Drosophila melanogaster and Drosophila simulans loci. These data are unique in that they include a large number of rapidly evolving genes, many of which are candidate male accessory gland proteins (Acps) and thus likely targets of sexual selection (19–21). We have reason to believe a priori not only that many of these substitutions are adaptive, but also that many of these genes may have experienced a long history of sexual selection possibly predating the D. melanogaster–D. simulans split. Such genes are especially good candidates for detecting the signature of interference. We therefore use these data to test for two kinds of limits imposed on adaptive evolution by linkage. First, we ask whether protein evolution is constrained by linkage. Second, we ask whether the efficacy of weak selection is compromised by a stream of strongly selected traffic at nearby sites.
Methods
The Data.
Coding sequences from 102 D. melanogaster and D. simulans genes were downloaded from GenBank, and aligned by eye using SE-AL v. 1.0. For genes with multiple transcripts of different lengths, we used the longest transcript; for those with multiple transcripts of the same length, we used an arbitrarily selected transcript. The remaining 153 genes (for which W. Swanson generously provided alignments) come from a D. simulans male-specific EST screen (20) and their D. melanogaster homologues, downloaded from Flybase (http://flybase.bio.indiana.edu). We used only coding ESTs from this screen that were either nonredundant or that were the most rapidly evolving of a set of redundant ESTs. Although ESTs are unreplicated single-pass sequences, none of the results in this paper are caused by a difference in sequencing error rate (and consequent elevated divergence estimates) between the EST and reference quality data, as all of our results are also found within the EST data set.
Estimates of Divergence, Recombination, and Codon Usage.
We used maximum likelihood estimates of the rates of amino acid (dN) and synonymous (dS) site divergence using paml (22). One anomalously high dS value (1.71) was excluded from the analysis (excluding this value does not affect the results). We used a likelihood ratio test of Goldman and Yang (23) to test for a significant excess of amino acid substitutions in those genes with dN/dS > 1. Briefly, we used paml to calculate the likelihood under the null model of equal dN and dS (L0) and the alternative model with dN and dS free to vary (L1). The likelihood test statistic (−2[ln L0 − ln L1]) was then compared with the χ2 distribution with 1 degree of freedom. dN/dS > 1 is strong evidence for adaptive evolution, as this test is conservative.
We estimated GC-content and the frequency of optimal codon usage (Fop) by using the online program codonw (http://bioweb.pasteur.fr/seqanal/interfaces/codonw.html). Fop is the proportion of codons in a gene that are optimal codons, defined as those used most frequently in highly expressed genes (24). Optimal codons for both species were assumed to be those of D. melanogaster. Although we estimate Fop from a single sequence from each species, these estimates should accurately reflect population levels of optimal codon usage (25).
We estimated recombination rates by using the data and standard method of Kliman and Hey (26). For the X, second, and third chromosomes, we fit least-squares polynomial curves relating recombination rate to DNA content per interval on the cytological map (all curves have R2 > 0.989), and used equations from these curves (available upon request) to predict recombination rates. Estimates for X-linked loci were multiplied by 4/3 to correct for the absence of recombination in Drosophila males. For the Y and dot-fourth chromosomes, we assume recombination is absent. The mean recombination rate in this data set is c = 0.0029 centimorgan (cM)/kb, close to the global mean for the D. melanogaster genome (see supplimentary information for ref. 27). We therefore defined “low” recombination as c < 0.0029 and “high” as c > 0.0029. We used recombination rate estimates from D. melanogaster for both species, as recombination data for D. simulans are sparse. Although recombination rates in D. simulans may differ (they are, on average, ≈30% higher than in D. melanogaster; see ref. 28), subtle local changes in recombination rate are unlikely to lead to misclassification of loci into high vs. low recombination regions. Changes in recombination rate that do result in misclassification likely contribute noise to our analysis, but this would obscure rather than create the patterns seen here.
Statistical Tests.
For nonnormally distributed variables, we used nonparametric Spearman rank correlations to test for associations and permutation t tests (with null distributions generated by ≥10,000 randomization of the data) to test for differences between means. For multivariate analyses, nonnormal variables were first normalized by log-transformation. All tests are two-tailed. Means are reported with ± 1 SE. All data are available online in Table 1, which is published as supporting information on the PNAS web site, www.pnas.org.
Results and Discussion
Does Linkage Limit Protein Evolution?
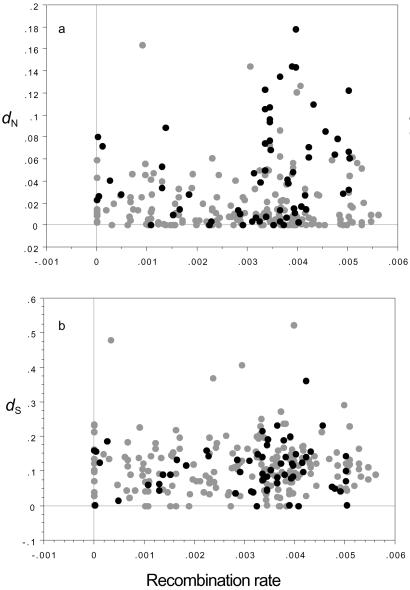
We first ask whether linkage limits protein adaptation. Rates of adaptive protein evolution, if limited by interference, should be relatively constrained in regions of low versus high recombination. We therefore test for an excess (paucity) of rapid evolution in regions of high (low) recombination. It is important to note, however, that slowly evolving genes (those mostly subject to purifying selection) should occur in all recombinational environments. A plot of dN vs. recombination should thus reveal a wedge-shaped distribution, with slowly evolving genes in regions of low recombination and both slowly and rapidly evolving genes in regions of high recombination. Fig. 1a confirms this prediction; genes in high recombination environments show both a higher mean and variance of dN values than those in low recombination environments (dN,high = 0.031 ± 0.003; dN,low = 0.019 ± 0.002; t test = 2.780, P = 0.007, F151,103 = 2.524, P < 0.0001; a qualitatively similar pattern appears in a plot of dN/dS vs. recombination rate, data not shown).
Figure 1.
dN, the rate of amino acid substitution (a), and dS, the rate of silent substitution (b), vs. recombination rate (in cM/kb). Black and gray circles are putative Acp's and non-Acp's, respectively.
We can eliminate three alternative explanations for this regional difference in evolutionary rates. First, this wedge pattern is not caused by regional differences in mutational input (as might be seen, e.g., if recombination were mutagenic) as such differences would produce a similar pattern for dS. The distribution of dS is not, however, wedge-shaped; neither the means nor the variances differ between regions of high and low recombination (Fig. 1b; dS,high = 0.118 ± 0.006, dS,low = 0.108 ± 0.007; t test = 1.081, P = 0.282; F150,103 = 1.012, P = 0.947). Second, this result is also inconsistent with most rapid evolution being caused by the fixation of many slightly deleterious mutations. In that case, we would expect higher rates of evolution in regions of low recombination (where Ne is reduced by interference). Other studies have noted such elevated rates of protein divergence in low recombination regions, likely because, in contrast to our data set, most of the genes in these studies are relatively conserved and therefore subject mainly to purifying selection (29–32). Although many of the genes in this study are also relatively conserved, the signal of interference on adaptive evolution detected here (elevated dN in high recombination regions) is detectable despite the opposing signal from relaxed purifying selection (elevated dN in low recombination regions). Third, although candidate Acp's constitute a large fraction of the data (24.4%), our results do not depend on some peculiarity of these genes other than their rapid evolution. Not surprisingly, if all candidate Acp's are excluded from the analysis, too few rapidly evolving genes remain to detect a pattern. But within Acp's, rapid protein evolution is also largely confined to regions of high recombination (dN,high = 0.057 ± 0.007; dN,low = 0.029 ± 0.007; t test = 2.369, P = 0.017; F44,18 = 2.884, P = 0.013; dS,high = 0.111 ± 0.011; dS,low = 0.101 ± 0.013; t test = 0.514, P = 0.601; F44,18 = 1.800, P = 0.155). We therefore conclude that rates of protein adaptation are constrained in the low recombination environments of the Drosophila genome, as predicted by population genetic theory.
Does Rapid Protein Evolution Limit Weak Selection at Linked Sites?
We now turn to a second test of the effect of linkage on adaptation. We ask whether evolution at strongly selected (amino acid) sites limits the efficacy of selection at tightly linked, weakly selected (synonymous) sites. Synonymous sites are not selectively equivalent, as certain synonymous codons (“optimal” or “preferred” codons) are used more frequently than others, apparently because of selection for translational efficiency and accuracy (33–36). But selection coefficients acting on alternative synonymous codons are on the order of 1/Ne, i.e., approaching the limits of selection (36). Such weakly selected sites are especially susceptible to interference from selection at linked sites (11, 37–39). Two particularly relevant studies (40, 41) have shown, using different models, that a series of strongly selected substitutions reduces the rate of substitution of linked weakly favored mutations. This effect is caused by genetic hitchhiking; as the frequency of hitchhiking increases, weakly selected linked sites behave more neutrally and thus come to more closely reflect the mutational spectrum. Because there are more ways, on average, to mutate to nonoptimal codons, the net effect of hitchhiking is to increase the fixation rate of nonoptimal mutations.
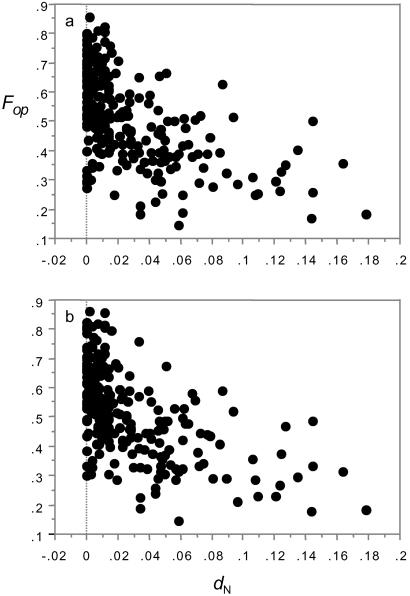
We test this prediction by asking whether the frequency of optimal codon usage (Fop) decays as the rate of protein evolution increases. As Fig. 2 shows, Fop declines sharply as dN increases (statistics identical for both species: rs = −0.559, P < 0.0001; the same patterns appear using dN/dS ratios, not shown). dS shows a weak positive correlation with Fop (D. melanogaster: rs = 0.172, P < 0.0061; D. simulans: rs = 0.224, P = 0.0004), but, as partial correlation analysis shows, the relationship between Fop and dN is independent of dS and twice as strong (D. melanogaster: partial r = −0.505 vs. 0.251; D. simulans: partial r = −0.491 vs. 0.180). The effect of strongly selected traffic on linked synonymous sites can be illustrated in another way. In Drosophila, AT-biased mutation pressure causes GC-content at mutational equilibrium to approach ≈35% (42, 43). Coding sequences are nevertheless highly GC-biased, at least partly because of constraints on codon usage as virtually all preferred codons end in C or G. If a stream of strongly selected amino acid traffic depresses Ne at weakly selected linked synonymous sites, GC-content at those sites should more closely reflect the mutational spectrum. We find that this is indeed the case. As the rate of amino acid substitution increases, third position GC-content decreases significantly (dN, D. melanogaster: rs = −0.540, P < 0.0001; dN, D. simulans: rs = −0.549, P < 0.0001; dN/dS, D. melanogaster: rs = −0.569, P < 0.0001; dN/dS, D. simulans: rs = −0.584, P < 0.0001).
Figure 2.
Fop, the frequency of optimal codon usage, vs. dN in D. melanogaster (a) and D. simulans (b).
Because Acp's as a class show both low optimal codon usage (19) and rapid protein evolution (19, 20), it is possible that the above findings are entirely due to some special property of these genes. We can rule out this possibility, however. The correlation between dN and optimal codon usage remains even when Acp's are excluded from the analysis (D. melanogaster: rs = −0.452, P < 0.0001; D. simulans: rs = −0.431, P < 0.0001). Moreover, the correlation exists, and is in fact stronger, within Acp's (D. melanogaster: rs = −0.695, P < 0.0001; D. simulans: rs = −0.711, P < 0.0001). This stronger correlation probably reflects the fact that other factors known to contribute to variation in optimal codon usage—tissue specificity, gene expression level, and gene length (reviewed in refs. 34 and 35)—are partially controlled within Acp's, as these genes share a common tissue type (male accessory glands), similar (high) expression levels, and similar (short) gene lengths.
Because there is reason to believe that both protein evolution and optimal codon usage are related to gene length (44, 45) and recombination rate (refs. 25 and 45, and see above), we tested the possibility that the correlation between dN and Fop is an artifact of one of these other relationships. We find that optimal codon usage is significantly correlated with gene length (D. melanogaster: rs = −0.142, P = 0.0287; D. simulans: rs = −0.129, P = 0.0463), as seen in previous studies (42, 43), but not with recombination (D. melanogaster: rs = −0.066, P > 0.05; D. simulans: rs = 0.081, P > 0.05). [The previously reported correlation between optimal codon usage and recombination is weak and detected in a much larger data set than ours (26, 45).] To distinguish the effects of gene length and protein evolution on optimal codon usage, we estimated partial correlation coefficients. Both relationships persist, but the correlation between dN and Fop is much stronger (partial r for gene length vs. dN in D. melanogaster: −0.163 vs. −0.505; in D. simulans: −0.152 vs. −0.518; gene length and dN are log-transformed; P < 0.05 for all).
Several workers (19, 46, 47) have observed high nonoptimal codon usage in rapidly evolving genes, but suggested relaxation of selective constraints as the cause. These relaxed constraints explanations come in two flavors. The first invokes codon-specific constraints. Akashi (48) has argued that selection for translational accuracy (i.e., selection against misincorporation of amino acids) should be strongest at functionally important residues. Consistent with this, he found that evolutionarily conserved residues tend to use preferred codons, which are less often mistranslated (48). This explanation alone cannot account for the results reported here, because even conserved sites in our rapidly evolving genes show poor optimal codon usage. After expunging all divergent codons from the genes in our data, we find that the relationship between Fop and dN is unchanged (D. melanogaster: rs = −0.546, P < 0.0001; D. simulans: rs = −0.555, P < 0.0001).
The second relaxed constraints explanation invokes gene-specific constraints, in which constraints on amino acid sites and synonymous sites are correlated within a gene, i.e., genes evolving rapidly because of relaxed selection on amino acid composition are likely to also have relaxed selection for optimal codon usage (49). There are three reasons to think this explanation does not explain our results. First, the rapid evolution in our data set does not appear to be caused by relaxed purifying selection, as evidenced by the relationship between rates of recombination and protein evolution (barring some spurious relationship between high recombination rate and relaxed constraint). Second, if the rapid evolution in our data were mostly caused by relaxed constraints, GC content in both synonymous and amino acid sites should decay in those loci with high dN/dS because of AT-biased mutation pressure. Although dN/dS is negatively related to GC content of synonymous sites (see above), no such negative relationship exists for amino acid sites (D. melanogaster: rs = 0.204, P = 0.0013; D. simulans: rs = 0.214, P = 0.0007). Third, interference is the only explanation for why genes with dN/dS > 1 (i.e., genes whose rapid evolution is caused by positive selection, not relaxed constraints) should show depressed codon usage: we find that mean Fop for loci with dN/dS > 1 is significantly lower than that for loci with dN/dS < 1 (D. melanogaster: Fop, dN/dS < 1 = 0.534 ± 0.009 vs. Fop, dN/dS > 1 = 0.355 ± 0.023, t test = 6.645, P < 0.0001; D. simulans: Fop, dN/dS < 1 = 0.544 ± 0.009 vs. Fop, dN/dS > 1 = 0.356 ± 0.022, t test = 7.077, P < 0.0001). Even when we consider only those loci with dN/dS significantly greater than 1 (a conservative standard), Fop remains significantly depressed (D. melanogaster: Fop, dN/dS < 1 = 0.516 ± 0.009 vs. Fop, dN/dS > 1 = 0.411 ± 0.084, t test = 2.999, P = 0.0020; D. simulans: Fop, dN/dS < 1 = 0.523 ± 0.009 vs. Fop, dN/dS > 1 = 0.415 ± 0.077, t test = 2.878, P = 0.0044). Taken together, these lines of evidence suggest that neither flavor of relaxed constraint hypothesis accounts for the correlation between rapid protein evolution and low optimal codon usage. Instead, it seems that interference from strongly selected traffic compromises weakly selected codon usage at tightly linked sites.
Concluding Remarks.
We find evidence that linkage constrains—and recombination facilitates—adaptation in Drosophila. We have shown that (i) the rate of protein adaptation appears limited by interference in low recombination regions, and (ii) strong directional selection on proteins interferes with weak selection for optimal codon usage at linked sites. That such limits on adaptation are detectable even in a recombining genome is surprising and has several implications. It follows, for example, that the limiting effects of linkage on protein adaptation may be manifest in the genetic basis of phenotypic evolution. As Birky and Walsh (50) point out in their classic study of the theory of linkage and selection, “recombination enhances the rate of phenotypic evolution, to the extent that phenotypic evolution is driven by the fixation of advantageous mutations.” We might expect, therefore, that adaptive species differences should map disproportionately to high recombination regions of genomes.
It does not necessarily follow, however, that genes in low recombination regions are maladapted. If genomes are luxuriant, so that there are many ways to adapt to new environments, adaptation will simply proceed via substitutions in regions of high recombination. There is some indication, though, that genomes are not luxuriant. A comparative quantitative trait locus study in domesticated cereal species found that convergent traits map to homeologous genomic regions (and therefore possibly to the same genes), suggesting that there may be a limited number of ways to construct these traits genetically (51). More convincingly, high resolution molecular and experimental evolution studies have uncovered convergence at the DNA sequence level (52–54), suggesting that selection at least sometimes uses the same nucleotide repeatedly. If the Drosophila genome is not luxuriant, then our results imply that flies are not perfectly adapted because of the slower average response of genes in regions of low recombination to directional selection. The weakly selected silent sites of rapidly evolving genes, in contrast, seem more clearly maladapted. With perfect recombination, rapidly evolving genes could both substitute beneficial amino acids and maintain optimal codon usage.
The detrimental effects of interference appear hierarchical in that linkage constrains protein adaptation, which in turn constrains codon adaptation. Formally, either clonal or ruby-in-the rubbish interference can limit protein evolution. But ruby-in-the-rubbish seems likely more important as the rate of deleterious mutation (55–57), and so the opportunity for interference from deleterious mutations, far exceeds the rate of favorable mutation. Furthermore, low recombination regions in Drosophila may suffer an additional load of deleterious mutations because of the higher numbers of transposable element insertions found there (58). Assuming that adaptive protein divergence is mostly limited by linkage to deleterious mutations, the hierarchical effects of interference may reflect the relative magnitudes of selection coefficients in Drosophila: mean selection against deleterious mutations is probably stronger than that favoring beneficial amino acid mutations, which in turn is larger than that favoring preferred codons (i.e., sd ≈ 10−2 > sb,amino acid ≈ 10−3 to 10−4 > sb,codon ≈ 10−6; see refs. 34, 56, and 59). This does not, however, mean that nonoptimal codon usage has a negligible effect on genomes. Although selection on individual preferred codons is weak (33), the cumulative effect of many unpreferred codons may be considerable (33, 37).
Supplementary Material
Acknowledgments
We thank P. Andolfatto, D. Begun, J. Bollback, Y. Chen, J. Gillespie, J. Huelsenbeck, C. Jones, Y. Kim, W. Stephan, two anonymous reviewers, and especially A. Orr for helpful comments and discussion. This work was supported by National Institutes of Health Grant GM526738 and by funding from the David and Lucile Packard Foundation (to A. Orr), and by Caspari Fellowships and Messersmith Fellowships (to A.J.B. and D.C.P.).
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Fisher R A. The Genetical Theory of Natural Selection. Oxford: Oxford Univ. Press; 1930. [Google Scholar]
- 2.Manning J T, Thompson D J. Acta Biotheor. 1984;33:219–225. [Google Scholar]
- 3.Peck J. Genetics. 1994;137:597–606. doi: 10.1093/genetics/137.2.597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Orr H A. Genetics. 2000;155:961–968. doi: 10.1093/genetics/155.2.961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Muller H J. Am Nat. 1932;66:118–138. [Google Scholar]
- 6.Crow J F, Kimura M. Am Nat. 1965;99:439–450. [Google Scholar]
- 7.Gerrish P J, Lenski R E. Genetica. 1998;102/103:127–144. [PubMed] [Google Scholar]
- 8.de Visser J A G M, Zeyl C W, Gerrish P J, Blanchard J L, Lenski R L. Science. 1999;283:404–406. doi: 10.1126/science.283.5400.404. [DOI] [PubMed] [Google Scholar]
- 9.Miralles R, Gerrish P J, Moya A, Elena S F. Science. 1999;285:1745–1747. doi: 10.1126/science.285.5434.1745. [DOI] [PubMed] [Google Scholar]
- 10.Miralles R, Moya A, Elena S F. J Virol. 2000;74:3566–3571. doi: 10.1128/jvi.74.8.3566-3571.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hill W G, Robertson A. Genet Res. 1966;8:269–294. [PubMed] [Google Scholar]
- 12.Felsenstein J. Genetics. 1974;78:737–756. doi: 10.1093/genetics/78.2.737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lewontin R C. The Genetic Basis of Evolutionary Change. New York: Columbia Univ. Press; 1974. [Google Scholar]
- 14.Begun D J, Aquadro C F. Nature. 1992;356:519–520. doi: 10.1038/356519a0. [DOI] [PubMed] [Google Scholar]
- 15.Nachman M. Genetics. 1997;147:1303–1316. doi: 10.1093/genetics/147.3.1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stephan W, Langley C H. Genetics. 1998;150:1585–1593. doi: 10.1093/genetics/150.4.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nachman M W, Bauer V L, Crowell S L, Aquadro C F. Genetics. 1998;150:1133–1141. doi: 10.1093/genetics/150.3.1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dvorak J, Luo M-C, Yang Z-L. Genetics. 1998;148:423–434. doi: 10.1093/genetics/148.1.423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Begun D J, Whitley P, Todd B L, Waldrip-Dail H M, Clark A G. Genetics. 2000;156:1879–1888. doi: 10.1093/genetics/156.4.1879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Swanson W J, Clark A G, Waldrip-Dail H M, Wolfner M F, Aquadro C F. Proc Natl Acad Sci USA. 2001;98:7375–7379. doi: 10.1073/pnas.131568198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wolfner M F. Heredity. 1997;88:85–93. doi: 10.1038/sj.hdy.6800017. [DOI] [PubMed] [Google Scholar]
- 22.Yang Z. Comput Appl Biosci. 1997;13:555–556. doi: 10.1093/bioinformatics/13.5.555. [DOI] [PubMed] [Google Scholar]
- 23.Goldman N, Yang Z. Mol Biol Evol. 1994;11:725–736. doi: 10.1093/oxfordjournals.molbev.a040153. [DOI] [PubMed] [Google Scholar]
- 24.Ikemura T. J Mol Biol. 1981;151:389–409. doi: 10.1016/0022-2836(81)90003-6. [DOI] [PubMed] [Google Scholar]
- 25.McVean G A T, Charlesworth B. Genet Res. 1999;74:145–158. [Google Scholar]
- 26.Kliman R M, Hey J. Mol Biol Evol. 1993;10:1239–1258. doi: 10.1093/oxfordjournals.molbev.a040074. [DOI] [PubMed] [Google Scholar]
- 27.Marais G, Mouchiroud D, Duret L. Proc Natl Acad Sci USA. 2001;98:5688–5692. doi: 10.1073/pnas.091427698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.True J R, Mercer J M, Laurie C C. Genetics. 1996;142:507–523. doi: 10.1093/genetics/142.2.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hilton H R, Kliman R M, Hey J. Evolution (Lawrence, Kans) 1994;48:1900–1913. doi: 10.1111/j.1558-5646.1994.tb02222.x. [DOI] [PubMed] [Google Scholar]
- 30.Pal C, Papp B, Hurst L D. Mol Biol Evol. 2001;18:2323–2326. doi: 10.1093/oxfordjournals.molbev.a003779. [DOI] [PubMed] [Google Scholar]
- 31.Comeron J, Kreitman M. Genetics. 2000;156:1175–1190. doi: 10.1093/genetics/156.3.1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bachtrog D, Charlesworth B. Nature. 2002;416:323–326. doi: 10.1038/416323a. [DOI] [PubMed] [Google Scholar]
- 33.Powell J R, Moriyama E N. Proc Natl Acad Sci USA. 1997;94:7784–7790. doi: 10.1073/pnas.94.15.7784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Akashi H, Eyre-Walker A. Curr Biol. 1998;8:688–693. doi: 10.1016/s0959-437x(98)80038-5. [DOI] [PubMed] [Google Scholar]
- 35.Kreitman M, Antezana M. In: Evolutionary Genetics: From Molecules to Morphology. Singh R S, Krimbas C B, editors. Vol. 1. New York: Cambridge Univ. Press; 2000. pp. 82–100. [Google Scholar]
- 36.Akashi H. Genetics. 1995;139:1067–1076. doi: 10.1093/genetics/139.2.1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Barton N H. Genetics. 1995;140:821–841. doi: 10.1093/genetics/140.2.821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stephan W, Charlesworth B, McVean G. Genet Res. 1999;73:133–146. [Google Scholar]
- 39.McVean G A T, Charlesworth B. Genetics. 2000;155:929–944. doi: 10.1093/genetics/155.2.929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Barton N H. Genet Res. 1994;64:199–208. [Google Scholar]
- 41.Gillespie J H. Evolution (Lawrence, Kans) 2001;55:2161–2169. doi: 10.1111/j.0014-3820.2001.tb00732.x. [DOI] [PubMed] [Google Scholar]
- 42.Moriyama E N, Hartl D L. Genetics. 1993;134:847–858. doi: 10.1093/genetics/134.3.847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Petrov D A, Hartl D L. Proc Natl Acad Sci USA. 1999;96:1475–1479. doi: 10.1073/pnas.96.4.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Marais G, Duret L. J Mol Evol. 2001;52:275–280. doi: 10.1007/s002390010155. [DOI] [PubMed] [Google Scholar]
- 45.Comeron J M, Kreitman M, Aguade M. Genetics. 1999;151:239–249. doi: 10.1093/genetics/151.1.239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schmidt P S, Duvernell D D, Eanes W F. Proc Natl Acad Sci USA. 2000;97:10861–10865. doi: 10.1073/pnas.190338897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schmid K J, Aquadro C F. Genetics. 2001;159:589–598. doi: 10.1093/genetics/159.2.589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Akashi H. Genetics. 1994;136:927–935. doi: 10.1093/genetics/136.3.927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Comeron J M, Kreitman M. Genetics. 1998;150:767–775. doi: 10.1093/genetics/150.2.767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Birky C W, Walsh J B. Proc Natl Acad Sci USA. 1988;85:6414–6418. doi: 10.1073/pnas.85.17.6414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Paterson A H, Yann-Rong L, Zhikang L, Schertz K F, Doebley J F, Pinson S R M, Liu S-C, Stansel J W, Irvine J E. Science. 1995;269:1714–1718. doi: 10.1126/science.269.5231.1714. [DOI] [PubMed] [Google Scholar]
- 52.Stewart C-B, Wilson A C. Cold Spring Harbor Symp Quant Biol. 1987;52:891–899. doi: 10.1101/sqb.1987.052.01.097. [DOI] [PubMed] [Google Scholar]
- 53.ffrench-Constant R H. Insect Biochem Mol Biol. 1994;24:335–345. doi: 10.1016/0965-1748(94)90026-4. [DOI] [PubMed] [Google Scholar]
- 54.Wichman H A, Badgett M R, Scott L A, Boulianne C M, Bull J J. Science. 1999;285:422–424. doi: 10.1126/science.285.5426.422. [DOI] [PubMed] [Google Scholar]
- 55.Mukai T, Chigusa S I, Mettler L E, Crow J F. Genetics. 1972;72:333–355. doi: 10.1093/genetics/72.2.335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Keightley P D, Eyre-Walker A. Genetics. 1999;153:515–523. doi: 10.1093/genetics/153.2.515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lynch M, Blanchard J, Houle D, Kibota T, Schultz S, Vassilieva L, Willis J. Evolution (Lawrence, Kans) 1999;53:645–663. doi: 10.1111/j.1558-5646.1999.tb05361.x. [DOI] [PubMed] [Google Scholar]
- 58.Charlesworth B. Genet Res. 1996;68:131–150. doi: 10.1017/s0016672300034029. [DOI] [PubMed] [Google Scholar]
- 59.Gillespie J H. The Causes of Molecular Evolution. Oxford: Oxford Univ. Press; 1991. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.