Abstract
Tetraodon nigroviridis is among the smallest known vertebrate genomes and as such represents an interesting model for studying genome architecture and evolution. Previous studies have shown that Tetraodon contains several types of tandem and dispersed repeats, but that their overall contribution is >10% of the genome. Using genomic library hybridization, fluorescent in situ hybridization, and whole genome shotgun and directed sequencing, we have investigated the global and local organization of repeat sequences in Tetraodon. We show that both tandem and dispersed repeat elements are compartmentalized in specific regions that correspond to the short arms of small subtelocentric chromosomes. The concentration of repeats in these heterochromatic regions is in sharp contrast to their paucity in euchromatin. In addition, we have identified a number of pseudogenes that have arisen through either duplication of genes or the retro-transcription of mRNAs. These pseudogenes are amplified to high numbers, some with more than 200 copies, and remain almost exclusively located in the same heterochromatic regions as transposable elements. The sequencing of one such heterochromatic region reveals a complex pattern of duplications and inversions, reminiscent of active and frequent rearrangements that can result in the truncation and hence inactivation of transposable elements. This tight compartmentalization of repeats and pseudogenes is absent in large vertebrate genomes such as mammals and is reminiscent of genomes that remain compact during evolution such as Drosophila and Arabidopsis.
The reasons for which some eukaryote genomes accommodate large quantities of apparently unnecessary DNA, whereas others seem to either engage in a more efficient “pruning” process or are equipped with better protective mechanisms against parasitic elements remains a mystery (1). It is, however, largely accepted that transposable elements and pseudogenes are two categories of such sequences that generate large amounts of “ballast” DNA and thus contribute to increasing genome size. The genomes of Caenorhabditis elegans, Drosophila melanogaster, and Arabidopsis thaliana comprise between 100 and 125 Mb of euchromatin, and this fraction has now been nearly fully sequenced (2–4). In the euchromatin of these small genomes, only between 3% and 10.5% of nucleotides are contributed by transposable elements (5). Only 10 pseudogenes have been documented in Flybase (6), 432 in Wormbase (7), and 750 in the A. thaliana genome (4). The heterochromatic fraction (centromeres, short chromosome arms or dispersed heterochromatin) varies between approximately 10 and 60 Mb for A. thaliana and D. melanogaster, respectively, and concentrates a large proportion of the transposable elements of these genomes (β heterochromatin) while containing few active genes (4, 8, 9), which are often interspersed with alternating blocks of tandem repeats (α heterochromatin). The majority of this sequence has not yet been sequenced and/or assembled, mostly because the repetitive nature of these regions is likely to make any shotgun sequence assembly extremely difficult.
The only repeat-rich genome that has been extensively sequenced and analyzed is the human genome (5). Here the density and distribution of transposable elements and pseudogenes in the euchromatin contrasts severely with the situation in sequenced small genomes. At least 45% of human euchromatin is contributed by transposable elements, and the annotation of chromosomes 20, 21, and 22 (10–12) shows that ≈20% of annotated features are pseudogenes, which provides an estimate of 6,000 pseudogenes in the entire genome. Repeats and pseudogenes are scattered in the euchromatin as well as in the heterochromatin, with variations in local densities but with a relative overall uniformity. Is this situation seen in human general for vertebrates or is it typical of a genome that is much less efficient at the pruning of transposable elements and pseudogenes than a compact genome?
To investigate this issue, we have studied the distribution of transposable elements and pseudogenes in a compact vertebrate genome. Among vertebrates, the pufferfish Tetraodon nigroviridis possesses the smallest known genome (13), and we have previously characterized the sequence and genomic localization of several tandem repeat sequences (including the centromeric and subtelocentric satellite sequences) and established a preliminary catalog of transposable elements (14). This initial analysis based on 50 Mb of random shotgun reads indicated that the genome contains very few repeats (≈7%, of which 1% is contributed by transposable elements) compared with other known vertebrates such as mammals, but that most families of mobile elements, including LTR retrotransposons, non-long interspersed elements (LINEs), and DNA transposons are represented. Tetraodon is 18–30 million years distant from Takifugu rubripes, a marine pufferfish for which a whole genome shotgun (WGS) assembly is now available (15).
Here, we show that the distribution of transposable elements and pseudogenes in Tetraodon is highly compartmentalized between heterochromatin and euchromatin. The genomic landscape is reminiscent of that seen in compact genomes across other phyla and contrasts with the larger mammalian genomes. Based on these results we discuss the implications that compact eukaryotic genomes may deal with transposable elements using common mechanisms.
Materials and Methods
Colony Filter Hybridizations.
The construction of two Tetraodon bacterial artificial chromosome (BAC) libraries has been described (14). An equal number of clones from library A (pBACe.3.6, EcoRI) and library B (pBeloBAC11, HindIII) were spotted on nylon membranes (Appligene, Strasbourg, France) by using a robotic arrayer (QBOT, Genetix, New Milton, U.K.). Membranes were placed on 2YT agar overnight at 37°C, and colonies were then lysed and DNA was fixed as described (16). Each membrane contains 27,648 clones in duplicate (55,296 colonies) or 10 equivalents of the Tetraodon genome. Transposable element hybridization probes were PCR products amplified with incorporation of digoxigenin-labeled nucleotides (Roche Diagnostics) from the Tetraodon BAC clone C0AA029L14 (GenBank accession no. AJ457054; see below) and correspond to the following positions on this sequence: Dm-Line, 5′ end of putative coding region between base pairs 62656 and 62902; TC1-like, nearly the entire coding sequence between base pairs 82039 and 83147; Dr-Line, 5′ end of putative coding region between base pairs 108856 and 109120; and Copia-like, central region of the putative coding region between base pairs 66851 and 67038. The 10-bp satellite probe was prepared as described (14). After denaturation, all probes from transposable elements were hybridized without competition by using a nonisotopic protocol as described (17).
BAC Sequencing and Assembly.
The insert of BAC clone C0AA029L14 was sequenced by using standard protocols for shotgun sequencing of BAC inserts (GenBank accession no. AJ457054). Because of the particular complexity of this sequence, each of the 1,299 reads was manually edited and assembled by using the Staden package (18), taking into account the pairing and estimated distances between end-clones. The assembly was verified and confirmed by restriction digest and electrophoresis of the BAC DNA with six enzymes expected to cut frequently (AatII, BglII, BsrGI, EagI, SpeI, SphI), in comparison with the patterns expected from the assembled sequence (data not shown). In addition, the total size of the assembled sequence is in perfect agreement with restriction digests obtained with three rare cutting enzymes (NotI, PmeI, PacI; data not shown).
Fluorescent in Situ Hybridization (FISH).
Biotin- and digoxigenin-labeled Dm-Line and TC1-like probes were mixed in Quantum-Appligene high stringency Hybrisol VI at 10 ng/μl each. Probes were hybridized and detected on Tetraodon freshly thawed chromosome preparations without any pretreatment, according to the Quantum-Appligene protocol for repetitive probes (19). Preparations were mounted in 1.2 ng/μl 4′,6-diamidino-2-phenylindole in Antifade (Vector Laboratories) and analyzed by using Genus FISH-imaging equipment and software for animal chromosomes from Applied Imaging (Santa Clara, CA).
Sequence Comparisons.
The sequence of BAC clone C0AA029L14 was annotated by comparisons to nucleotide and protein databases using blast (20). WGS sequencing of the Tetraodon genome at Genoscope and the Whitehead Institute for Genome Research, (Cambridge, MA) will be described in full elsewhere. To estimate the copy numbers of enhancer of Zeste homolog 2 (EZH2), translocom-associated protein α (TRAPα), Trapeze, and isolated SET (iSET), exons of ≈100 bp were selected and compared by using blastn to 2.6 and 2.9 genome equivalent of shotgun reads from Genoscope and the Whitehead Institute, respectively. Alignments that were retained contained at least 20 consecutive identical bases (W = 20) and covered the entire length of the exons, and consecutive mismatches were not allowed (X = 5). The number of alignments was then adjusted to one genome equivalent for comparison purposes. Substitutions and insertion/deletion analysis of the Trapeze sequence was performed by first identifying all shotgun sequences that contained exons 5–8 and exons 9–12. Each set of sequences was aligned by using pileup in GCG version 10–2 (21) and manually edited. Substitutions, deletions, and insertions were then automatically identified by comparison to the original EZH2 and TRAPα sequences with purpose-built scripts.
Results
Tetraodon Transposable Elements Are Highly Compartmentalized in Heterochromatin.
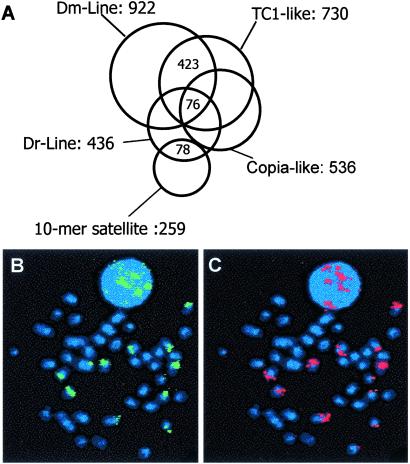
To investigate the distribution of transposable elements in the Tetraodon genome, we hybridized four probes to a 10-genome equivalent Tetraodon BAC library. Each probe was specific for a different element, which belongs to the two main classes of transposable elements: class I retrotransposons with and without LTR (probe copia-like, Dm-Line, and Dr-Line), and DNA transposons (probe TC1-like). Each probe identifies between 1.6% and 3.3% of the clones arrayed on the membranes (Fig. 1A), in agreement with previous results that showed that the Tetraodon genome contains few transposable elements compared with other vertebrate genomes (14). Strikingly, the four probes hybridize to an unexpectedly high fraction of clones in common. For instance, of 922 clones that hybridize to the Dm-Line probe (a retrotransposon), 423 clones (46%) also hybridize to the TC1-like probe (a DNA transposon). This finding shows that the four transposable elements are clustered in close proximity to each other in regions that cover ≈5% of the genome (1,562 clones of 27,648).
Figure 1.
One satellite and four transposable element probes were hybridized to a 10-genome equivalent Tetraodon BAC library, and a large fraction of clones in common were identified. (A) The four transposable elements include Dr-Line, a LINE element similar to a Danio rerio (zebrafish) LINE; Dm-Line, which is a different LINE element homologous to Drosophila factor I; a TC1-like, and a copia-like sequences. Together the five probes (10-mer satellite, Dr-Line, Dm-Line, TC1-like, and Copia-like) identify 1,743 clones, of which 1,562 are identified with at least one transposable element and 76 clones hybridize with all four elements. The Dm-Line (B) and the TC1-like (C) probes were cohybridized by FISH to the same Tetraodon metaphase chromosome preparations and show distinct signals in the same pairs of small subtelocentric chromosomes. (Magnification: ×750.)
The BAC library was then hybridized with a 10-bp tandem repeat that we previously localized specifically in short heterochromatic arms of approximately 10 pairs of subtelocentric chromosomes (19). Of the 259 positive clones, 30% are shared with the set of clones identified by the Dr-Line element, instead of the 0.015% expected if the two sequences were independently distributed in the genome. This finding clearly suggests that the satellite and the LINE element are located in the same regions of the genome. To confirm this result, we cohybridized the Dm-Line and the TC1-like probes to metaphase chromosomes by double-color FISH (Fig. 1 B and C). The two probes specifically hybridize to the same short heterochromatic arms of six pairs of subtelocentric chromosomes, where the 10-mer satellite resides.
Frequent Rearrangements in Heterochromatin.
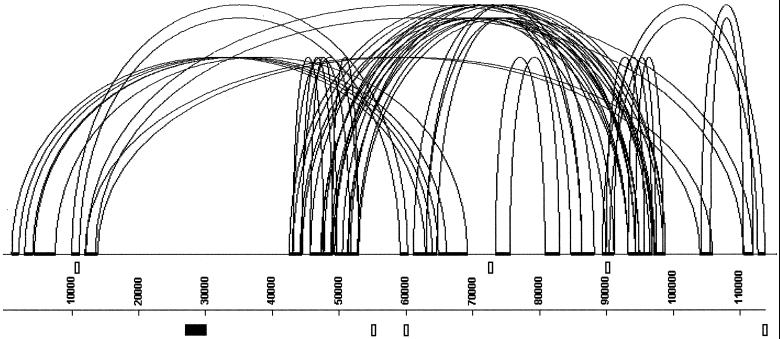
We further investigated the local organization of Tetraodon heterochromatin by sequencing one of the 76 BAC clones that was positive with all four probes tested on the library (BAC clone C0AA029L14, Fig. 1A). A comparison of the 115-kb insert sequence against itself shows that 34% is duplicated at least once either in tandem or inverted orientation, in multiple fragments ranging from a few hundred base pairs to 15 kb (Fig. 2). This organization suggests that frequent rearrangements take place between such sequences in this region. The insert contains 18 transposable elements (Table 1) of which only two are complete. These are duplicate copies of a TC1-like transposon with the same in-frame stop codon within the gene, indicating that both are nonfunctional and that one originates from a local duplication. The abundance of transposable elements in this BAC clone is in sharp contrast with their average frequency in the Tetraodon sequence database (0.9% of nucleotides) (14), thus supporting at the sequence level our observations based on hybridization results that indicate a high density of transposable elements in heterochromatic regions compared with the rest of the genome.
Figure 2.
Structure and annotation of BAC clone C0AA029L14 positive with all four transposable elements tested on the BAC library. Analysis of duplicated sequences larger than 1,000 bases with miropeats (45) shows that >34% of the sequence is not unique. Arched lines join the beginning and end of duplicated regions that are represented as thick horizontal lines. The Trapeze pseudogene (filled box) and the six iSET pseudogenes (open boxes) are represented on either side of the scale bar (forward and reverse strand).
Table 1.
Transposable elements, genes, and pseudogenes identified in BAC C0AA029L14
| Category | Name | Copies | Total | % of BAC DNA |
|---|---|---|---|---|
| Transposable elements | Copia-Ten | 3 | 18 | 16.6 |
| Rex1-Ten | 4 | |||
| TC1-Ten | 2 | |||
| Factorl-Ten | 3 | |||
| Maui-Ten | 1 | |||
| BUSTER2 like | 5 | |||
| Genes and pseudogenes | iSET domains | 6 | 13 | 8.2 |
| Protein kinases | 5 | |||
| Trapeze | 1 | |||
| Unknown ORF | 1 |
Trapeze, a Chimeric Pseudogene, Is Present in 50 Copies.
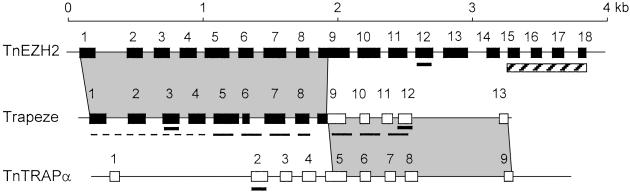
The 13 genes and gene fragments identified in the BAC insert can be grouped into three categories: (i) one highly conserved ORF; (ii) five gene fragments with homologies to kinase genes; and (iii) one Trapeze and six iSET pseudogenes. The highly conserved ORF finds similar sequences in metazoans, plants, and prokaryotes, and belongs to the Cluster of Orthologous Group (COG) of proteins COG1990 (22) whose members are of unknown function. The pseudogene that we have called Trapeze is made up of 13 exons based on similarities to known mammalian genes. The first eight exons are similar to the human EZH2 gene, and the last four exons are similar to the human TRAPα gene (Fig. 3), whereas exon 9 is chimeric, containing sequences similar to both EZH2 (exon 9) and TRAPα (exon 5). This fusion event introduces a frame shift that comes in addition to the absence of a start methionine and suggests that Trapeze is a pseudogene. In addition, no expression product of a potential Trapeze gene could be found by either RT-PCR or cDNA library screenings. Several shotgun plasmid clones from the WGS sequence project were identified as containing the native Tetraodon EZH2 and TRAPα genes and fully sequenced, revealing the complete structure of the two genes (Fig. 3). This result, in addition to the identification of a TRAPα cDNA clone in a Tetraodon cDNA library by hybridization, indicates that the native EZH2 and TRAPα genes still exist in the Tetraodon genome. Trapeze is thus the result of a fusion event between duplicated copies of the Tetraodon EZH2 and TRAPα genes.
Figure 3.
Exon–intron structure of the Trapeze pseudogene (GenBank accession no. AJ313483; Middle) and the native Tetraodon EZH2 gene (GenBank accession no. AJ313481, Top) and TRAPα genes (GenBank accession no. AJ313482, Bottom). Filled exons indicate EZH2 exons both in the native gene and Trapeze, and open exons indicate TRAPα exons, both in Trapeze and the native gene. Exon 9 of Trapeze is the result of a fusion between part of exon 9 of EZH2 and part of exon 5 of TRAPα. The Tetraodon EZH2 gene spans <4 kb, a 10-fold reduction in size compared with the human EZH2 gene (46). Underlined exons were used to evaluate the number of copies of each gene in the Tetraodon genome by sequence comparison. The region in Trapeze underlined with a thin dashed line was used as probe in FISH and BAC library hybridization, and the region underlined with a thick dashed line was used for deletion rate measurements. The dashed box spans the exons that code for the SET domain of EZH2.
In the process of exploring the Tetraodon sequence database for sequences homologous to either EZH2 or TRAPα, we identified an unexpectedly high number of matching copies. To confirm this finding, we selected one exon in each unique half of the EZH2 and TRAPα genes and two exons from Trapeze (exons 3 and 12, Fig. 3) and searched for matching sequences in two independent Tetraodon WGS databases. Results show a striking difference between exons that are in Trapeze and exons that are uniquely found in the single-copy genes EZH2 and TRAPα (Table 2). Exons 2 of TRAPα and exon 12 of EZH2 identify an average of one sequence as expected for single-copy exons (see Materials and Methods), whereas in contrast exons 3 and 12 of Trapeze identify an average of 51 sequences. This finding suggests that the fusion of EZH2 and TRAPα to create Trapeze was followed by an amplification process that results today in the presence of approximately 50 copies of this chimeric pseudogene. Indeed, because there is no evidence of more than one original complete copy of EZH2 and TRAPα the amplification of sequences derived from EZH2 and TRAPα must be subsequent to the fusion event.
Table 2.
Copies of EZH2, TRAPα, Trapeze, and iSET exons identified in Tetraodon shotgun data
| Sequence | Number of copies in 1× Genoscope shotgun | Number of copies in 1× WICGR shotgun |
|---|---|---|
| TRAP_exon2 | 0 | 1 |
| EZH2_exon12 | 3 | 0 |
| Trapeze_exon3 | 100 | 31 |
| Trapeze_exon12 | 75 | 28 |
| iSET | 192 | 318 |
WICGR, Whitehead Institute for Genome Research.
iSET, a Retrotransposed Pseudogene, Is Highly Amplified in the Genome.
The C-terminal region of human EZH2, mouse EZH1, and fly E(z) proteins contain a highly conserved SET domain (23). This domain spans the last five exons of the predicted Tetraodon EZH2 protein. Because Trapeze is similar only to the first eight exons of EZH2, it does not contain the SET sequence. However, the sequence of the BAC clone C0AA029L14 contains six dispersed copies of an iSET domain. The six 125-aa sequences are nearly identical to each other but only 57% similar and 39% identical to the Tetraodon EZH2 SET domain (see Fig. 5, which is published as supporting information on the PNAS web site, www.pnas.org). The iSET sequence is intronless and contains a stop codon in position 77, implying that it is not part of a functional protein. A comparison of iSET to TREMBL (24) shows that the best alignment is obtained with the SET domain of the Tetraodon EZH2 domain. Taken together, these observations suggest that iSET is a pseudogene that originates from the reverse transcription of a truncated 3′ EZH2 mRNA. The sequences flanking the six copies of iSET in the insert of the BAC clone are always identical up to at least a few hundred base pairs upstream and downstream, indicating that iSET does not multiply as an isolated sequence but rather as part of larger segmental duplications. The minimal region of sequence identity between the six loci containing iSET is 2.3 kb. Interestingly the sequences flanking iSET sequences always include the same 3′ segment of a LINE element 900 bp upstream, pointing toward a possible mechanism for the appearance of this pseudogene (see Discussion). No poly(A) tail in the immediate downstream region of iSET could be identified. The high level of divergence between iSET and the EZH2 SET domain indicates a very ancient retrotranscription event whereas the high similarity between six copies that are presumably under no selection indicate a very recent serial duplication event. When searched against the two available Tetraodon WGS databases, iSET identifies an average of 240 sequences per genome, and thus is ≈5 times more abundant than Trapeze (Table 2).
The Multiple Copies of Trapeze and iSET Are Tightly Linked to Transposable Elements and Heterochromatin.
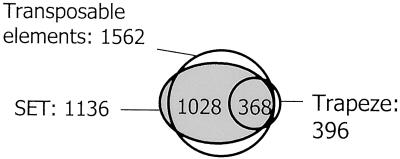
We hybridized one probe from Trapeze (spanning exons 1–4) and one from iSET to the Tetraodon BAC library. Trapeze identified 396 positive clones, of which 368 (93%) are included in the 1,562 clones that we previously identified with the four transposable element probes (Fig. 4), indicating that the 50 copies of Trapeze are not randomly distributed in the genome but are located mainly in heterochromatin, in close association with transposable elements. We hybridized the same probe onto metaphase chromosomes, revealing dispersed and abundant signals in short heterochromatic arms of ≈6–10 pairs of subtelocentric chromosomes (data not shown), i.e., at the same location where transposable elements and the 10-mer satellite repeat are preferentially located. No signals could be seen in the rest of the genome. The 1,136 positive clones identified by the iSET probe in the 10 X BAC library is consistent with approximately 240 copies in the genome. Notably, 1,028 (66%) of the positive clones are common with clones identified by transposable elements and include the 368 clones previously identified by Trapeze (Fig. 4).
Figure 4.
Hybridization results from the iSET domain and the Trapeze pseudogene, in comparison with those of transposable elements. Of the 1,136 BAC clones identified by iSET, 1,028 were already identified by one of the four transposable elements previously hybridized to the library. Similarly, of the 396 clones positive with Trapeze, 368 are also positive with iSET, indicating that both sequences are always physically associated in the genome.
These data indicate that iSET and Trapeze are invariably found in the same BAC clones, and that these clones represent a distinct subset of the entire library, which was also previously shown to concentrate most transposable elements. This is evidence, in a vertebrate genome, of tight linkage between two different pseudogenes that originally seem to derive from the same gene, are amplified to approximately 50 and 240 dispersed copies, respectively, and are confined to a very specific compartment of the genome, i.e., the heterochromatin.
The Trapeze Pseudogene Is Submitted to a High Rate of DNA Loss.
Pseudogenes are sequences that are generally thought to be unconstrained by natural selection and thus diverge neutrally by the accumulation of substitutions, insertions, and deletions. This feature makes it possible to use pseudogenes to study the fate of nonfunctional DNA in a genome (25, 26). For instance, it has been shown that pseudogenes in the small genomes of different species of Drosophila are submitted to a higher rate of deletions compared with the larger genomes of mammals (27) and to the 11 times larger genome of the Laupala cricket (28), thus providing evidence that DNA loss may be a long-term determinant of genome size.
Here, we have examined the pattern of substitutions, insertions, and deletions in multiple copies of two regions of the Trapeze pseudogene: 19 copies of the sequence encompassing exons 5–8 in comparison to the homologous region of the EZH2 gene (874 bp), and 47 copies of the sequence encompassing exon 9 and exon 12 in comparison to the homologous region in the TRAPα gene (690 bp) (Fig. 3 and Table 3). This set of 66 Trapeze sequences represents all of the single reads that spanned the required region of the pseudogene sampled from the entire Tetraodon WGS database. From the results it is striking that exons and introns are submitted to very different mutation rates. Substitutions, insertions, and deletions are 1.7, 2.1, and 9.9 times more frequent in exons, respectively, and the average deletion size in exons is more than three times larger than in introns. This limited dataset provides a glimpse at the rate of DNA loss in unconstrained DNA in the Tetraodon genome. The mutational rate of DNA loss through small deletions and insertions can be expressed per nucleotide per nucleotide substitution as D = [(rate of deletion per nucleotide substitution) × (average deletion size)] − [(rate of insertion per nucleotide substitution) × (average insertion size)] (29). We find that the rate of DNA loss for the Trapeze sequences examined here is D = 0.5619, suggesting that Tetraodon nonfunctional DNA is submitted to a higher rate of deletions than in human or mouse, but lower than in Drosophila (29). Overall it is striking that this value is in excellent agreement with the negative correlation between genome size and rate of DNA loss observed across genomes from dipterians, mammals, and nematodes, i.e., that smaller genomes tend to be submitted to a higher rate of DNA loss.
Table 3.
Substitutions, insertions, and deletions in exons and introns of Trapeze pseudogenes
| Substitutions | Insertions | Deletions | Average insertion sizes, bp | Average deletion sizes, bp | |
|---|---|---|---|---|---|
| Exons | 803 | 44 | 82 | 1.3 | 8.3 |
| Introns | 481 | 21 | 9 | 1.8 | 2.7 |
| Total | 1,282 | 65 | 91 | 1.5 | 7.7 |
To enable comparisons between exons and introns, values for introns are adjusted to correct for the fact that the total size of introns is smaller than the total size of exons. Values for exons are absolute values. The value of DNA loss rate (D; see text) was, however, calculated across all exons and introns by using uncorrected values.
Discussion
The distribution of transposable elements and pseudogenes in the compact pufferfish T. nigroviridis genome is in sharp contrast to the situation seen in the larger genome of other vertebrates such as human or mouse. In Tetraodon, transposable elements are tightly clustered in a specific compartment where abundant satellite sequences are found and they constitute a minimal fraction of the genome (<10%). These regions are the short heterochromatic arms of approximately 10 pairs of small subtelocentric chromosomes. In addition, although pseudogenes were previously thought to be an extremely rare feature in pufferfishes with a single documented case in Takifugu rubripes (30), we show that the Tetraodon genome contains several hundred copies of a few highly amplified pseudogenes, found in close association with transposable elements.
A number of studies have already shown that transposable elements are preferentially clustered in or around centric and telomeric heterochromatin in other sequenced genomes such as that of Drosophila and Arabidopsis (4, 8, 31–34). Although some plant and insect genomes may reach enormous sizes in some species, that of Arabidopsis and Drosophila each represent a very compact version of the genome in their respective phylum, as does the genome of Caenorhabditis elegans among nematodes. It is striking that the genome of Tetraodon, the smallest known vertebrate genome, shares a number of features with these compact genomes, such as a preferential clustering of transposable elements in heterochromatin and the paucity of euchromatic pseudogenes. This feature is not shared with other vertebrate genomes such as mouse or human, where transposable elements and pseudogenes are abundant and well dispersed in euchromatin (5). Thus, we can delineate across several phyla a common trend in four small genomes: they are not immune to repeat invasions but the amplification and dispersion of parasitic sequences are somehow contained and show similar distribution profiles.
In Tetraodon, we show that some pseudogenes are highly amplified but remain exclusively in heterochromatin. The sequencing of a BAC clone from such a region shows that the multiple copies amplify by duplications of large segments as opposed to being restricted to the pseudogene sequences only. In all copies found, the iSET pseudogene, which corresponds to the last four exons of the EZH2 gene, is systematically downstream of and on the same segment as the 3′ end of a LINE element highly similar to the Drosophila factor I element (35) (Dm-Line in the hybridization experiments). It is thus possible that this LINE element was once inserted in intron 14 (Fig. 3) of the EZH2 gene and generated the iSET pseudogene via a transduction or “read through” mechanism where the LINE RNA polymerase skipped its own 3′ end to instead use the poly(A) signal of the EZH2 gene (36–38). Because the current Tetraodon EZH2 gene does not show any trace of this LINE element, this event would have been followed by the excision of the LINE copy from the intron of the EZH2 gene. We believe that the emergence of the first copy of the iSET pseudogene and the amplification of the different copies seen in the genome today are two independent events separated by a long evolutionary distance, judging by the high sequence similarity between the different copies (98% amino acid similarity), but the high divergence between these sequences and the original EZH2 exons (57% similarity).
Trapeze, the second highly prevalent pseudogene in T. nigroviridis heterochromatin, is not retroprocessed but is a chimeric structure between part of the EZH2 gene and part of the TRAPα gene. The much higher sequence similarity between the homologous parts of Trapeze and EZH2 compared with iSET and EZH2 suggests that the emergence of Trapeze occurred later than that of iSET.
The analysis of the spectrum of substitutions, insertions, and deletions in Trapeze is in agreement with previous studies showing that the number of deletions is higher than insertions in neutrally evolving DNA (27, 39). However, although most studies have focused on defunct transposable elements or retroprocessed pseudogenes, we have studied a pseudogene that retains an intron/exon structure. Strikingly, across all types of mutations (deletions, insertions, and substitutions) the mutational rate is significantly higher in exons than in introns. Assuming a model of uniform mutation frequencies in neutrally evolving DNA, these data suggest that mutations affecting exons are more likely to be retained than those affecting introns, implying that natural selection may be acting to reduce the potential of a pseudogene to produce a functional mRNA, let alone a functional protein. Overall, the deletion rate, in terms of frequency and size of deletions, is higher than in mammals and supports the view that small, but constant, deletions over long evolutionary periods may be one of the forces that tend to decrease the size of a genome (1, 40). Because Trapeze and iSET sequences seem to reside almost exclusively in heterochromatin, one question is whether their absence from euchromatin is the result of (i) a more intense deletion rate of parasitic sequences in this compartment of the genome, (ii) a transfer of such sequences from euchromatin to heterochromatin, or (iii) an amplification that is itself confined to heterochromatin. Any of these hypotheses would require a mechanism that is not yet known to occur in eukaryotic cells.
It is intriguing that the EZH2 gene is at the origin of both Trapeze and iSET pseudogenes, albeit via different routes. Strong experimental evidence links EZH2 to a conserved mechanism of eukaryotic gene silencing (41), and this protein has been shown to form a complex with the embryonic ectoderm development protein (42) and histone deacetylases that mediate the repression of gene transcription (43). More generally, SET domain proteins are seen as multifunctional chromatin regulators with roles that include telomeric and centromeric gene silencing (23) and possibly the determination of chromosome architecture. This observation raises the question of a potential link between the presence of high copy number EZH2-derived pseudogenes in transcriptionally repressed heterochromatin and the known function of EZH2 in eukaryotic cells.
In the human (5), Drosophila (44), and Takifugu (15) genome assembly projects, heterochromatin has been left out from the initial assembly for technical reasons, although many clones and shotgun sequences are available. This compartment is, however, likely to be a dynamic and vital part of the nucleus, and essential for understanding many aspects of genome evolution and nuclear organization. We would like to draw attention to the vast amount of shotgun sequences that remain to be studied in human, Drosophila, Takifugu, and probably also mouse after the initial assembly, as a way to access and study the heterochromatic compartment in different eukaryotic genomes as was done here for Tetraodon. In particular, comparisons with Takifugu heterochromatic sequences should reveal whether the same clustering of transposable elements occurs and whether Trapeze and iSET pseudogenes also exist and are similarly amplified.
Our results for the Tetraodon genome contrast with well-known mammalian genomes and underline the fact that organisms from different phyla (plants, invertebrates, and vertebrates) with small genomes appear to share common features in terms of transposable element and pseudogene content and distribution. This finding may suggest common mechanisms in dealing with such sequences and may guide further studies on the molecular evolution of pseudogenes and transposable elements for better understanding of how the smallest known vertebrate genome achieves this extraordinary efficiency in packaging genetic information.
Supplementary Material
Acknowledgments
We thank Fiona Francis for critical reading of the manuscript, Frederic Brunet, Pierre Capy, and an anonymous reviewer for insightful comments on an earlier version of the text, and the Whitehead Institute Center for Genome Research for early access to part of the Tetraodon shotgun sequences used in this study.
Abbreviations
- WGS
whole genome shotgun
- BAC
bacterial artificial chromosome
- FISH
fluorescent in situ hybridization
- iSET
isolated SET
- TRAP
translocom-associated protein
- E2H2
enhancer of Zeste homolog 2
- LINE
long interspersed element
Footnotes
References
- 1.Hartl D L. Nat Rev Genet. 2000;1:145–149. doi: 10.1038/35038580. [DOI] [PubMed] [Google Scholar]
- 2.The C. elegans Sequencing Consortium. Science. 1998;282:2012–2018. doi: 10.1126/science.282.5396.2012. [DOI] [PubMed] [Google Scholar]
- 3.Adams M D, Celniker S E, Holt R A, Evans C A, Gocayne J D, Amanatides P G, Scherer S E, Li P W, Hoskins R A, Galle R F, et al. Science. 2000;287:2185–2195. doi: 10.1126/science.287.5461.2185. [DOI] [PubMed] [Google Scholar]
- 4.The Arabidopsis Genome Initiative. Nature. 2000;408:796–815. doi: 10.1038/35048692. [DOI] [PubMed] [Google Scholar]
- 5.International Human Genome Sequencing Consortium. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 6.The Flybase Consortium. Nucleic Acids Res. 1999;27:85–88. doi: 10.1093/nar/26.1.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stein L, Sternberg P, Durbin R, Thierry-Mieg J, Spieth J. Nucleic Acids Res. 2001;29:82–86. doi: 10.1093/nar/29.1.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pimpinelli S, Berloco M, Fanti L, Dimitri P, Bonaccorsi S, Marchetti E, Caizzi R, Caggese C, Gatti M. Proc Natl Acad Sci USA. 1995;92:3804–3808. doi: 10.1073/pnas.92.9.3804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dimitri P, Junakovic N. Trends Genet. 1999;15:123–124. doi: 10.1016/s0168-9525(99)01711-4. [DOI] [PubMed] [Google Scholar]
- 10.Deloukas P, Matthews L H, Ashurst J, Burton J, Gilbert J G, Jones M, Stavrides G, Almeida J P, Babbage A K, Bagguley C L, et al. Nature. 2001;414:865–871. doi: 10.1038/414865a. [DOI] [PubMed] [Google Scholar]
- 11.Hattori M, Fujiyama A, Taylor T D, Watanabe H, Yada T, Park H S, Toyoda A, Ishii K, Totoki Y, Choi D K, et al. Nature. 2000;405:311–319. doi: 10.1038/35012518. [DOI] [PubMed] [Google Scholar]
- 12.Dunham I, Shimizu N, Roe B A, Chissoe S, Hunt A R, Collins J E, Bruskiewich R, Beare D M, Clamp M, Smink L J, et al. Nature. 1999;402:489–495. doi: 10.1038/990031. [DOI] [PubMed] [Google Scholar]
- 13.Hinegardner R. Am Naturalist. 1968;102:517–523. [Google Scholar]
- 14.Roest Crollius H, Jaillon O, Dasilva C, Ozouf-Costaz C, Fizames C, Fischer C, Bouneau L, Billault A, Quetier F, Saurin W, et al. Genome Res. 2000;10:939–949. doi: 10.1101/gr.10.7.939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Aparicio S, Chapman J, Stupka E, Putnam N, Chia J M, Dehal P, Christoffels A, Rash S, Hoon S, Smit A F, et al. Science. 2002;297:1301–1310. doi: 10.1126/science.1072104. [DOI] [PubMed] [Google Scholar]
- 16.Nizetic D, Drmanac R, Lehrach H. Nucleic Acids Res. 1991;19:182. doi: 10.1093/nar/19.1.182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Maier E, Roest Crollius H, Lehrach H. Nucleic Acids Res. 1994;22:3423–3424. doi: 10.1093/nar/22.16.3423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Staden R. Mol Biotechnol. 1996;5:233–241. doi: 10.1007/BF02900361. [DOI] [PubMed] [Google Scholar]
- 19.Fischer C, Ozouf-Costaz C, Roest Crollius H, Dasilva C, Jaillon O, Bouneau L, Bonillo C, Weissenbach J, Bernot A. Cytogenet Cell Genet. 2000;88:50–55. doi: 10.1159/000015484. [DOI] [PubMed] [Google Scholar]
- 20.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 21.Devereux J, Haeberli P, Smithies O. Nucleic Acids Res. 1984;12:387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tatusov R L, Galperin M Y, Natale D A, Koonin E V. Nucleic Acids Res. 2000;28:33–36. doi: 10.1093/nar/28.1.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jenuwein T, Laible G, Dorn R, Reuter G. Cell Mol Life Sci. 1998;54:80–93. doi: 10.1007/s000180050127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bairoch A, Apweiler R. Nucleic Acids Res. 2000;28:45–48. doi: 10.1093/nar/28.1.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li W H, Gojobori T, Nei M. Nature. 1981;292:237–239. doi: 10.1038/292237a0. [DOI] [PubMed] [Google Scholar]
- 26.Petrov D A, Hartl D L. J Hered. 2000;91:221–227. doi: 10.1093/jhered/91.3.221. [DOI] [PubMed] [Google Scholar]
- 27.Petrov D A, Lozovskaya E R, Hartl D L. Nature. 1996;384:346–349. doi: 10.1038/384346a0. [DOI] [PubMed] [Google Scholar]
- 28.Petrov D A, Sangster T A, Johnston J S, Hartl D L, Shaw K L. Science. 2000;287:1060–1062. doi: 10.1126/science.287.5455.1060. [DOI] [PubMed] [Google Scholar]
- 29.Petrov D. Theor Popul Biol. 2002;61:531–543. doi: 10.1006/tpbi.2002.1605. [DOI] [PubMed] [Google Scholar]
- 30.Clark M S, Pontarotti P, Gilles A, Kelly A, Elgar G. J Immunol. 2000;165:4446–4452. doi: 10.4049/jimmunol.165.8.4446. [DOI] [PubMed] [Google Scholar]
- 31.Biessmann H, Champion L E, O'Hair M, Ikenaga K, Kasravi B, Mason J M. EMBO J. 1992;11:4459–4469. doi: 10.1002/j.1460-2075.1992.tb05547.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Le M-H, Duricka D, Karpen G H. Genetics. 1995;141:283–303. doi: 10.1093/genetics/141.1.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Junakovic N, Terrinoni A, Di Franco C, Vieira C, Loevenbruck C. J Mol Evol. 1998;46:661–668. doi: 10.1007/pl00006346. [DOI] [PubMed] [Google Scholar]
- 34.Kapitonov V V, Jurka J. Genetica. 1999;107:27–37. [PubMed] [Google Scholar]
- 35.Bucheton A, Paro R, Sang H M, Pelisson A, Finnegan D J. Cell. 1984;38:153–163. doi: 10.1016/0092-8674(84)90536-1. [DOI] [PubMed] [Google Scholar]
- 36.Moran J V, DeBerardinis R J, Kazazian H H., Jr Science. 1999;283:1530–1534. doi: 10.1126/science.283.5407.1530. [DOI] [PubMed] [Google Scholar]
- 37.Goodier J L, Ostertag E M, Kazazian H H., Jr Hum Mol Genet. 2000;9:653–657. doi: 10.1093/hmg/9.4.653. [DOI] [PubMed] [Google Scholar]
- 38.Esnault C, Maestre J, Heidmann T. Nat Genet. 2000;24:363–367. doi: 10.1038/74184. [DOI] [PubMed] [Google Scholar]
- 39.Ophir R, Graur D. Gene. 1997;205:191–202. doi: 10.1016/s0378-1119(97)00398-3. [DOI] [PubMed] [Google Scholar]
- 40.Petrov D A. Trends Genet. 2001;17:23–28. doi: 10.1016/s0168-9525(00)02157-0. [DOI] [PubMed] [Google Scholar]
- 41.Laible G, Wolf A, Dorn R, Reuter G, Nislow C, Lebersorger A, Popkin D, Pillus L, Jenuwein T. EMBO J. 1997;16:3219–3232. doi: 10.1093/emboj/16.11.3219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sewalt R G, van der Vlag J, Gunster M J, Hamer K M, den Blaauwen J L, Satijn D P, Hendrix T, van Driel R, Otte A P. Mol Cell Biol. 1998;18:3586–3595. doi: 10.1128/mcb.18.6.3586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.van der Vlag J, Otte A P. Nat Genet. 1999;23:474–478. doi: 10.1038/70602. [DOI] [PubMed] [Google Scholar]
- 44.Myers E W, Sutton G G, Delcher A L, Dew I M, Fasulo D P, Flanigan M J, Kravitz S A, Mobarry C M, Reinert K H, Remington K A, et al. Science. 2000;287:2196–2204. doi: 10.1126/science.287.5461.2196. [DOI] [PubMed] [Google Scholar]
- 45.Parsons J D. Comput Appl Biosci. 1995;11:615–619. doi: 10.1093/bioinformatics/11.6.615. [DOI] [PubMed] [Google Scholar]
- 46.Cardoso C, Mignon C, Hetet G, Grandchamps B, Fontes M, Colleaux L. Eur J Hum Genet. 2000;8:174–180. doi: 10.1038/sj.ejhg.5200439. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.