Abstract
The SIR2 (silent information regulator 2) gene family has diverse functions in yeast including gene silencing, DNA repair, cell-cycle progression, and chromosome fidelity in meiosis and aging. Human homologues, termed sirtuins, are highly conserved but are of unknown function. We previously identified a large imprinted gene domain on 11p15.5 and investigated the 11p15.5 sirtuin SIRT3. Although this gene was not imprinted, we found that it is localized to mitochondria, with a mitochondrial targeting signal within a unique N-terminal peptide sequence. The encoded protein was found also to possess NAD+-dependent histone deacetylase activity. These results suggest a previously unrecognized organelle for sirtuin function and that the role of SIRT3 in mitochondria involves protein deacetylation.
In eukaryotes, transcriptional domains are divided into active euchromatin and silenced heterochromatin. Mechanisms of active chromatin transcription have been studied extensively (1). However, factors that render silenced chromatin inaccessible to the transcriptional machinery are much less clear. One of the factors involved in silencing in yeast is the SIR2 gene (2). The yeast SIR2 gene belongs to a highly conserved family found in both prokaryotes and eukaryotes (3). The yeast-encoded protein Sir2p maintains the heterochromatic state at the mating-type loci, telomeres, and rRNA-encoding DNA repeats (2, 4). Increased dosage of the SIR2 gene increases life span in yeast (5) and Caenorhabditis elegans (6) because of a failure to repress recombination generated by the Fob1p-mediated replication block in the rRNA-encoding DNA in yeast and coupling longevity to nutrient availability in C. elegans (6). Sir2p is also involved in repair of DNA double-strand breaks (7) and progression of the cell cycle through anaphase (8) and in a meiotic checkpoint that ensures chromosome fidelity during segregation (9). However, the biochemical mechanisms by which Sir2p functions are largely unknown, although histones are excellent substrates in vitro and probably in vivo as well. Recently it was shown that yeast SIR2 possesses at least two NAD-dependent enzymatic activities, a putative ribosylation activity (10) and a well established protein deacetylase activity (11–13).
Yeast has four additional Sir2-like genes termed “HST genes” (homologues of Sir two), whereas seven homologues termed “sirtuins” have been identified in human (14, 15). It is not clear what the distinct functions of the SIR2 homologues are; however, yeast hst3 and hst4 mutants have multiple effects on telomeric silencing, chromosome stability, and cell growth (14). Overexpression of HST2 in yeast depresses subtelomeric silencing and increases repression of rRNA-encoding DNA sequences (16). Consistent with its effect on silencing in the rRNA-encoding DNA and at other loci, the yeast Sir2p protein has been localized to the nucleus, with the bulk of the protein in the nucleolus and telomeric foci (17). Recently, the human tumor suppressor gene p53 was shown to be down-regulated modestly when SIRT1 was overexpressed (18, 19). Mouse Sirt1 deacetylated TAFI68, leading to repression of RNA polymerase I transcription in vitro (20). However, yeast Hst2p, human hSirT2p, and human SirT1p have been localized to the cytoplasm (16, 21). The presence of a sirtuin in the cytoplasm is intriguing and suggests that these genes may have a more global protein deacetylase activity or other function. Identification of substrates targeted by the Sir2 homologues remains a formidable task.
Our laboratory has identified a domain of imprinted genes on 11p15, i.e., genes that show silencing of a specific parental allele. As part of our analysis of 11p15 imprinting, we focused on the sirtuin SIRT3 in this region. Although the gene was not imprinted, it showed surprising subcellular localization and biochemical activity.
Materials and Methods
Isolation, Mapping, and Cloning of SIRT3.
Chromosome 11-mapped UniGene collection (www.ncbi.nlm.nih.gov/UniGene) of ESTs were obtained from Genome Systems (St. Louis). We used chromosome 11p15 somatic hybrid DNA, isolated from either J1 cell lines or a subchromosomal transferable fragment cell line for Southern blots (22, 23). ESTs were mapped onto Southern blot mapping panels generated after EcoRI DNA digests. In addition, similarly treated DNA samples from human placenta and the two parental somatic hybrid cell lines A9 and Chinese hamster ovary were included as controls. We sequenced the inserts of the ESTs that mapped to 11p15.5 and selected EST9 (our designation; GenBank accession no. R55106) for detailed analysis. We performed 5′ RACE to identify the missing 5′ region, but we were unsuccessful probably because of a secondary structure in template mRNA, preventing the RT enzyme access. We therefore developed a technique we termed 5′ prediction RT-PCR to identify the missing 5′ region. The 5′ prediction RT-PCR involved using 5′ anchor primers designed from the available genomic sequence (GenBank accession no. AF015416), which were located at various distances 5′ of EST9 sequence. In each 5′ prediction RT-PCR analysis, primer RIS2FP-R was used in combination with the following anchor primers: 1–5′ RIS (5′-TTAACCACCGCGGTGGCGGCCGCTGGTTGCTCAGTTACAGC-3′), 2–5′ RIS (5′-TTAACCACCGCGGTGGCGGCCGCACTTGGCTGTGCCATTG-3′), 3–5′ RIS (5′-TTAACCACCGCGGTGGCGGCCGCGAGAGATGAGACACCAGACT-3′), and 4–5′ RIS (5′-TTAACCACCGCGGTGGCGGCCGCACTCCTCGGACTGCGCGGAAC-3′). To obtain full-length cDNA for this EST we performed 5′ RACE by using Invitrogen's kit. The gene-specific primer used for 5′ RACE was RIS2FP-R (5′-CATGGCGATGGATCCAGCTTCCCAGTTTCCCGCTG-3′). RT-PCR was performed by using Invitrogen's superscript first-strand synthesis system kit for RT-PCR according to supplied protocols. PCR conditions were as follows: 95°C for 2.5 min; 40 cycles of 95°C for 1 min, 60°C for 30 sec, 72°C for 3 min; final extension at 72°C for 9 min. RT-PCR products were sequenced as described below. Thus, we obtained a complete cDNA that was directionally cloned into the BamHI–EcoRI sites of pEGFP.N3 vector (CLONTECH).
Database Sequence Analysis.
Sequence searches were done online by using the BLAST family of programs on the National Center for Biotechnology Information web site (www.ncbi.nlm.nih.gov/blast/). Multiple sequence alignment was done by using CLUSTALV and shaded by using BOXSHADE (BCM Launcher, http://searchlauncher.bcm.tmc.edu/). Protein database searches were performed at the BCM Launcher web site. The SEQUENCHER program (Gene Codes, Ann Arbor, MI) was used for contig assembly, cDNA-to-protein translation, and DNA-sequence comparison. Exon predication were done by using GENSCAN (http://genes.mit.edu/GENSCAN.html) and the Gene Recognition and Assembly Internet Link (GRAIL) software (http://grail.lsd.ornl.gov/Grail-1.3/), version 1.3. The PSORT prediction program (http://psort.nibb.ac.jp) was used to predict the cellular localization of SIRT3 and its homologues.
Cellular Localization.
Full-length SIRT3 and truncated partial-length PLSIRT3 (residues 1–142 missing) cDNA sequences were amplified by RT-PCR from normal liver, kidney, muscle, brain, heart, and placenta by using primers 5′ RIS-RI-GFP (5′-TTTAGAATTCTGCCGCCATGGCACGAGGGTTCTGGGGTT-3′) and RIS-1221 (5′-GGCGATGGATCCTTTGTCTGGTCCATCAAGCTTCC-3′) for full-length and 5′ PL RISGFP (5′-TTAATTGAATTCGCCGCCATGGTGGGGGCCGGCATCAGCACA-3′) and RIS1221 for PLSIRT3. Amplified cDNA sequences were cloned in-frame and N terminus to the enhanced GFP (EGFP) coding sequence in the BamHI–EcoRI sites of pEGFP. N3 vector. Constructs were transfected into cell lines purchased from the American Type Tissue Culture Collection. Cells were grown on glass cover slips in high-glucose DMEM supplemented with 10% FBS/100 units/ml penicillin/100 mg/ml streptomycin/0.1 mM nonessential amino acids. Bovine serum was obtained from Sigma, and all other cell culture media and additives were purchased from Invitrogen. Transformation of the cDNA into Cos7 (African green monkey simian virus 40-transformed, kidney), SiHa (squamous carcinoma, cervix, human), G401 (Wilms' tumor, human) and RD (rhabdomyosarcoma, human) cell lines was achieved by using lipofectamine according to recommended protocols (Invitrogen). Stable cell lines were generated by selection using geneticin (G418 sulfate, 0.4–0.9 mg/ml) and subsequently maintained in 0.2–0.45 mg/ml G418 sulfate. For subcellular localization of SIRT3, transfected cells were analyzed after 48 h. The organelle markers MitoTracker red (mitochondria), concanavalin A, Alexa 594 conjugate (Golgi tracker), and endoplasmic reticulum tracker N-({4-[4,4-difluoro-5-(2-thienyl)-4-bora-3a,4a-diaza-s-indacene-3-yl]phenoxy}acetyl) sphingosine were purchased from Molecular Probes. Organelles were labeled according to manufacture-supplied protocols. Cells were fixed to coverslips by using 1% formaldehyde in 1× PBS. To detect the nucleus, fixed cells were counterstained with 4′,6-diamidino-2-phenylindole (DAPI, Roche Molecular Biochemicals) and mounted in antifade (Glycerol/PBS, 10:1) containing 5 mM 4-phenylenediamine (Sigma), pH 8.5, onto slides. Digital images were acquired on a Photometrics PXL-1400 charge-coupled device camera attached to a Zeiss fluorescence Axiovert microscope 135TV using IP LAB SPECTRUM software (Signal Analytics, Vienna, VA).
Histone Deacetylase Assay and Protein Purification.
Histone deacetylase activity assay was carried out as described (13) with the following modifications. Each reaction contained 40.74 μg/μl 3H-acetylated chicken erythrocyte histones (specific activity, 672.6 cpm/μg) and 20 μl of immunoprecipitated SIRT3 or EGFP on protein G-Sepharose. Purified Sir2 homologue from Archaeoglobus fulgidus (SIR2AF; ref. 13) was used as a positive control at a final concentration of 0.04 μg/μl. All reactions were incubated at 37°C for 19.5 h. Stably transfected SiHa cell clones expressing either a full-length SIRT3 fused to EGFP or only EGFP were harvested at confluence. A total of 6.5 × 108 cells were used in each case to purify the expressed proteins following immunoprecipitation protocols supplied by the manufacture of the rabbit anti-GFP antibodies used (CLONTECH). In each case, 700 μl of protein G-Sepharose 4 Fast Flow (Amersham Pharmacia) was used to precipitate the complex containing expressed proteins bound to anti-GFP antibodies.
Mitochondrial Fractionation and Immunoelectron Microscopy.
Mitochondria were fractionated and prepared as described (24). To purify SIRT3 from the mitochondria, we immunoprecipitated SIRT3 after lysing the mitochondria as described. Immunoelectron microscopy was performed as described (25). Briefly, for immunogold labeling of ultrathin cryosections, cells were variously fixed in either periodate-lysine-paraformaldehyde, (75 mM phosphate buffer containing 2% formaldehyde, 70 mM lysine, and 10 mM sodium periodate, pH 6.2), for 4 h or 4/8% paraformaldehyde contained in 1× PBS, pH 7.4, for 1 h at room temperature. The cells were washed, scraped, pelleted, cryoprotected in 2.3 M sucrose containing 20% polyvinylpyrrolidone (1 h), mounted on aluminum cryopins, and frozen in liquid N2. Ultrathin cryosections then were cut on a Reichert UCT ultramicrotome equipped with FCS cryostage, and sections were collected onto 300 mesh, formvar/carbon-coated nickel grids. Grids were washed through several drops of PBS containing 5% FCS and 0.01 M glycine, pH 7.4, blocked in 10% FCS, and incubated overnight with mixed primary antibodies containing ≈10 μg/ml specific antibody. After washing, grids were incubated with mixed secondary gold conjugates (available from Jackson ImmunoResearch), diluted 1:50, for 2 h. The grids were washed several times in PBS, washed through several drops of double distilled H2O, and subsequently embedded in a mixture containing 3.2% polyvinyl alcohol (molecular mass, 10 kDa), 0.2% methyl cellulose (400 centipoises), and 0.2% uranyl acetate. The sections were observed and photographed on a Philips EM 410 transmission electron microscope.
Results
Initial Cloning of SIRT3.
Because of the incompleteness of ordering and assembly of the human 11p15 sequence even at this date, we performed fine mapping of UniGene sequences on a panel of J1 and subchromosomal transferable fragment somatic hybrids containing various portions of 11p15.5 (data not shown). One EST was found to be a homologue of SIR2 and was located within a somatic-cell hybrid containing the most telomeric 500 kb of 11p. The completed sequence of the EST9 clone, revealed after BLAST analysis, matches exactly to the human chromosome 11p15.5 sequence (GenBank accession no. U73637). Closer examination of the sequence indicated that the EST represented a partial cDNA, because an initiating methionine and an upstream in-frame stop codon were missing. By performing 5′ prediction RT-PCR, we isolated a 2.1-kb cDNA with a complete ORF. The cDNA encoded a predicted protein of 399 aa, with a transcription initiation site that matched the Kozak consensus sequence (26) and polyadenylation signal at position 1,221–1,226. A database search revealed that the gene was homologous to yeast SIR2 and also several homologues in other organisms. The cDNA sequence is identical to a partial EST sequence previously reported for SIRT3 (15). Genomic sequence alignment with the cDNA showed that the cDNA was encoded in seven exons.
Sequence Analysis of SIRT3.
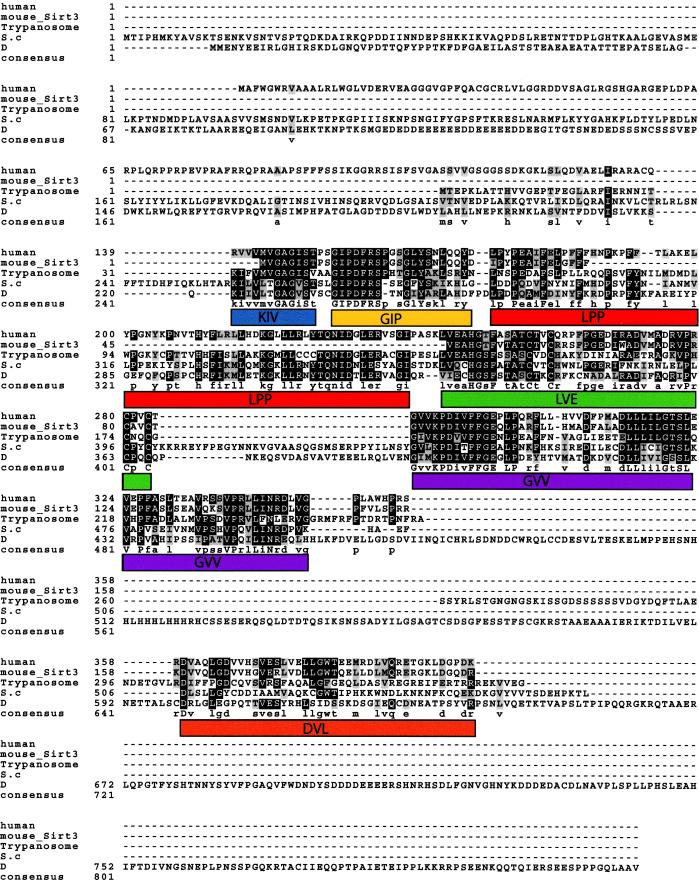
We performed multiple sequence alignment of yeast SIR2 (GenBank accession no. XO1419) and its homologues from Drosophila (accession no. AF068758), trypanosome (accession no. AF102869), and mouse (accession no. NM_022433) using the CLUSTALV program (Fig. 1). We discerned six putative motifs shown in Fig. 1. The KIV and GIP motifs span the core domain previously shown to be essential for silencing (27). Second, we searched the PIR pattern database and identified one potential glycosylation site at amino acids 207–210; three potential protein kinase C phosphorylation sites at amino acids 94–96, 118–120, and 391–393; four potential casein kinase II phosphorylation sites at amino acids 124–127, 320–323, 329–332, and 372–375; seven potential N-myristoylation sites at amino acids 49–54, 97–102, 113–118, 145–150, 163–168, 238–243, and 249–254; and one potential amidation site at amino acids 97–100. These results suggest that SIRT3 might be subject to posttranslational modification at some or all of the predicted sites.
Figure 1.
Multiple sequence alignment of the mouse and human SIRT3 and trypanosoma, yeast (S.c), and Drosophila (D) SIR2 proteins. Identities are shaded black, and conserved substitutions are shown in gray. The cutoff for inclusion in the consensus is a 50% match in all the aligned proteins at a given position. Note the relative loss in conservation at the first 142 aa of SIRT3. The overall similarity between the mouse and the human gene is the best, understandably because of a more recent divergence in evolution, compared with the other organisms included in this analysis. Deduced motifs are represented by colored boxes and are given names derived from the first three letters shared from the consensus in all the proteins or, if conserved, in at least three of the five sequences. The motifs are: KIV, blue; GIP, yellow; LPP, red; LVE, green; GVV, purple; and DVL, orange. The KIV and GIP motifs span the conserved Sir2p core domain previously shown to be important in silencing (27).
Analysis of the PRINTS database (28) for protein similarity identified six motifs. Two of these motifs were HTHLACI1 (bacterial regulatory protein lacI family HTH signature, amino acids 355–365, DNA- or RNA-associated protein) and PNDRDTASEI6 (pyridine nucleotide disulphide reductase class I signature, 359–366 aa, oxidoreductases), suggesting that SIRT3 protein may have enzymatic activity and could bind nucleic acids. Two motif matches were to GLUTRNSPORT (glucose transporter signature) at amino acids 337–345 and 370–379. Additional matches were to zinc finger motifs at amino acids 252–266 and 350–360, indicating that SIRT3 may interact with DNA or other proteins or functions as a dimer or multimer. Finally, a BLAST search of the nonredundant nucleotide database using the SIRT3 protein sequence identified matched to prokaryotic sequences with a known function. The match was 40% identity covering 201 aa (residues 139–335) to cobB, an enzyme with an enigmatic role in the biosynthesis of cobalamin that is in fact a bona fide SIR2 gene (refs. 13 and 29; GenBank accession no. AAC78722). These results suggest that SIRT3 has a common ancestor with these prokaryotic genes and may well have similar substrates.
Expression Pattern of Human SIRT3 in Human Tissue.
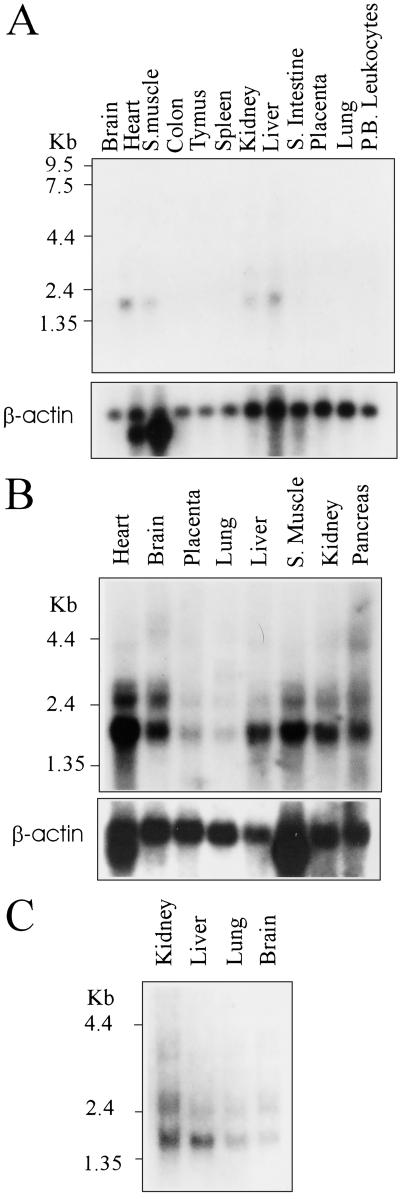
Multiple-tissue Northern blots were probed with the human SIRT3 DNA to determine the tissues in which the gene is expressed, and a β-actin probe was used to control for RNA loading. All the tissues tested of human fetal or adult origin showed detectable expression of a 2.1-kb transcript (Fig. 2 A–C). In the muscle, liver, kidney, and heart, elevated expression levels were evident when corrected for loading differences (Fig. 2A). These are metabolically active tissues, suggesting a potential role for SIRT3 in metabolism.
Figure 2.
Expression profile of SIRT3 on a panel of multiple human tissue Northern blot. (A) A 2.1-kb SIRT3 transcript detectable in the heart, skeletal muscle, kidney, and liver. β-actin probe is included as a loading control. (B) Expression of SIRT3 is detectable in all the tissues tested, although only a low level of expression can be seen is the placenta and lung. (Lower) β-actin was used again as a loading control. (C) Expression analysis of SIRT3 in fetal tissues. From loading controls not included, we deduced that the expression level in fetal tissues was equal in all the tested tissues.
SIRT3 Protein Is Targeted to the Mitochondria.
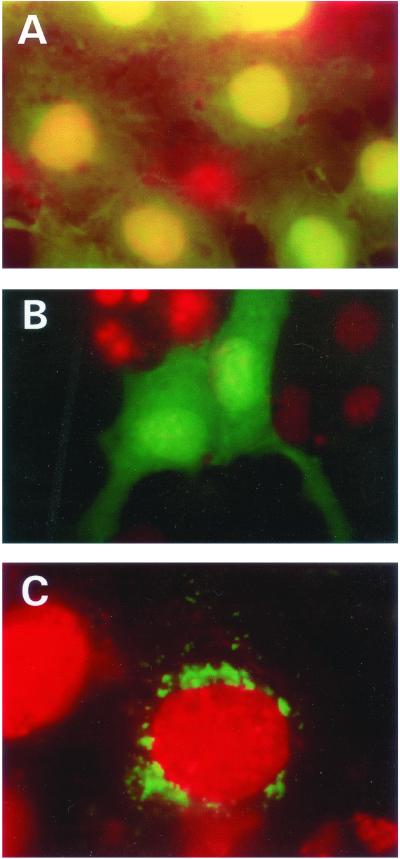
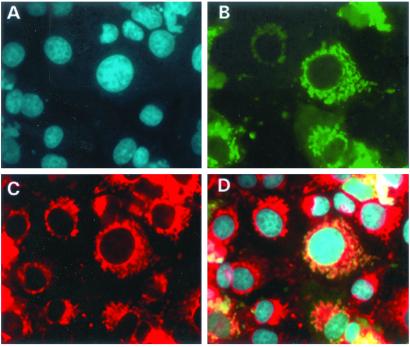
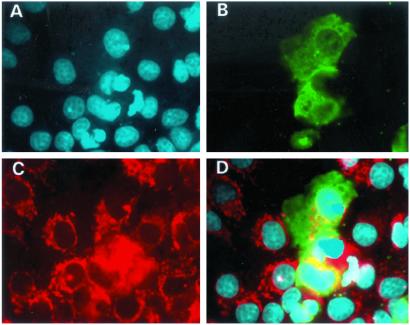
As a clue to the function of SIRT3, we investigated the subcellular location of the protein. We engineered a GFP fusion vector with the entire SIRT3 coding sequence. In addition, we cloned a truncated protein, PLSIRT3, that did not contain the upstream 142 aa that is distinct from other sirtuins (Fig. 1). We were able to localize the SIRT3 full-length protein to cytoplasmic organelles of unknown nature, while the partial-length protein showed diffuse homogeneous staining throughout the cytoplasm (Fig. 3 B and C). Thus, the unique N terminus of SIRT3 is responsible for targeting to these organelles. To identify the organelle to which the SIRT3 protein was targeted, we performed dual localization experiments using a second fluorescent signal marker specific to either the mitochondria, endoplasmic reticulum, or Golgi apparatus. Fig. 4 shows that the SIRT3 protein is localized to the mitochondria. None of the other organelle markers showed identical patterns to SIRT3 (data not shown). The N terminus was also shown to be required for the correct targeting of SIRT3 to the mitochondria, because PLSIRT3 failed to be targeted correctly to the mitochondria (Fig. 5).
Figure 3.
Cellular localization of SIRT3 full-length and truncated proteins in Cos7 cells. (A) Shown are cells transfected with vector plasmid. Strong expression of the EGFP in the nucleus and also expression in the cytoplasm is evident. Nuclei were stained with DAPI and colored red, hence the yellow coloration when the green and red signals are superimposed in the nucleus. (B) The N-terminal (1–142 aa) deleted SIR2T3 gene fused in-frame to the EGFP reading frame was transfected into these cells. Note the diffused expression of the chimeric EGFP protein throughout the cell. Unlike with the EGFP shown above, there was no preferential expression of the chimeric protein in the nucleus. Nuclei were detected by counterstaining as described for A. (C) SIR2T3 fused to EGFP (green), localized predominantly around the nuclear periphery. Red shows the nucleus detected by counterstaining with DAPI .
Figure 4.
Subcellular localization of the full-length SIRT3 gene in Cos7 cells. A shows DAPI staining of the nuclei, and B depicts the localization of the SIRT3-EGFP protein. Shown in red in C is the mitochondrial marker, Mitotracker. D shows superimposed images of A–C. It is clear from D that SIRT3 localizes to the mitochondria.
Figure 5.
Subcellular localization of truncated SIRT3 in Cos7 cells. (A) DAPI staining of cell nuclei (blue). (B) SIRT3–EGFP fusion construct transfected into the cells. Green shows the localization of the protein predominantly throughout the cytoplasm. (C) Cells stained with the mitochondrial marker, Mitotracker (red). (D) Superimposition of A–C. Note that the fusion construct does not colocalize with the mitochondrial marker.
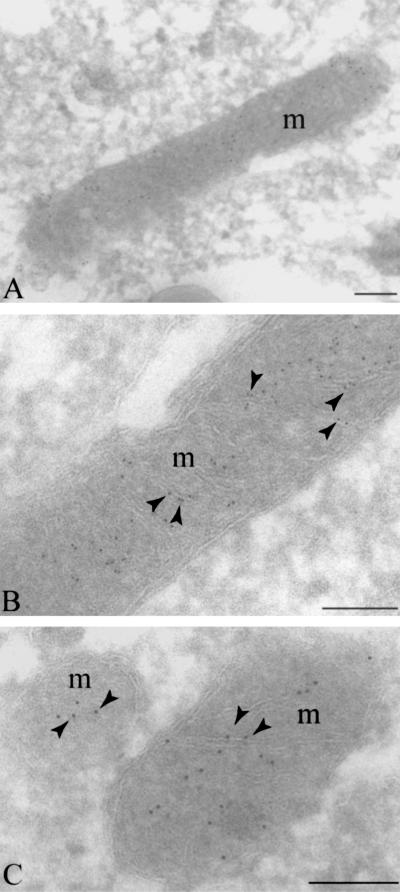
To confirm SIRT3 mitochondrial localization, we performed immunoelectron microscopy. These experiments revealed clear association of the SIRT3–EGFP fusion protein with the mitochondria (Fig. 6A). It is evident at high magnification that the SIRT3–EGFP localized to the mitochondrial cristae (Fig. 6 B and C). Finally, we used the PSORT program (30) to identify putative mitochondrial localization motifs in SIRT3. Interestingly, this analysis revealed a mitochondrial targeting sequence motif beginning at residue 19 (GRVVER) as well as a potential signal cleavage site located at residue 58. The PSORT program also predicted that SIRT3 was localized to the inner membrane of the mitochondria.
Figure 6.
Immunoelectron microscopy showing localization of SIRT3–EGFP to the mitochondrial cristae. (A) Indirect immunogold labeling of ultrathin cryosections of human cervical carcinoma (SiHa) cells demonstrates SIRT3–EGFP label (5 nm gold) in plain association with mitochondria in three independent images (A–C). High-magnification images reveal label clearly associated with mitochondrial cristae (B and C). m, mitochondria; arrowheads, cristae labeling. (Bars, 0.5 μm.)
SIRT3 Has a Protein Deacetylase Activity.
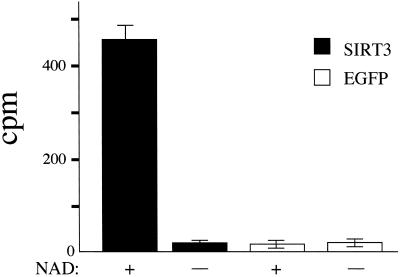
To determine whether SIRT3 possesses a protein deacetylase enzymatic activity (12, 13), we purified expressed SIRT3 by immunoprecipitation from five lines of mammalian SiHa cells stably overexpressing the protein fused to GFP at its C terminus. As negative controls, we similarly immunoprecipitated the vector-expressed EGFP, and as a positive control we used purified SIR2Af2, which has been shown previously to have protein deacetylase activity (13). We used purified chicken histones labeled with [3H]acetate as a substrate and assayed the release of free acetate. Only SIRT3 and SIR2Af2 showed robust in vitro histone deacetylase activity, and this activity was completely NAD+-dependent (Fig. 7). In addition, only SIRT3 and not EGFP purified from fractionated mitochondrial lysates showed NAD+-dependent protein deacetylase activity. Thus, SIRT3 is a protein deacetylase localized to mitochondria.
Figure 7.
Protein deacetylase activity of purified SIRT3. Assay for histone deacetylase activity in SIR2T3 and in plasmid vector expressing EGFP only in the presence and absence of 200 mM NAD+. The histone deacetylase activity (cpm) is plotted on the y axis. Experiments were performed in triplicate.
Discussion
Studies in yeast and C. elegans have implicated the SIR2 gene in gene silencing and in extension of life span (6, 31). Recently it was shown that the human SIRT1 homologue may be involved in negative control of p53 during cell survival and stress (18). Despite these advances, little is known about the mechanism of Sir2 gene function in yeast, and even less is known about the role of mammalian Sir2 homologues. In this study we characterized a human Sir2 homologue, SIRT3, using molecular approaches and bioinformatics. We show that (i) SIRT3 is conserved in prokaryotes and eukaryotes, (ii) the encoded product of SIRT3 is targeted to the mitochondrial cristae, (iii) the unique domain located at the N terminus contains a mitochondrial localization signal and is necessary for mitochondrial localization, (iv) SIRT3 is ubiquitously expressed, particularly in metabolically active tissues, and (v) SIRT3 has an NAD+-dependent protein deacetylase activity. These results suggest that SIRT3 functions in mitochondria in NAD+-dependent protein deacetylation.
Interestingly, NAD is required as an electron carrier in the mitochondria, a function that is critical for ATP generation. We postulate that SIRT3 may be critical in sensing NAD+ level (or NAD+/NADH ratios) in the mitochondria such that increased NAD+ levels would trigger a regulatory pathway that would activate SIRT3, leading to deacetylation of specific targets. It is unclear at this time what these targets are or what the physiological consequences might be. Mutation of the NPT1 gene, which is required for NAD+ synthesis through a salvage pathway, reduces both intracellular NAD+ levels and gene silencing (13), suggesting that cellular energy levels are critical in modulating silencing. Another possibility is that SIRT3 might deacetylate mitochondrial proteins directly involved in apoptosis, mimicking the function of SIRT1 in inhibiting apoptosis by deactylating p53. Because ADP-ribosyl transfer has been reported for some sirtuins, it is formally possible that SIRT3 could be involved in mitochondrial ADP ribosylation.
We also document an effective approach of complementing bioinformatics analysis and molecular characterization of SIRT3. We show by molecular approaches that SIRT3 is localized subcellularly to the mitochondria and show similar results predicted by the PSORT program. In addition, we were able to identify a unique domain at the N terminus of SIRT3 that is absent in other human Sir2 homologues, and deletion of this domain in SIRT3 abrogates the targeting of the protein product to the mitochondria. Interestingly, the same algorithms predict an exodeoxynuclease activity and glucose transporter motif within SIRT3. Furthermore, analysis of other mammalian Sir2 homologues by using the PSORT program suggest unique localization signals to nuclei and mitochondria (P.O., unpublished observation). We report protein deacetylase activity in the mitochondria and suggest an important previously unrecognized function of a SIR2 homologue in mitochondria biology.
Acknowledgments
We are grateful to Doug Murphy and Maria Mancini for reagents. This work was supported by National Institutes of Health Grants R37 CA54358 (to A.P.F.) and GM62385 (to J.D.B.) and National Science Foundation Grant NSF-DBI-0099706 (to J.M.M.).
Abbreviations
- EGFP
enhanced GFP
- DAPI
4′,6-diamidino-2-phenylindole
References
- 1.Kuo M H, Allis C D. BioEssays. 1998;20:615–626. doi: 10.1002/(SICI)1521-1878(199808)20:8<615::AID-BIES4>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- 2.Aparicio O M, Billington B L, Gottschling D E. Cell. 1991;66:1279–1287. doi: 10.1016/0092-8674(91)90049-5. [DOI] [PubMed] [Google Scholar]
- 3.Dutnall R N, Pillus L. Cell. 2001;105:161–164. doi: 10.1016/s0092-8674(01)00305-1. [DOI] [PubMed] [Google Scholar]
- 4.Smith J S, Boeke J D. Genes Dev. 1997;11:241–254. doi: 10.1101/gad.11.2.241. [DOI] [PubMed] [Google Scholar]
- 5.Kaeberlein M, McVey M, Guarente L. Genes Dev. 1999;13:2570–2580. doi: 10.1101/gad.13.19.2570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tissenbaum H A, Guarente L. Nature. 2001;410:227–230. doi: 10.1038/35065638. [DOI] [PubMed] [Google Scholar]
- 7.Tsukamoto Y, Kato J, Ikeda H. Nature. 1997;388:900–903. doi: 10.1038/42288. [DOI] [PubMed] [Google Scholar]
- 8.Straight A F, Shou W, Dowd G J, Turck C W, Deshaies R J, Johnson A D, Moazed D. Cell. 1999;97:245–256. doi: 10.1016/s0092-8674(00)80734-5. [DOI] [PubMed] [Google Scholar]
- 9.San-Segundo P A, Roeder G S. Cell. 1999;97:313–324. doi: 10.1016/s0092-8674(00)80741-2. [DOI] [PubMed] [Google Scholar]
- 10.Tanny J C, Dowd G J, Huang J, Hilz H, Moazed D. Cell. 1999;99:735–745. doi: 10.1016/s0092-8674(00)81671-2. [DOI] [PubMed] [Google Scholar]
- 11.Imai S, Armstrong C M, Kaeberlein M, Guarente L. Nature. 2000;403:795–800. doi: 10.1038/35001622. [DOI] [PubMed] [Google Scholar]
- 12.Landry J, Sutton A, Tafrov S T, Heller R C, Stebbins J, Pillus L, Sternglanz R. Proc Natl Acad Sci USA. 2000;97:5807–5811. doi: 10.1073/pnas.110148297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Smith J S, Brachmann C B, Celic I, Kenna M A, Muhammad S, Starai V J, Avalos J L, Escalante-Semerena J C, Grubmeyer C, Wolberger C, Boeke J D. Proc Natl Acad Sci USA. 2000;97:6658–6663. doi: 10.1073/pnas.97.12.6658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brachmann C B, Sherman J M, Devine S E, Cameron E E, Pillus L, Boeke J D. Genes Dev. 1995;9:2888–2902. doi: 10.1101/gad.9.23.2888. [DOI] [PubMed] [Google Scholar]
- 15.Frye R A. Biochem Biophys Res Commun. 1999;260:273–279. doi: 10.1006/bbrc.1999.0897. [DOI] [PubMed] [Google Scholar]
- 16.Perrod S, Cockell M M, Laroche T, Renauld H, Ducrest A L, Bonnard C, Gasser S M. EMBO J. 2001;20:197–209. doi: 10.1093/emboj/20.1.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gotta M, Strahl-Bolsinger S, Renauld H, Laroche T, Kennedy B K, Grunstein M, Gasser S M. EMBO J. 1997;16:3243–3255. doi: 10.1093/emboj/16.11.3243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Luo J, Nikolaev A Y, Imai S, Chen D, Su F, Shiloh A, Guarente L, Gu W. Cell. 2001;107:137–148. doi: 10.1016/s0092-8674(01)00524-4. [DOI] [PubMed] [Google Scholar]
- 19.Vaziri H, Dessain S K, Ng Eaton E, Imai S I, Frye R A, Pandita T K, Guarente L, Weinberg R A. Cell. 2001;107:149–59. doi: 10.1016/s0092-8674(01)00527-x. [DOI] [PubMed] [Google Scholar]
- 20.Muth V, Nadaud S, Grummt I, Voit R. EMBO J. 2001;20:1353–1362. doi: 10.1093/emboj/20.6.1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Afshar G, Murnane J P. Gene. 1999;234:161–168. doi: 10.1016/s0378-1119(99)00162-6. [DOI] [PubMed] [Google Scholar]
- 22.Glaser T, Housman D, Lewis W H, Gerhard D, Jones C. Somatic Cell Mol Genet. 1989;15:477–501. doi: 10.1007/BF01534910. [DOI] [PubMed] [Google Scholar]
- 23.Koi M, Johnson L A, Kalikin L M, Little P F, Nakamura Y, Feinberg A P. Science. 1993;260:361–364. doi: 10.1126/science.8469989. [DOI] [PubMed] [Google Scholar]
- 24.Vander Heiden M G, Chandel N S, Williamson E K, Schumacker P T, Thompson C B. Cell. 1997;91:627–637. doi: 10.1016/s0092-8674(00)80450-x. [DOI] [PubMed] [Google Scholar]
- 25.McCaffery J M, Farquhar M G. Methods Enzymol. 1995;257:259–279. doi: 10.1016/s0076-6879(95)57031-4. [DOI] [PubMed] [Google Scholar]
- 26.Kozak M. J Mol Biol. 1987;196:947–950. doi: 10.1016/0022-2836(87)90418-9. [DOI] [PubMed] [Google Scholar]
- 27.Sherman J M, Stone E M, Freeman-Cook L L, Brachmann C B, Boeke J D, Pillus L. Mol Biol Cell. 1999;10:3045–3059. doi: 10.1091/mbc.10.9.3045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Attwood T K, Flower D R, Lewis A P, Mabey J E, Morgan S R, Scordis P, Selley J, Wright W. Nucleic Acids Res. 1999;27:220–225. doi: 10.1093/nar/27.1.220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tsang A W, Escalante-Semerena J C. J Biol Chem. 1998;273:31788–31794. doi: 10.1074/jbc.273.48.31788. [DOI] [PubMed] [Google Scholar]
- 30.Nakai K, Kanehisa M. Genomics. 1992;14:897–911. doi: 10.1016/S0888-7543(05)80111-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Guarente L. Genes Dev. 2000;14:1021–1026. [PubMed] [Google Scholar]