Abstract
Tetraspanins encode a large conserved family of proteins that span the membrane four times and are expressed in a variety of eukaryotic tissues. They are part of membrane complexes that are involved in such diverse processes as intracellular signaling, cellular motility, metastasis, and tumor suppression. The single fly tetraspanin characterized to date, late bloomer (lbm), is expressed on the axons, terminal arbors, and growth cones of motoneurons. In embryos lacking Lbm protein, motoneurons reach their muscle targets, but initially fail to form synaptic terminals. During larval stages, however, functional contacts are formed. The newly available genomic sequence of Drosophila melanogaster indicates the existence of 34 additional members of the tetraspanin family in the fly. To address the possibility that other tetraspanins with functions that might compensate for a lack of lbm exist, we determined the expression domains of the Drosophila tetraspanin gene family members by RNA in situ analysis. We found two other tetraspanins also expressed in motoneurons and subsequently generated a small chromosomal deletion that removes all three motoneuron-specific tetraspanins. The deletion results in a significant enhancement in the lbm phenotype, indicating that the two additional motoneuron-expressed tetraspanins can, at least in part, compensate for the absence of lbm during the formation of the embryonic synapse.
The recent determination of the worm, fly, and human genomic sequences has created exciting new possibilities for a better understanding of the biology of the higher eukaryotes. It has become apparent over the last few decades that many genes are part of highly conserved extended families that encode proteins that share related domain structures. In many cases, gene family members have overlapping expression domains, suggesting they are functionally redundant or can compensate for each other's roles in the specification of tissue identity and other processes.
Although genetic redundancy likely provides essential safeguards during development and adult life (1, 2), it can confound the identification of genes via classical genetic screens, where usually only a single gene is mutated. One-third of the ≈13,600 genes encoded in the Drosophila genome are predicted to lack easily discernable phenotypes on mutagenesis (3), suggesting that redundancy or compensation will likely complicate our abilities to understand the functions of many thousands of genes. The identification of genes with similar activities and expression domains and analyses of their combined functions is among the challenges of the functional genomic studies lying ahead.
Studies of the developing nervous system have provided evidence for functional redundancy, for example, in studies of the Drosophila receptor tyrosine protein phosphatases (RTP) and mouse receptor tyrosine kinase (RTK) families (reviewed in ref. 4) and the interactions of the mammalian ephrins with their receptors, the EpH tyrosinase kinase receptors (5). Mutants lacking only a single RTP or RTK display little or no apparent abnormalities, whereas increasingly severe phenotypes are observed in animals with the elimination of the expression of more family members that are expressed in overlapping expression domains.
The tetraspanin family encodes a large number of four-transmembrane-spanning proteins. They are involved in numerous intracellular interactions, e.g., cellular activation, proliferation and motility, metastasis, and tumor suppression (reviewed in refs. 6–9). Tetraspanins differ from other four-transmembrane proteins by the presence of specific short motifs in their second extracellular loop including highly conserved cysteine residues. Their apparent diversity of function, the lack of strong phenotypes associated with the elimination of single tetraspan genes, and data suggesting that tetraspanin proteins can act as adjunct, but nonessential, components of signaling complexes led to their designation as “molecular facilitators” (6).
Tetraspanins are associated with a variety of other proteins, notably integrins, other tetraspanins, and receptor complexes (reviewed in refs. 6–9). Evidence points to roles of tetraspanins in facilitating the assembly or function of signal transduction complexes (reviewed in refs. 6, 8, 10, and 11; see also refs. 12 and 13). The complexity of tetraspanin interactions and the existence of multiple tetraspanins in single cell types has led to the “tetraspanin web” model (11), in which the cell membrane can be envisioned as containing a matrix of diverse tetraspanin-containing multiprotein complexes.
Several tetraspanins are expressed in the mammalian and avian adult and developing nervous system, including CD9 (14, 15), Tspan-2 (16, 17), cnag, chCD9, and neurospanin (18), Tspan-5 (19, 20), CD151 and CD81 (21, 22), TM4SF2 (23), and OAP-1 (24). Although the roles and interactions of these tetraspanins in nervous system function and development remain largely unexplored, evidence presented thus far indicates that antibodies recognizing specific tetraspanins may either inhibit (CD151 and CD81; ref. 22) or promote (CD9; ref. 14) α3β1 integrin-dependent neurite outgrowth from cultured neurons.
Several of the nontetraspanin transmembrane 4 super family-encoding genes are associated with human disease (9); however, only recently has tetraspanin dysfunction been correlated with disease. Mutations in the peripherin/retinal degeneration slow (RDS) tetraspanin gene underlie several human retinal diseases (25) that are mimicked by the mouse peripherin/RDS knockout model (ref. 26 and references therein). Recently, disruption of the transmembrane 4 super family 2 tetraspanin gene by translocation or point mutation was described in ≈10% of families with nonsyndromic X-linked mental retardation and it may therefore represent a causative or contributing factor for this disease (23).
Latebloomer, (lbm), the first identified Drosophila tetraspanin gene, was cloned and characterized (27) in the course of a high-throughput screen for secreted and transmembrane proteins (28). Lbm, which is expressed on motoneurons, was named so because of the delay in the establishment of synaptic contacts at the neuromuscular junction observed in the null mutant. Although delayed, normal electrically active larval synapses eventually form and the homozygous mutant line is fully viable and fertile, suggesting the existence of functionally redundant gene products or other compensatory mechanisms. The uncovering of a large number of Drosophila tetraspanin family members in the fly genome (35 members in total; ref. 29 and this study) and the reversibility of the lbm phenotype indicated to us that other members of this family might encode proteins whose functions are redundant to the Lbm protein.
In this study, we have evaluated the degree to which other tetraspanin proteins, expressed at a similar place and time to Lbm, might compensate for a lack of Lbm. We determined the RNA in situ expression pattern for each of the flybase-annotated Drosophila tetraspanin genes and eliminated expression of the subset of three genes expressed on motoneurons by creation of a precise chromosomal deletion. The results of these studies indicate the existence of tetraspanin subfamilies whose multiple members are expressed in tissue-specific manners, i.e., in overlapping subsets of the CNS and in partially overlapping subdomains of the foregut, midgut, and hindgut. We found that removal of the two additional tetraspanins expressed on motoneurons enhance the lbm-delayed synaptogenesis phenotype.
Strikingly, elimination of fully 25% of the Drosophila tetraspanin gene family does not noticeably perturb progression of the Drosophila life cycle, suggesting that significant molecular compensation exists for most tetraspanin functions and/or that the roles of tetraspanins are likely to be, as suggested previously, accessories to other primary cell surface signaling pathway interactions. These studies lay the groundwork for genetic approaches for furthering our understanding of tetraspanin functions during the development of the nervous system and other tissues.
Materials and Methods
Cloning ESTs Representing the 35 Drosophila Tetraspanin Genes.
Subsequent to the discovery of lbm (27), two other tetraspanin members were found in the same screen (28), CK02527 (CG10106), expressed in motoneurons and the ventral unpaired midline neurons and CK02579 (CG8666), expressed in midline glia. A total of 20 other family members were identified by iterative blast analyses of the Berkeley Drosophila Genome Project EST collections. These clones were obtained, verified by 5′ and 3′ prime end sequencing, and used to determine gene expression patterns by RNA in situ analysis. blast analyses of the total Drosophila genome sequence led to the uncovering of another 12 predicted tetraspanins. Two cDNAs were obtained by screening four cDNA libraries by PCR with gene-specific primers: the KZ embryonic library (K. Zinn and C.S.G., unpublished work) and the Larval-Pupal, the Good Head Adult, and LD embryonic cDNA libraries (Berkeley Drosophila Genome Project). The remaining 10 ESTs were PCR-amplified from first-strand cDNA generated from total Drosophila embryonic RNA (0–24 h egg collection) by using gene-specific primers and subsequently cloned into pGEMT-EASY (Promega). Two potential additional tetraspanins reported by others (29), Dm.68C and Dm.40A, were not included in this study as we were unable to recover their ESTs from embryonic first-strand cDNA. Furthermore, they are not present in the EST database set, nor have they been annotated by the Berkeley Drosophila Genome Project. A list of the Drosophila tetraspanins, their aliases, and chromosomal locations are shown in Table 2, which is published as supporting information on the PNAS web site, www.pnas.org.
RNA in Situ Analysis and Immunohistochemistry.
RNA in situs (30), antibody stainings (31), and embryo staging (32) were performed as described. Description of the expression patterns was done by using a standardized vocabulary (http://flybase.bio.indiana.edu/) and according to Hartenstein (33). Motoneuron projections were studied by using an antibody against Fasciclin II (mAb 1D4, G. Helt and C.S.G., unpublished work). To further analyze the location of the RNA expression domains double RNA in situ and antibody labelings were performed with a number of tetraspanin antisense RNAs in combination with mAb BP102, which labels all CNS neurons (A. Bieber, N. Patel, and C.S.G., unpublished work). A mouse polyclonal anti-Lbm antibody (27) was used to examine the expression of Lbm protein in deficiencies described below, Df(2)19TET and Df(2)27TET.
Generation of the Tetraspanin Deficiency Stocks.
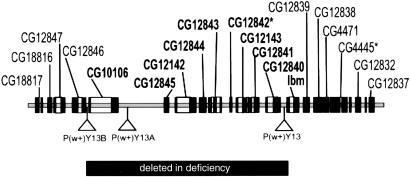
To create a deficiency that removes a large number of tetraspanin family members in the 42D-E interval, including all tetraspanins expressed in motoneuron populations, the original P element, P(w+)Y13, located 151 bp from the start of translation of lbm (27), was used in a local P element hopping approach (34). After one round of transposition, a fly line that contains an additional P element [P(w+)Y13A], located 7 kb distal to the 3 prime end of CG10106, was identified by long-range PCR of genomic DNA with region-specific primers. This stock was used again for mobilization and a line harboring three P elements, P(w+)Y13, P(w+)Y13A, and P(w+)Y13B, was created. The most distal P element was 5.2 kb upstream from the 5′ end of CG10106 (see Fig. 4). In the final round of P element mobilization, flies with white eyes, indicating the loss of all three P elements, were selected after introduction of transposase. Two lines, Df(2)19TET and Df(2)27TET, were characterized in detail by long-range PCR and sequencing of PCR products.
Figure 4.
Schematic representation of the P element mobilization strategy and the generation of the small deficiency in the 42D-E chromosomal interval. The P element locations and extent of the deletion are shown. The sequence of mobilization steps is given in Materials and Methods. The genes deleted in Df(2)TET19 and Df(2)TET27 are shown in bold. The two genes indicated by an asterisk are not related to the tetraspanin gene family.
Approximately 37.6 kb of genomic sequence between the exact insertion sites of P(w+)Y13 and P(w+)Y13B was deleted in both lines. This deletion removes nine genes (see Fig. 4) and virtually all of the 5′ flanking sequences of lbm. Both deficiency stocks are homozygous viable. Eleven partially overlapping PCR primers sets that cover the entire genomic region were used to confirm that the apparently deleted sequences were not transposed elsewhere in the genome (data not shown). The sequences of all gene- and region-specific PCR primers are available on request. Loss of expression of the tetraspanins within the deficiencies was confirmed by RNA in situ analysis using probes for CG12143 and CG12845 (data not shown). No Lbm protein was detected by using the anti-Lbm mouse polyclonal antibody (data not shown).
Results
Identification of Tetraspanin RNA Expression Domains.
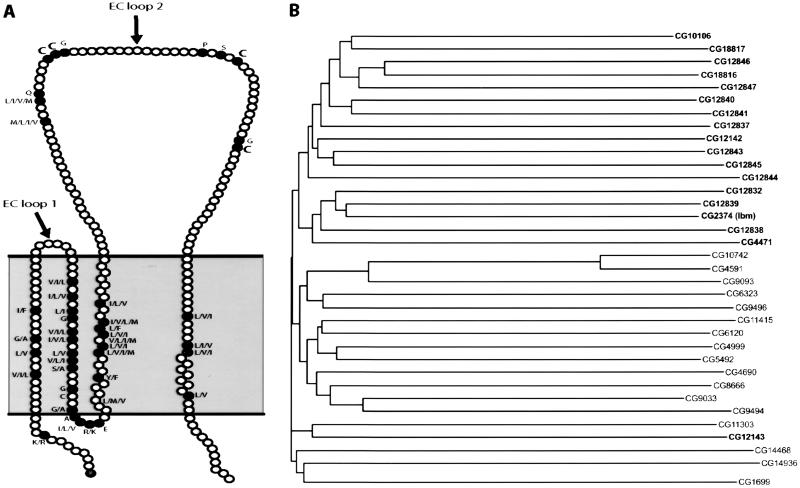
Tetraspanin proteins are defined by the presence of four transmembrane domains with the larger of the two extracellular loops falling between the third and fourth transmembrane domains and the presence of conserved cysteine residues in the second extracellular loop (Fig. 1A). We have analyzed the 35 tetraspanin family genes in Drosophila for which we could recover EST clones from public collections or by gene-specific PCR amplification of phage libraries and total embryonic first-strand cDNA. The genes are found dispersed throughout the fly genome (Table 2 and ref. 29); a cluster of 18 exists in a 63-kb region on the second chromosome. Analysis of the evolutionary divergence of the fly tetraspanins indicates that all but one of the 18 proteins in this cluster group together; however, as the predicted points of divergence between tetraspanins within the cluster are not significantly different from between any two tetraspanins, the evolutionary relationships of the clustered tetraspanins remains unclear (Fig. 1B).
Figure 1.
Primary structural conservation of the Drosophila tetraspanin proteins and their phylogenetic relationships. (A) The conserved positions and the predominant amino acids at those positions are shown overlaid on a schematic of a prototypic tetraspanin structure indicating the predicted intracellular and extracellular domains. The conserved cysteine residues are indicated in bold. (B) The phylogenetic relationships between the tetraspanin proteins were calculated by using the neighbor-joining algorithm (41) and plotted. The genes indicated in bold are present in a cluster of 18 genes on the second chromosome.
To examine the expression patterns of the tetraspanins during embryonic development, ESTs representing the predicted Drosophila tetraspanin genes were obtained or generated (Materials and Methods). RNA in situ analysis of whole-mount embryos representing all stages of embryonic development was performed by using each EST (Materials and Methods).
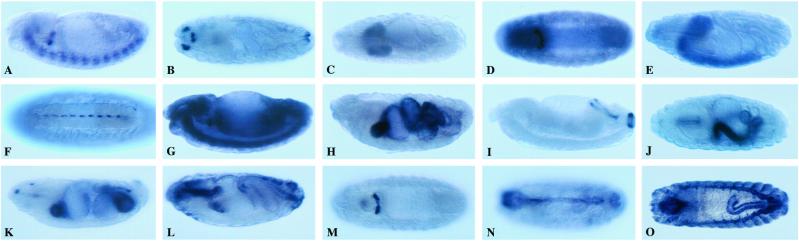
The 35 tetraspanins show strikingly different patterns of expression, but most of the family members have expression domains in one or more of the following three tissues: the central or peripheral nervous system (14 tetraspanins), the gut (12 tetraspanins), and low or high overall expression, predominantly in epidermis (19 tetraspanins) (Fig. 2; Table 2). The tetraspanins expressed in the nervous system can be divided into five groups: two are expressed in midline glia (CG4690 and CG8666), three are expressed in motoneurons (CG12143, CG10106, and CG2374/lbm; Fig. 3), six are predominantly expressed throughout the ventral cord and brain (CG10742, CG4471, CG11303, CG4591, CG6120, and CG6323), a single gene is expressed in sensilla in the head (CG9033), and two other genes are expressed in the peripheral nervous system (CG12142 and CG18817) (Table 2, Fig. 2). The three tetraspanins expressed in motoneurons, CG2374/lbm, CG10106, and CG12143, are located in the second chromosome cluster.
Figure 2.
A selection of the RNA expression domains of the Drosophila tetraspanins during embryonic development. (A–F) Expression of specific tetraspanins in the nervous system. (G–O) Tetraspanin expression domains in other parts of the embryo. Anterior is to the left. (A, E, G–I, K, and L) Lateral views are shown, dorsal is up. (B, C, J, N, and O) Dorsal views are shown. (D, F, and M) Ventral views. CG12143 (A) is expressed in the ventral cord and the hypocerebral ganglion; CG9033 (B) in the hypopharyngeal sense organs; CG6323 (C) in the two brain lobes; CG4591 (D) throughout the ventral cord and in the stomatogastric nervous system; CG6323 (E) in the brain and ventral cord; CG4690 (F) in the midline glia; CG10742 (G) throughout the whole embryo; CG12846 (H) in midgut compartments; CG11415 (I) in the hindgut and anal plate; CG12847 (J) in a midgut compartment; CG9496 (K) in the proventriculus and midgut; CG12840 (L) in the pharynx, esophagus, and hindgut; CG12840 (M) the garland cells (secretory cells); CG12840 (N) in the lymph gland; and CG4999 (O) in the tracheal system and hindgut. (Magnification: ×57.)
Figure 3.
Three tetraspanins are expressed in motoneuron populations during embryonic development. A (lbm), B (CG12143), and C (CG10106) show the RNA expression patterns in the cell bodies of motoneuron populations and brain. (D–F) Double labelings of dissected embryonic ventral cords with the BP102 mAb (brown) and lbm (D), CG12143 (E), or CG10106 (F) RNAs (blue). Anterior is to the left. (A–C) Lateral views, dorsal is up. All three tetraspanins are expressed in the RP3 motoneurons that innervate muscles 6 and 7 and are also expressed in the RP1 and RP4 motoneurons and in a number of more lateral motoneuron populations. Only CG10106 is expressed in the ventral unpaired midline neurons. (Magnification: A–C, ×70; and D–F, ×340.)
Deletion of the Motoneuron-Specific Subset of Tetraspanins.
To investigate the function of the three motoneuron-specific cDNAs, we examined the consequences of the lack of their expression. A P element insertion at the 5′ end of the lbm gene, P(w+)Y13, was used as a starting point to engineer a small precise deletion in the second chromosome tetraspanin cluster (Fig. 4). Three subsequent transposase-mediated P element mobilizations led to the generation of several deficiency stocks that removed a portion of the 42D-E chromosomal interval (Materials and Methods).
Specific genome sequence-derived primer sets were used to perform PCR to establish the extent of the deletions in these lines (Materials and Methods). Two lines, Df(2)TET19 and Df(2)TET27, were studied in detail after eliminating the possibility that the genes between the P elements had transposed elsewhere in the genome (data not shown). Sequence analysis of the Df(2)TET19 chromosome allowed precise definition of the deletion endpoints. Approximately 37.6 kb of genomic sequence was deleted. The DNA sequence across the deletion junction and extensive test PCRs in the genomic regions flanking the deletion indicate that the deletion occurred between the most distal and proximal P elements and did not affect chromosomal structure outside of the immediate deletion (data not shown). Nine of the 18 tetraspanins in the cluster, including the three expressed in motoneurons, cannot be expressed as a consequence of their removal from the genome. The two deficiency stocks are homozygous viable and fertile.
One nontetraspanin intervening putative gene of unknown function, CG12842, was also removed in these deficiencies. We isolated an EST representing this gene and determined that its mRNA is expressed at a low level throughout the embryo (data not shown). Protein database searches (National Center for Biotechnology Information nonredundant and Berkeley Drosophila Genome Project predicted proteins datasets) reveals that it contains a short amino acid sequence of unknown function also present in another Drosophila ORF, CG13617, present on the third chromosome. No significantly related protein sequences were found in other species.
Enhancement of the lbm Phenotype in the Homozygous Deficiency Stock.
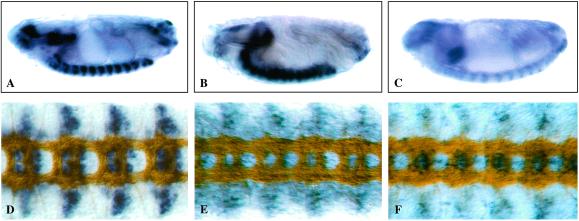
The RNA expression domains of the three tetraspanin members in subsets of motoneuron populations is shown in Fig. 3. All three genes are expressed in the RP1, RP3, and RP4 motoneurons. In WT embryos, the RP3 motoneuron establishes a synaptic terminal on muscles 6 and 7 at the end of embryogenesis (stage 17, Fig. 5A). The RP3 synapse is the best studied in the Drosophila embryo and can be visualized by staining all motoneurons with a mAb recognizing Fasiculin II (1D4).
Figure 5.
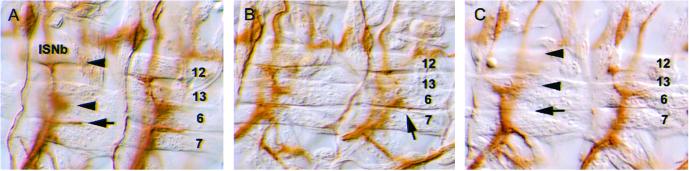
The projection of the RP3 synaptic terminal is often absent or abnormal when three tetraspanins expressed on RP3 motoneurons are not present. The projection of the RP3 terminal into the cleft between muscle fibers 6 and 7 was visualized with an antibody against Fasciclin II that labels all motoneurons. The WT RP3 innervation of muscles 6 and 7 in a precise excision control embryo is shown (A, arrow). In lbm, RP3 innervation is often absent (B, arrow), whereas in the Df(2)TET19 embryos in which all three motoneuron tetraspanins (lbm, CG10106, and CG12143) are deleted, the innervation of muscles 6 and 7 is, in most cases, absent (C, arrow). In addition, the innervation of muscles 12 and 13 is often abnormal (C, arrowheads). (Magnification: ×550.)
The original P(w+)Y13 element insert near lbm results in loss of lbm transcription. Embryos lacking the lbm gene product display delayed formation of mature synaptic contacts, as exemplified by the failure of innervation of muscle fibers 6 and 7 by motoneuron RP3 in ≈ one-third of the segments (ref. 27, Fig. 5B, and Table 1). This synaptic defect is transient during development and is fully restored by the third larval instar (27).
Table 1.
Enhancement of Ibm RP3 innervation defect in Df(2)TET19 and Df(2)TET27
| Genotype | Percentage of segments lacking correct RP3 innervations | No. of segments counted |
|---|---|---|
| P(w+)Y13 precise excision | 3 | 120 |
| Lbm P(w+)Y13 | 32 | 110 |
| Lbm P(w+)S64 | 36 | 111 |
| Df(2)TET19 | 64 | 120 |
| Df(2)TET27 | 60 | 115 |
Mature and delayed RP3 innervations of muscles 6 and 7 were counted in the indicated number of segments.
To evaluate possible redundant functions of the tetraspanins CG10106 and CG12143 that overlap in expression in motoneuron populations with lbm, we studied the arborizations of the RP3 motoneuron in the lines with deficiencies that remove all three genes. The deficiency stocks were back-crossed extensively to remove any modifiers of the mutant phenotype. Homozygous embryos derived from the Df(2)19TET, Df(2)27TET, the original P(w+)Y13 lbm allele, a second lbm allele [P(w+)S64] (27), and the control P(w+)Y13 precise excision stocks were stained and the number of failed RP3 motoneuron innervations on muscle 6 and 7 were counted at stage 17 (Table 1; Fig. 5).
The lbm homozyogotes, P(w+)Y13 lbm and P(w+)S64, lack the RP3 innervation in 32% and 36% of the segments scored, respectively. The WT control, a precise excision of P(w+)Y13 P element, lacks only 3% of possible synapses at muscles 6 and 7; however, the terminal arbor of the RP3 motoneuron is absent in 60% and 64% of the segments counted in Df(2)19TET and Df(2)27TET, respectively. Thus, the elimination of the two additional tetraspanins expressed in motoneurons leads to a significant increase in the lbm phenotype, suggesting that one or both of these genes at least partially shares lbm function in synapse formation. Ultimately, however, as in the Lbm mutant, third larval instar synapses appear normal in animals homozygous for the deficiencies that uncover all three motoneuron-expressed tetraspanins (data not shown).
In addition to the three genes that are expressed in the motoneurons, six other tetraspanins are deleted in the two deficiencies: CG12142 and CG12840, predominantly expressed in the gut, and CG12841, CG12843, CG12845, and CG12844, expressed in the epidermis or expressed throughout the embryo at low levels (Table 2, Fig. 2). Inspection of the general morphology of the embryonic segmentation, the gut, and musculature of the deficiency embryos did not show any obvious abnormalities in comparison with the precise excision P(w+)Y13 homozygous embryos.
Discussion
The genomic sequence of several of the higher eukaryotes has been determined; the task ahead is to understand the functions of individual genes. Chemical, radiation, and transposon-based genetic screens, although powerful tools for forward genetics (35), are unlikely to be useful for identifying the functions of all genes because in part of the inability of some of these agents to mutate all parts of the genome, and also because of the existence of partially or fully redundant or compensatory genes or processes that mask the phenotypes associated with the mutation of a single gene. Furthermore, loss of a single component of a multiple component complex might impair but not abolish its function. Reverse genetics, where related genes are identified initially through bioinformatic approaches with the subsequent analysis of their expression domains and functions (36), will therefore play an important role in furthering our understanding of the roles played by related and possibly redundantly functioning genes. In the studies presented in this article, we have taken such a reverse genetic approach to further our understanding of the Drosophila tetraspanin genes, a large gene family with 35 members.
Roughly half of the tetraspanin genes are dispersed throughout the genome, and half are clustered in a 63-kb region on the second chromosome that contains three tetraspanins expressed on motoneurons. There is no strong evidence for a duplicative origin of the clustered tetraspans in our and others' phylogenetic analysis of the Drosophila tetraspanin family (ref. 29 and this study). The available data regarding the chromosomal locations of tetraspanin genes in the human genome does not indicate any such similar clustering (11). As the genomic sequence of closely related insects becomes available, it will be interesting to ascertain whether physical grouping of tetraspanins is conserved.
Our phylogenetic analysis does not indicate the three motoneuron-expressed tetraspanins are more related to each other than other tetraspanins expressed in other tissues. Genomic clustering of related genes can reflect, among other selective forces, an evolutionary advantage in coordinate tissue-specific control of closely linked genes (37). The three tetraspanin genes expressed highly in motoneurons are, however, not adjacent to each other. Other tetraspanins expressed in different regions of the embryo intervene, suggesting that coordinate control of the motoneuron tetraspanins does not rely solely on regionwide mechanisms. Examination of the 5′ regions of the tetraspanins expressed in motoneurons as compared with those expressed in other tissues does not reveal any apparent common constellation of transcription factor binding sites responsible for motoneuron-specific expression (data not shown).
RNA in situ analysis of the Drosophila tetraspanin expression domains indicates that most tetraspanins are expressed in highly specific manners either in the nervous system or the gut or have a low-level ubiquitous expression. Their expression at later stages of development has not been examined.
The fortuitous clustering of half of the Drosophila tetraspanins at the 42D-E interval of the second chromosome allowed us to create a precise chromosomal deletion, removing nine of them. In addition to the three tetraspanins specifically expressed in the motoneurons, two of the deleted tetraspanins are expressed in the gut and four are uniformly expressed at low level throughout the embryo.
Strikingly, elimination of ≈25% of all fly tetraspanins does not affect viability or fertility under laboratory conditions. It remains possible that other more ubiquitously expressed tetraspanins compensate for the roles of the deleted tetraspanins. However, the most straightforward interpretation of these data is that tetraspanins perform nonessential accessory functions, in line with previous reports of tetraspanin function (reviewed in refs. 6–8 and 11). The deletion removes the three tetraspanins that are highly expressed in motoneurons, which allowed our subsequent observation that one or both of the additional motoneuron-specific tetraspanins provides functions similar to that of lbm.
What are the functions of tetraspanins in motoneurons? From analogy to numerous previous studies on the function of mammalian tetraspanins, we think they are likely part of receptor complexes that function in signaling and/or have roles in neural adhesion and motility, probably via members of the integrin family of extracellular matrix receptors that form a bridge between the cytoskeleton and the extracellular matrix. We do not observe a requirement for tetraspanins in initial neurite outgrowth, as suggested by two mammalian tissue culture studies (14, 22), because the motoneuron axons do reach their targets, but are delayed in making the final synaptic contact point (Fig. 5).
The strongest connection of tetraspanins to a specific signaling pathway to date has been the demonstration that five of 10 human tetraspanins examined, including CD63, are physically associated with phosphoinositide 4-kinase (PI 4-kinase) (38, 39), but not with other PI kinases (39). As shown for CD63 (38), these complexes can include the α3β1 integrin, suggesting that the roles of tetraspanins may include recruiting PI 4-kinase to the cell cytoskeleton to facilitiate phosphoinositide-dependent signaling (40). Initial reports that tetraspanins may also signal through other pathways, e.g., CD82 through Rho-GTPases in T cells (13) and CD53 through the c-Jun N-terminal kinase in several cell types (12), have recently been made.
The deficiency that removes nine tetraspanins described in this study provides a starting point for using the power of Drosophila genetics to identify tetraspanin-interacting proteins acting at the neuromuscular junction. Members of the signaling pathways mentioned above and integrins expressed at the neuromuscular junction are likely candidates to test in directed approaches.
Supplementary Material
Acknowledgments
We thank Monique Radjkoemar-Bansraj, Wei-Yu Chen, and Peter Chiu for technical assistance. We are grateful to Dr. Volker Hartenstein for his help in identifying the expression domains of several of the tetraspanins and thank Dr. Silvere van de Maarel for providing first-strand cDNA and Dr. Andrea Page-McCaw for the gift of four unpublished tetraspanin cDNA clones. C.S.G. was an Investigator for the Howard Hughes Medical Institute. A.D. is a McKnight and Keck Scholar and was supported by the Whitehall and Mallinkrodt foundations. This work is supported by ASPASIA Grant 015-000-056 (to J.N.N.) and Programma Genomics Grant 050-10-155 (to L.G.F. and J.N.N.) from the Nederlandse Organisatie voor Wetenschappelijk Onderzoek (NWO).
Abbreviation
- lbm
late bloomer
References
- 1.Cooke J, Nowak M A, Boerlijst M, Maynard-Smith J. Trends Genet. 1997;13:360–364. doi: 10.1016/s0168-9525(97)01233-x. [DOI] [PubMed] [Google Scholar]
- 2.Thomas J H. Trends Genet. 1993;9:395–399. doi: 10.1016/0168-9525(93)90140-d. [DOI] [PubMed] [Google Scholar]
- 3.Miklos G L, Rubin G M. Cell. 1996;86:521–529. doi: 10.1016/s0092-8674(00)80126-9. [DOI] [PubMed] [Google Scholar]
- 4.Desai C J, Sun Q, Zinn K. Curr Opin Neurobiol. 1997;7:70–74. doi: 10.1016/s0959-4388(97)80122-5. [DOI] [PubMed] [Google Scholar]
- 5.Gale N W, Holland S J, Valenzuela D M, Flenniken A, Pan L, Ryan T E, Henkemeyer M, Strebhardt K, Hirai H, Wilkinson D G, et al. Neuron. 1996;17:9–19. doi: 10.1016/s0896-6273(00)80276-7. [DOI] [PubMed] [Google Scholar]
- 6.Maecker H T, Todd S C, Levy S. FASEB J. 1997;11:428–442. [PubMed] [Google Scholar]
- 7.Hemler M E. J Cell Biol. 2001;155:1103–1107. doi: 10.1083/jcb.200108061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Berditchevski F. J Cell Sci. 2001;114:4143–4151. doi: 10.1242/jcs.114.23.4143. [DOI] [PubMed] [Google Scholar]
- 9.Bronstein J M. Curr Opin Neurobiol. 2000;10:552–557. doi: 10.1016/s0959-4388(00)00125-2. [DOI] [PubMed] [Google Scholar]
- 10.Levy S, Todd S C, Maecker H T. Annu Rev Immunol. 1998;16:89–109. doi: 10.1146/annurev.immunol.16.1.89. [DOI] [PubMed] [Google Scholar]
- 11.Boucheix C, Rubinstein E. Cell Mol Life Sci. 2001;58:1189–1205. doi: 10.1007/PL00000933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yunta M, Oliva J L, Barcia R, Horejsi V, Angelisova P, Lazo P A. Eur J Biochem. 2002;269:1012–1021. doi: 10.1046/j.0014-2956.2001.02741.x. [DOI] [PubMed] [Google Scholar]
- 13.Delaguillaumie A, Lagaudriere-Gesbert C, Popoff M R, Conjeaud H. J Cell Sci. 2002;115:433–443. doi: 10.1242/jcs.115.2.433. [DOI] [PubMed] [Google Scholar]
- 14.Banerjee S A, Hadjiargyrou M, Patterson P H. J Neurosci. 1997;17:2756–2765. doi: 10.1523/JNEUROSCI.17-08-02756.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kaprielian Z, Cho K O, Hadjiargyrou M, Patterson P H. J Neurosci. 1995;15:562–573. doi: 10.1523/JNEUROSCI.15-01-00562.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Todd S C, Doctor V S, Levy S. Biochim Biophys Acta. 1998;1399:101–104. doi: 10.1016/s0167-4781(98)00087-6. [DOI] [PubMed] [Google Scholar]
- 17.Birling M C, Tait S, Hardy R J, Brophy P J. J Neurochem. 1999;73:2600–2608. doi: 10.1046/j.1471-4159.1999.0732600.x. [DOI] [PubMed] [Google Scholar]
- 18.Perron J C, Bixby J L. FEBS Lett. 1999;461:86–90. doi: 10.1016/s0014-5793(99)01429-5. [DOI] [PubMed] [Google Scholar]
- 19.Garcia-Frigola C, Burgaya F, Calbet M, de Lecea L, Soriano E. NeuroReport. 2000;11:3181–3185. doi: 10.1097/00001756-200009280-00027. [DOI] [PubMed] [Google Scholar]
- 20.Garcia-Frigola C, Burgaya F, de Lecea L, Soriano E. Mech Dev. 2001;106:207–212. doi: 10.1016/s0925-4773(01)00436-1. [DOI] [PubMed] [Google Scholar]
- 21.Kelic S, Levy S, Suarez C, Weinstein D E. Mol Cell Neurosci. 2001;17:551–560. doi: 10.1006/mcne.2000.0955. [DOI] [PubMed] [Google Scholar]
- 22.Stipp C S, Hemler M E. J Cell Sci. 2000;113:1871–1882. doi: 10.1242/jcs.113.11.1871. [DOI] [PubMed] [Google Scholar]
- 23.Zemni R, Bienvenu T, Vinet M C, Sefiani A, Carrie A, Billuart P, McDonell N, Couvert P, Francis F, Chafey P, et al. Nat Genet. 2000;24:167–170. doi: 10.1038/72829. [DOI] [PubMed] [Google Scholar]
- 24.Tiwari-Woodruff S K, Buznikov A G, Vu T Q, Micevych P E, Chen K, Kornblum H I, Bronstein J M. J Cell Biol. 2001;153:295–305. doi: 10.1083/jcb.153.2.295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kohl S, Giddings I, Besch D, Apfelstedt-Sylla E, Zrenner E, Wissinger B. Acta Anat. 1998;162:75–84. doi: 10.1159/000046471. [DOI] [PubMed] [Google Scholar]
- 26.Jansen H G, Sanyal S. J Comp Neurol. 1992;316:117–125. doi: 10.1002/cne.903160110. [DOI] [PubMed] [Google Scholar]
- 27.Kopczynski C C, Davis G W, Goodman C S. Science. 1996;271:1867–1870. doi: 10.1126/science.271.5257.1867. [DOI] [PubMed] [Google Scholar]
- 28.Kopczynski C C, Noordermeer J N, Serano T L, Chen W Y, Pendleton J D, Lewis S, Goodman C S, Rubin G M. Proc Natl Acad Sci USA. 1998;95:9973–9978. doi: 10.1073/pnas.95.17.9973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Todres E, Nardi J B, Robertson H M. Insect Mol Biol. 2000;9:581–590. doi: 10.1046/j.1365-2583.2000.00222.x. [DOI] [PubMed] [Google Scholar]
- 30.Tautz D, Pfeifle C. Chromosoma. 1989;98:81–85. doi: 10.1007/BF00291041. [DOI] [PubMed] [Google Scholar]
- 31.Patel N H. Methods Cell Biol. 1994;44:445–487. doi: 10.1016/s0091-679x(08)60927-9. [DOI] [PubMed] [Google Scholar]
- 32.Wieschaus E, Nusslein-Volhard C. In: Drosophila: A Practical Approach. Roberts D B, editor. Oxford, U.K.: IRL; 1986. pp. 199–227. [Google Scholar]
- 33.Hartenstein V. Atlas of Drosophila Development. Plainview, NY: Cold Spring Harbor Lab. Press; 1993. [Google Scholar]
- 34.Tower J, Karpen G H, Craig N, Spradling A C. Genetics. 1993;133:347–359. doi: 10.1093/genetics/133.2.347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.St. Johnston D. Nat Rev Genet. 2002;3:176–188. doi: 10.1038/nrg751. [DOI] [PubMed] [Google Scholar]
- 36.Adams M D, Sekelsky J J. Nat Rev Genet. 2002;3:189–198. doi: 10.1038/nrg752. [DOI] [PubMed] [Google Scholar]
- 37.Trowsdale T. Genome Biol. 2002;3:1–5. [Google Scholar]
- 38.Berditchevski F, Tolias K F, Wong K, Carpenter C L, Hemler M E. J Biol Chem. 1997;272:2595–2598. doi: 10.1074/jbc.272.5.2595. [DOI] [PubMed] [Google Scholar]
- 39.Yauch R L, Hemler M E. Biochem J. 2000;351:629–637. [PMC free article] [PubMed] [Google Scholar]
- 40.Fruman D A, Meyers R E, Cantley L C. Annu Rev Biochem. 1998;67:481–507. doi: 10.1146/annurev.biochem.67.1.481. [DOI] [PubMed] [Google Scholar]
- 41.Saitou N, Nei M. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.