Abstract
When a replication fork collides with a DNA topoisomerase I (Top1) cleavage complex, the covalently bound enzyme must be removed from the DNA 3′ end before recombination-dependent replication restart. Here we report that the tyrosyl-DNA phosphodiesterase Tdp1 and the structure-specific endonuclease Rad1-Rad10 function as primary alternative pathways of Top1 repair in Saccharomyces cerevisiae. Thus, tdp1 rad1 cells (including the catalytic point mutant rad1-D869A) not only are highly sensitive to the Top1 poison camptothecin but also exhibit a TOP1-dependent growth delay. Extensive genetic analysis revealed that both Tdp1 and Rad1-Rad10 repair proceed through recombination that equally depends on RAD52, RAD51, and RAD50. The Rad1-Rad10 pathway further particularly depends on RAD59 and SRS2 but is independent of other nucleotide excision repair genes. Although this pattern is consistent with Rad1-Rad10 removing Top1 in a manner similar to its removal of nonhomologous tails during gene conversion, these differ in that Top1 removal does not require Msh2-Msh3. Finally, we show that yeast lacking the Rad1-Rad10-related proteins Mus81-Mms4 display a unique pattern of camptothecin sensitivity and suggest a concerted model for the action of these endonucleases.
DNA topoisomerase I (Top1) acts to prevent the buildup of superhelical strain around the elongating replication fork by transiently cleaving and religating a single strand of duplex DNA via a covalent 3′ phosphotyrosyl enzyme–DNA intermediate (1). Normally, Top1–DNA cleavage complexes are short-lived catalytic intermediates. In the presence of the antineoplastic drug and reversible inhibitor camptothecin, however, the half-life of the complex is increased due to slowing of the rate of DNA religation (2). Cytotoxic lesions are thought to occur when the advancing replication machinery encounters a drug-stabilized enzyme–DNA complex, because treatment of cells with the DNA replication inhibitor aphidicolin abrogates the cytotoxicity of camptothecin (3). Additionally, Saccharomyces cerevisiae rad52 mutants are hypersensitive to the drug (4).
Collision between a Top1-DNA intermediate and an advancing replication fork represents a unique form of replicative damage because the enzyme remains covalently bound to the 3′ end of the break and must be excised during replication restart (see Fig. 5). Tdp1, which hydrolyses the phosphotyrosine linkage between Top1 and DNA, has been identified as one mechanism of Top1 removal (5). The product of this reaction is a 3′ phosphate, so further processing is required to convert the end to a 3′ hydroxyl. Yeast cells lacking the DNA 3′ phosphatase activities encoded by TPP1, APN1, and APN2 are consequently hypersensitive to camptothecin, and this sensitivity is suppressed by the loss of TDP1 (6). This result established a Tdp1-initiated pathway as one mechanism for removing Top1 from DNA, but it also demonstrated that efficient alternative pathways for Top1 removal must exist.
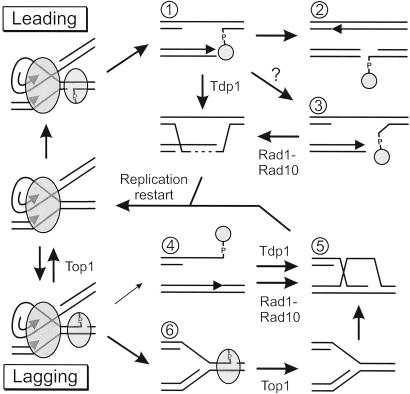
Figure 5.
Model for repair of camptothecin-induced replicative damage. The drawing depicts selected DNA lesions and repair intermediates (numbered) that might occur after a replisome (large oval) collides with a Top1 cleavage complex (small oval). The outcomes vary depending on whether Top1 was attached to the leading strand template (Top) or lagging strand template (Bottom). Possible repair steps are indicated by arrows, including enzymatic reversal of the Top1 cleavage complex by Top1 itself, repair that includes Top1 removal by Tdp1, and repair that includes Top1 removal by Rad1-Rad10. Smaller pathway arrows indicate less favored possibilities. Arrowheads in the drawings indicate DNA synthesis by a replication fork. The dashed line represents repair synthesis that must occur before formation of the recombination D-loop. See text for further discussion.
Recombinational repair of a double-strand break (DSB) involves 5′-3′ resection of the broken end to generate a 3′ nucleoprotein filament, which then invades a homologous donor duplex (7). To use the invading strand as a primer to initiate new DNA synthesis, any nonhomologous sequences at the 3′ end must be removed. In S. cerevisiae, this removal requires the nucleotide excision repair endonuclease Rad1-Rad10 as well as the mismatch repair proteins Msh2-Msh3 (8, 9). Although the exact role of Msh2-Msh3 is not understood, it is suggested to recognize and stabilize branched structures that are then cleaved by Rad1-Rad10 (9). Similarly, Rad1-Rad10-dependent tail removal is reduced in rad59 and srs2 mutants (9, 10). Rad59, a homologue of Rad52, and the Srs2 helicase may stabilize D-loops formed during strand invasion, thereby allowing sufficient time for tail removal.
We have systematically investigated the mechanisms for conferring camptothecin resistance in S. cerevisiae and identified the Rad1-Rad10 endonuclease as a major repair alternative to Tdp1. This tdp1 rad1 synthetic phenotype is conferred by both rad1 deletion and catalytic point mutants and is also manifest as a TOP1-dependent growth delay. By screening an array of mutants deficient in various DNA repair pathways, we further found that the function of Rad1-Rad10 in Top1 lesion repair is facilitated by Rad59 and Srs2, similar to its role in cleaving nonhomologous tails during gene conversion. Unlike tail removal, Top1 removal occurs without the involvement of Msh2-Msh3. We argue that Rad1-Rad10 can remove 3′ lesions from unwound but fully homologous sequences during replication restart. Finally, our genetic analysis indicates a role for Mus81-Mms4 in Top1 repair that is separate from the combined action of Tdp1 and Rad1-Rad10.
Methods
Strains and Plasmids.
Yeast strains used in this study are listed in Table 2, which is published as supporting information on the PNAS web site, www.pnas.org. Gene disruptions were made by using the one-step PCR-mediated replacement technique (11) and confirmed by PCR. Further derivatives were made by standard techniques of mating and tetrad dissection. Strains in the DNA repair array are haploid deletions of the parent strain BY4741 and were generated by the Saccharomyces Genome Deletion Project (12). Detailed methods for the construction of multiple mutants by array mating are published as Supporting Methods on the PNAS web site. The rad1-D869 mutation was made by first inserting the URA3 gene at position 869 of the RAD1 coding sequence. This strain was then transformed with a 1-kb DNA fragment bearing the desired rad1 point mutation (changing codon 869 from GAC to GCA), created by fusion PCR by using the high-fidelity HF Advantage PCR kit (CLONTECH). Reversion to Ura− (i.e., 5-fluoroorotic acid resistance), allele-specific PCR, and sequencing identified appropriately mutated strains. Strains JKM146, YFP103, and YFP255 as well as plasmids pFP122 and pFP120 were kindly provided by James Haber (Brandeis University) (9).
Measurement of Camptothecin Sensitivity.
Overnight cultures were diluted into YAPD (1% yeast extract/2% peptone/2% dextrose/40 μg/ml adenine/200 μg/ml G418) containing 2% DMSO and allowed to pregrow for a further 5 h. Cultures were then diluted to a calculated OD600 of 0.001 in YPAD and added to an equal volume of YPAD containing varying concentrations of drug and 4% DMSO (2% DMSO final). Cultures were shaken at 30°C until the OD600 of the untreated control reached 0.5 ± 0.15, corresponding to 10 ± 0.5 doublings (typically 16–20 h; note that camptothecin-induced damage is a function of the number of cell divisions, not time). After incubation, the OD600 of all cultures was determined. The slope of a plot of ln(A/A0) vs. the number of doublings of the untreated culture (nc = 0) proved the most reproducible measurement of growth inhibition. The effective slope at the n = 10 endpoint was calculated for all cultures based on the calculated beginning and measured ending OD600 values (slopec). The slope of a treated culture was then divided by the slope of the no-drug-control culture (slopec = 0) to yield the final measurement. The IC50 value corresponds to a relative slope of 0.9.
Determination of Nonhomologous Tail Removal Efficiency During Gene Conversion.
DSB repair efficiency was determined by using a slight modification of the procedure described by Paques and Haber (13). Briefly, cells were pregrown sequentially in plasmid-selective synthetic-defined dextrose medium followed by YPA containing 3% glycerol. Cells were then diluted in water and plated on plasmid-selective plates containing either dextrose or galactose. After 3–4 days, the DSB repair efficiency was calculated as the ratio of colonies on galactose versus dextrose plates.
Growth Rate Determination.
Overnight cultures grown in YPAD were diluted to OD600 = 0.04 in the same media and pregrown for a further 2–3 h. The density of the samples was then measured at 30-min intervals by using a genesys20 spectrophotometer for at least three doubling periods.
Results
Tdp1 and Rad1-Rad10 Function as Redundant Pathways of Top1 Repair.
We considered that the Tdp1-independent method of Top1 removal might occur during recombination after strand invasion to directly generate a 3′ hydroxyl (Fig. 1A), which suggested a role for Rad1-Rad10 in a fashion analogous to nonhomologous tail removal. To test this possibility, the camptothecin IC50 for rad1 and tdp1 rad1 mutant strains was determined on the basis of exposure to the drug for a period of 10 doubling times as described in Methods. The sensitivity of tdp1 and rad1 single mutants to camptothecin was comparable to the wild-type strain up to 10 μg/ml, the highest soluble drug concentration in 2% DMSO (Fig. 1B). In contrast, tdp1 rad1 mutant cells exhibited a marked increase in sensitivity to the drug, confirming the initial hypothesis that Tdp1 and Rad1 function as overlapping pathways for the repair of Top1 damage, although importantly this result does not itself establish the Rad1-Rad10 target lesion (see Discussion).
Figure 1.
Tdp1 and Rad1-Rad10 represent redundant pathways for repair of Top1 DNA damage. (A) The drawing depicts a simplified model of repair of camptothecin-induced replicative damage to illustrate the pathway relationships explored in B. (B) Camptothecin sensitivity was determined by growing strains isogenic to YW465 in the presence of varying concentrations of drug. Relative growth inhibition was measured as a ratio of the slopes of the growth curves at drug concentration c relative to the control (c = 0), as described in Methods. The dashed line corresponds to the position of the IC50 for each strain. Curves represent the mean ± standard deviation of at least two independent experiments for each strain. Strains and symbols are: wild-type, ■; tdp1, ♦; rad1, ▿; tpp1 apn1 apn2, Δ; tdp1 rad1, ●; rad52, □; tdp1 rad1 rad52, ▾.
Genetic Relationships of the Top1 Repair Pathways.
We next sought to compare the camptothecin sensitivities of all remaining combinations of tdp1, rad1, rad52, and tpp1 apn1 apn2 (i.e., 3′ phosphatase deficient) mutations to further delineate the pathway relationships illustrated in Fig. 1A. Many of these mutants proved to be inviable, however. For example, we have previously reported that loss of RAD52 in a tpp1 apn1 apn2 mutant is lethal (6). We further noted that loss of RAD1 caused lethality or severe growth defects in 3′ phosphatase-deficient backgrounds, phenotypes that will be described in detail elsewhere.
Among the viable multiple mutants, the following relationships were observed (Fig. 1B). First, mutating TDP1 in the tpp1 apn1 apn2 strain suppressed its camptothecin sensitivity to the level of the tdp1 mutant as previously described (not shown; ref. 6). This suppression is presumably due to shuttling of Top1 lesions normally handled by Tdp1 into the Rad1-dependent pathway, which in turn indicates that Top1 lesions are a better substrate for Rad1-Rad10 than 3′ phosphates. Importantly, though, the tdp1 rad1 mutant strain was clearly more camptothecin sensitive than the tpp1 apn1 apn2 3′ phosphatase-deficient strain, which is consistent with Rad1-Rad10 having some ability to remove 3′ phosphates (dashed arrow in Fig. 1A). This pattern might also be due to a basal level of Rad1-Rad10 cleavage of Top1 lesions even in wild-type cells, however. Unfortunately, the inviability of the rad1 tpp1 apn1 apn2 quadruple mutant made it impossible to explore these possibilities further. We next noted that, despite the marked sensitivity of the tdp1 rad1 strain to camptothecin, these cells were still significantly less sensitive than the rad52 mutant. Finally, mutation of rad52 was epistatic to tdp1 and rad1 mutations in that no multiple mutant was more sensitive than rad52 alone. Together, these results suggest that all repair of camptothecin-induced DNA damage must proceed through recombination, and that Tdp1 and Rad1-Rad10 can account for much, but not all, of the requisite Top1 removal.
Repair of Spontaneous Top1 Lesions.
We also observed a consistent increase in the doubling time in three different strain backgrounds when TDP1 and RAD1 were both mutated (Table 1). Loss of TDP1 or RAD1 alone had no such effect. This is the same pattern observed for camptothecin sensitivity, which supports the interpretation that delayed growth of tdp1 rad1 mutants is due to inefficient repair of spontaneous Top1-mediated damage. This interpretation would predict that the growth delay should be prevented by further mutation of TOP1, which was indeed the case (Table 1). Mutation of TOP1 itself increased the doubling time, as expected, but there was no further significant increase caused by tdp1 rad1 mutation in the top1 background.
Table 1.
TOP1-dependent growth delay of tdp1 rad1 cells
| Genotype | Doubling time, min
|
|||
|---|---|---|---|---|
| BY4741 TOP1 | JKM146 TOP1 | YW465 TOP1 | YW465 top1 | |
| TDP1 RAD1 | 83 ± 0.2 | 96 ± 0.4 | 95 ± 2.1 | 105 ± 3.3 |
| tdp1 RAD1 | 82 ± 0.3 | 96 ± 0.6 | 94 ± 3.3 | 105 ± 2.7 |
| TDP1 rad1 | 83 ± 0.9 | 96 ± 1.5 | 94 ± 1.7 | 108 ± 1.7 |
| tdp1 rad1 | 89 ± 1.1* | 113 ± 1.4* | 103 ± 1.7* | 107 ± 3.1 |
| rad52 | 110 ± 1.6* | ND | ND | ND |
Doubling times were determined in the indicated wild-type and top1 strain backgrounds; all are representative of multiple independent isolates, except JKM146. All doubling times represent the mean ± standard deviation of at least three independent measurements. ND, not determined.
, P < 0.001 compared to the TDP1 RAD1 strain for that background.
Top1 Repair Requires Rad1 Catalytic Function.
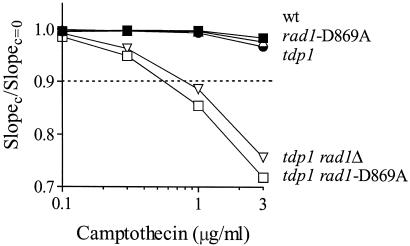
Because the above experiments used deletion mutants, it was possible that the phenotypes observed could be due to a pleiotropic effect distinct from the presumed catalytic function of Rad1-Rad10. To address this issue, we mutated aspartate 869 of Rad1 to alanine, an amino acid that is part of the nearly invariant V/IERKX3D motif of the Rad1/Mus81 class of repair endonucleases (14). The analogous mutant of the human Rad1 homologue XPF is known to lack catalytic activity while maintaining wild-type DNA and metal-binding activity, consistent with this amino acid playing a critical and selective role in the nuclease active site (14). The tdp1 rad1-D869A strain was indeed highly UV sensitive (not shown). Moreover, this strain showed camptothecin hypersensitivity nearly identical to the tdp1 rad1Δ strain (Fig. 2), demonstrating that Rad1 catalytic function is indeed required for Top1 repair.
Figure 2.
Rad1 catalytic activity is required for Top1 repair. Camptothecin sensitivity of the indicated strains was determined as described in Fig. 1, to compare the phenotypic effect of RAD1 deletion (rad1Δ) with a catalytic point mutation (rad1-D869A). All points represent the average of three independent experiments; error bars are within the symbols for each point. Strains and symbols are: wild-type, ■; rad1-D869A, Δ; tdp1, ●; tdp1 rad1Δ, ▿; tdp1 rad1-D869A, □.
Identification of Genes Involved in Top1 Repair by Using an Array-Based Approach.
We next assembled a 96-well array of mutant yeast strains, each disrupted for a different gene thought to be involved in the response to DNA damage. The camptothecin sensitivity of these mutants was then determined semiquantitatively in TDP1 RAD1, tdp1 RAD1, and tdp1 rad1 backgrounds. Twenty-three strains showed a basal hypersensitivity to camptothecin, although the level of sensitivity varied considerably among the mutants (Fig. 6 and Table 3, which are published as supporting information on the PNAS web site). In addition, loss of several genes, including RAD59, SRS2, TOP3, SGS1, RAD9, RAD17, RAD24, and MEC3, resulted in increased spot sensitivity to camptothecin in the absence of TDP1 and/or RAD1. The camptothecin IC50 of many of these mutants, as well as additional control mutants, was next determined with all possible combinations of tdp1 and rad1 to further elucidate the function of Rad1-Rad10 in Top1 repair (Fig. 3). Certain mutants were selected to be representative of known functionally related groups, e.g., rad50 was chosen to represent the Mre11-Rad50-Xrs2 complex. The IC50 for wild-type, tdp1, and rad1 cells (IC50 > 10 μg/ml), as well as mutants of tdp1 rad1 (IC50 = 0.96 ± 0.06 μg/ml) and rad52 (IC50 = 0.11 ± 0.03 μg/ml), serve as reference points for comparing the relative sensitivities. Results are discussed by relevant gene group.
Figure 3.
Camptothecin sensitivity of DNA repair mutants in combination with tdp1 and/or rad1. Strains derived from the BY4741 array were assayed for camptothecin sensitivity. Bars represent the mean ± standard deviation of the IC50 of at least two independent isolates. Horizontal lines indicate the IC50 values of wild-type, tdp1 rad1, and rad52 strains for comparison. One mutant series is shown in A–K in the order: mutx, tdp1 mutx, rad1 mutx, tdp1 rad1 mutx (where “mutx ” varies between panels). ND, not determined.
DSB Repair.
No mutants were more sensitive to camptothecin than those of the Rad52 epistasis group (excluding rad59, see below), which were themselves equally sensitive, including rad52 itself, rad50 (Fig. 3A), rad51 (Fig. 3B), and rad54 (not shown). As with the rad52 mutants described above, disruption of TDP1 and/or RAD1 did not lead to a further increase in sensitivity in these backgrounds. Loss of DNA ligase IV (DNL4) disables DSB repair through the nonhomologous end-joining pathway (15). Unlike the exquisite sensitivity of recombination mutants to camptothecin, a deficiency of DNL4 alone or in combination with tdp1 and/or rad1 did not confer additional hypersensitivity (Fig. 3C). Thus, repair ultimately requires recombination after removal of Top1 by either the Tdp1- or Rad1-Rad10-dependent pathway.
Nucleotide Excision Repair (NER).
Rad2 and Rad14, like Rad1-Rad10, are known to be required for NER (16), but unlike Rad1-Rad10 are not involved in the removal of nonhomologous tails during recombination (8). Neither rad2 (Fig. 3D) nor rad14 (not shown) mutants alone or in combination with tdp1 and/or rad1 were more sensitive to camptothecin. The role of the Rad1-Rad10 endonuclease in Top1 repair is thus separate from NER. This distinction is important, because Sastry and Ross (17) have reported that human cell-free extracts can cleave topoisomerase–DNA conjugates in a manner dependent on xeroderma pigmentosum complementation group A protein (XPA), which corresponds to Rad14 in yeast. Our rad14 data indicate that such cleavage has no role in repair of Top1 replicative damage, at least in yeast.
Rad59 and Srs2.
Several mutants in the array including rad59 (Fig. 3E), srs2 (Fig. 3F), sgs1 (Fig. 3I), rad17 (Fig. 3G), rad9 (not shown), and rad24 (not shown) produced very similar patterns of camptothecin sensitivity. Of these, the rad59 and srs2 mutants were of the most interest here. By themselves, both were significantly hypersensitive to camptothecin. The additional loss of TDP1 resulted in a further decrease in the IC50, whereas loss of RAD1 had little or no effect. Adding both tdp1 and rad1 mutations resulted in the greatest level of sensitivity. This pattern suggests that Rad1-Rad10 might act in Top1 removal in the context of D-loop formation (see Discussion). Although not discussed further, the similar pattern observed for the rad17, rad9, and rad24 checkpoint mutants likely reflects a delay in the repair of Top1 damage in the absence of Tdp1, such that the G2/M checkpoint must be activated (5, 18).
Msh2-Msh3.
Neither tdp1 msh2 nor tdp1 msh3 double mutants displayed an increase in sensitivity to camptothecin in the array spotting analysis, a finding confirmed for msh2 in the quantitative analysis (Fig. 3H). Given that gene conversion events requiring Rad1-Rad10 also depend on Msh2-Msh3 (9), this result was not expected. To confirm this by direct comparison, we used strains and plasmids developed by Haber and colleagues. Briefly, the HO endonuclease is expressed under the control of a galactose-inducible promoter in the JKM146 strain background to generate a site-specific DSB in a plasmid substrate that is repaired efficiently by gene conversion (13). In our hands, the efficiency of DSB repair of a plasmid with 308 and 610 nucleotide nonhomologous tails was specifically reduced approximately 30-fold in both rad1 (YFP103) and msh2 (YFP255) mutants (Fig. 4A). As expected, disruption of TDP1 in the wild-type, rad1, or msh2 strains had no effect on the efficiency of plasmid repair. In contrast, loss of TDP1 in the rad1, but not the msh2, strains again resulted in a 5-fold decrease in the camptothecin IC50 as compared with the tdp1 mutant alone (Fig. 4B). Thus, the role of Rad1-Rad10 in Top1 repair is independent of Msh2-Msh3.
Figure 4.
Differential requirement for Rad1-Rad10 and Msh2-Msh3 in repair of nonhomologous tails and Top1 damage. (A) The indicated JKM146-derived strains were assayed in triplicate for repair of a HO endonuclease-induced gene conversion event as described in Methods. Black bars represent repair of the control plasmid pFP122, whereas hatched bars correspond to repair of plasmid pFP120 containing 3′ nonhomologous ends. Mutants of rad1 and msh2 were equally deficient in the repair of plasmid pFP120 and were not affected by the loss of TDP1. (B) Using these same strains, tdp1 rad1, but not tdp1 msh2, cells were significantly more sensitive to camptothecin than the tdp1 mutant alone. Strains and symbols are: wild type, ■; rad1, Δ; msh2, ●; tdp1, □; tdp1 msh2, ♦; tdp1 rad1, ▿.
This different JKM146 strain background also provided insight into the relationship between the Tdp1 and Rad1 repair pathways because, unlike the array-derived strains used in Fig. 3, even wild-type JKM146 had a measurable IC50. It could thus be determined that disruption of tdp1, but not rad1, caused a minor degree of camptothecin hypersensitization (Fig. 4B), suggesting that Tdp1 is used in preference to Rad1-Rad10.
Mus81-Mms4.
The sensitivity pattern for mus81 was unique (Fig. 3J). The mus81 mutant was itself very sensitive, but this sensitivity was not increased by mutation of TDP1, in contrast to rad59, srs2 and others. The tdp1 rad1 mus81 triple mutant displayed a highly reproducible 2-fold lower camptothecin IC50, however. One explanation for this pattern might be that the Mus81-Mms4 endonuclease acts as a third pathway for the removal of Top1 (see Discussion). To begin to explore this hypothesis, we tested mus81 and rad1 mus81 strains in the nonhomologous tail removal assay described above and found no incremental defect in the mus81 strains (not shown). This result argues against a role of Mus81-Mms4 in 3′ processing during recombination, although nonhomologous tails are clearly only one of many possible types of lesion.
Asf1, Ctf4, Pol32, and Rad27.
Mutants of ctf4 (Fig. 3K), asf1, pol32, and rad27 (not shown) exhibited hypersensitivity to camptothecin, yet the loss of TDP1 and/or RAD1 in these mutants did not lead to a further increase in drug sensitivity. Although not discussed further, these mutants were important because they effectively served as controls, demonstrating the specificity of the tdp1 rad1 synthetic phenotypes presented above.
Discussion
Recombination-Dependent Repair of Top1 Replicative Lesions.
Accumulated data, most strongly the abrogation of camptothecin cytotoxicity by aphidicolin (3), have suggested that Top1 DNA damage is realized during replication when cleavage complexes lead to fork stalling, collapse, or breakage (Fig. 5). The finding here that the combined Tdp1/Rad1-Rad10 repair pathways are required for normal cellular growth even in the absence of a Top1 reversal inhibitor provides further evidence in support of this notion (Table 1). In such cells, there is clearly no disturbance of DNA topology or Top1 function, and yet Top1-dependent lesions in need of repair are occasionally created. Irreversible replication-dependent disruption of the Top1 cleavage complex also accounts for the fact that all Top1 repair seems to require RAD52, because even a single replicative break would be lethal in a rad52 mutant cell. It is quite interesting that both RAD50 and RAD51 were as required as RAD52 (Fig. 3), because these genes appear to represent alternative forms of RAD52-dependent recombination as revealed by both telomere maintenance (19) and repair of DSBs by RAD51-independent break-induced replication (BIR) (20). It is clear that this distinction is not evident in repair of camptothecin-induced damage, so that each Top1 break likely requires the concerted action of all three proteins. Indeed, RAD51-independent BIR apparently requires rare “facilitator” chromosomal sequences (21) and so may not be generally applicable to all forms of replication restart.
Multiple Mechanisms for Top1 Lesion Processing Before Recombination.
Tdp1 initially seemed an attractive candidate for therapeutic targeting to improve the efficacy of camptothecin and related antineoplastic topoisomerase poisons. Unfortunately, our results demonstrate that in at least one eukaryotic cell, yeast, there is substantial redundancy in Top1 lesion processing before recombination, such that significant sensitization to camptothecin occurs only when both TDP1 and RAD1-RAD10 are inactivated. Even then maximal sensitivity is not realized, demonstrating the presence of yet a third Top1 removal mechanism. The slight camptothecin sensitivity of tdp1 but not rad1 mutants (Fig. 4) argues that at least the Rad1-Rad10 mechanism is a compensatory response to damage that persists in the absence of Tdp1, but it is nonetheless highly efficient. Importantly, our array experiment provided no evidence that any of these mechanisms are related to DNA repair pathways other than homologous recombination, including nonhomologous end-joining, NER (see more below), base excision repair, and mismatch repair. Pichierri et al. have observed that mammalian MSH2−/− cells are in fact measurably hypersensitive to camptothecin (22). Our results support their interpretation that dysregulated recombination is responsible for this effect, rather than direct processing of Top1 damage by mismatch repair.
Rad1-Rad10-Dependent Recombinational Repair of Fully Homologous Sequences.
Rad1-Rad10 is a structure-specific endonuclease that cleaves Y-form branched DNA structures near the double- to single-stranded junction when tracing the cleaved strand from 5′ to 3′ (23). It has three known substrates in mitotic cells: the 5′ side of the unwound damaged strand during NER (16), the homology–nonhomology junction generated when Rad51 is unable to complete strand exchange of a 3′ nonhomologous tail during recombination, and similar nonhomologous tail junctions created during single-strand annealing (8). This work adds to this list a lesion generated when replication forks collide with Top1 cleavage complexes in the absence of Tdp1. Processing of this lesion is clearly a distinct function for Rad1-Rad10. NER is not known to process strand-break lesions, and indeed, no NER proteins other than Rad1-Rad10 were required for camptothecin resistance (Fig. 3 and Fig. 6). Further, there is no reason for Top1 damage to have nonhomologous tails, because the broken strands would be completely homologous with their sister chromatid (see Fig. 5). This last distinction is important because recombinational repair of homologous strand breaks is a quantitatively large burden in mitotically dividing cells (24), as compared with events with 3′ nonhomology.
What Is the Camptothecin-Induced Rad1-Rad10 Substrate?
Rad1-Rad10 must cleave DNA to participate in Top1 repair (Fig. 2), but this activity is completely dispensable in cells expressing Tdp1. It is thus highly likely that the role of Rad1-Rad10 is to remove the bound Top1 in the absence of Tdp1. Because yeast extracts prepared from tdp1 apn1 cells (i.e., still containing Rad1-Rad10) lack any residual ability to remove 3′ phosphotyrosyl lesions (Jeff Pouliot and Howard Nash, personal communication), it is further likely that Rad1-Rad10, in contrast to Tdp1, cleaves a branched structure more distantly from the Top1 break, consistent with its established substrate requirements (23). Our initial hypothesis was that Rad1-Rad10 acts at the D-loop predicted to be created when a Top1-terminated 3′ strand invades its sister chromatid (Fig. 5, intermediate 5), in direct analogy to nonhomologous tail removal during gene conversion. Although this would not generate a nonhomologous tail, we hypothesized that the covalently linked Top1 fragment would be bulky enough to prevent completion of strand invasion leaving a Y structure suitable for cleavage by Rad1-Rad10. Factors in favor of this D-loop model include the observed sensitivity pattern for rad59 and srs2 mutants. Each of these are known to show a partial recombination defect, but to be particularly inefficient in the gene conversion of DSBs when nonhomologous tails are present (10, 13). This pattern parallels the camptothecin sensitivity of rad59 and srs2 mutants in that Rad59 and Srs2 are especially important for the Rad1-Rad10-dependent pathway (Fig. 3), although this could certainly reflect aspects of Rad59 and Srs2 function separate from D-loop stabilization. An observation that initially appeared to argue against the D-loop hypothesis was that Rad1-Rad10-dependent Top1 repair proceeded perfectly well in the absence of Msh2-Msh3 (Fig. 4), proteins known to be required for removal of nonhomologous 3′ tails (9). This difference might simply reflect the fact that Top1-associated DSBs are completely homologous with their donor template, however, alleviating the need for mismatch recognition.
The greatest problem with the D-loop model is the nature of the collision needed to create this Top1 intermediate. Replication runout at a lagging strand cleavage complex would yield a DSB with Top1 bound to its 3′ end (Fig. 5, intermediate 4) that would require removal of Top1 during strand invasion in the absence of Tdp1 (Fig. 5, intermediate 5). However, it is generally believed that Top1 complexes bound to the lagging strand template preferentially cause fork stalling (Fig. 5, intermediate 6) instead of breakage, based on the clear asymmetry of Top1 binding to DNA (25–27). In contrast, runout at leading strand complexes clearly occurs (27) but yields a lesion in which Top1 remains bound to the daughter strand gap and not the DSB end (Fig. 5, intermediate 1). The gap is not predicted to be a substrate for Rad1-Rad10, however. It is possible that the 3′ terminus is unwound by the Srs2 or Sgs1 helicase or perhaps even by the replicative helicase, so that Rad1-Rad10 can act (Fig. 5, intermediate 3). Alternatively, proteolysis of Top1 might release its grip on the DNA so that a second fork ultimately colliding from the other direction would run out. The resulting lesion would be a DSB in a fully replicated chromosome, which could be repaired by gene conversion (Fig. 5, intermediate 2).
The Role of Mus81-Mms4.
Mus81-Mms4 is an endonuclease that is homologous to Rad1-Rad10 and has a similar, but distinct, ability to cleave branched substrates (28, 29). It is an intriguing candidate for the third Top1-removing enzyme, made evident by the partial camptothecin sensitivity of tdp1 rad1 mutants. Indeed, mus81 cells became more sensitive to camptothecin only when TDP1 and RAD1 were both mutated (Fig. 3). Loss of Mus81-Mms4 by itself conferred great sensitivity to camptothecin, however, and so, unlike Rad10-Rad10, this enzyme must play some role other than Top1 removal. Indeed, it has been proposed that Mus81-Mms4 and Sgs1-Top3 act as alternative mechanisms for restarting stalled replication forks, which in the case of Mus81-Mms4 is believed to proceed via fork cleavage to create a DSB that directly supports strand invasion (28, 30) (Fig. 5, intermediate 5). Mus81-Mms4 would thus be expected to facilitate the restart of replication forks that have irreversibly stalled at lagging-strand Top1 complexes without strand dissociation and formation of a DSB (Fig. 5, intermediate 6). Here, the third mechanism of Top1 removal is expected to be Top1 itself via its normal religation mechanism, which is possible because camptothecin is a reversible inhibitor. The synergistic sensitization to camptothecin in the mus81 tdp1 rad1 triple mutant thus suggests a model in which there are two classes of fork failure. In this model, Tdp1 and Rad1-Rad10 act redundantly to remove Top1 from collapsed forks with DSBs, whereas Mus81-Mms4 creates DSBs at stalled forks where Top1 reversal has occurred spontaneously.
Supplementary Material
Acknowledgments
We thank Mats Ljungman, James Haber, and Dindial Ramotar for comments on the manuscript. This work was supported in part by the Pew Scholars Program in the Biomedical Sciences of the Pew Charitable Trusts and Public Health Service Grant CA-90911 (to T.E.W.) and Training Grant 5T32HL07157 (to J.R.V.).
Abbreviations
- DSB
double-strand break
- NER
nucleotide excision repair
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Champoux J J. Annu Rev Biochem. 2001;70:369–413. doi: 10.1146/annurev.biochem.70.1.369. [DOI] [PubMed] [Google Scholar]
- 2.Svejstrup J Q, Christiansen K, Gromova I I, Andersen A H, Westergaard O. J Mol Biol. 1991;222:669–678. doi: 10.1016/0022-2836(91)90503-x. [DOI] [PubMed] [Google Scholar]
- 3.Hsiang Y H, Lihou M G, Liu L F. Cancer Res. 1989;49:5077–5082. [PubMed] [Google Scholar]
- 4.Nitiss J, Wang J C. Proc Natl Acad Sci USA. 1988;85:7501–7505. doi: 10.1073/pnas.85.20.7501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pouliot J J, Yao K C, Robertson C A, Nash H A. Science. 1999;286:552–555. doi: 10.1126/science.286.5439.552. [DOI] [PubMed] [Google Scholar]
- 6.Vance J R, Wilson T E. Mol Cell Biol. 2001;21:7191–7198. doi: 10.1128/MCB.21.21.7191-7198.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Paques F, Haber J E. Microbiol Mol Biol Rev. 1999;63:349–404. doi: 10.1128/mmbr.63.2.349-404.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ivanov E L, Haber J E. Mol Cell Biol. 1995;15:2245–2252. doi: 10.1128/mcb.15.4.2245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sugawara N, Paques F, Colaiacovo M, Haber J E. Proc Natl Acad Sci USA. 1997;94:9214–9219. doi: 10.1073/pnas.94.17.9214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sugawara N, Ira G, Haber J E. Mol Cell Biol. 2000;20:5300–5309. doi: 10.1128/mcb.20.14.5300-5309.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brachmann C B, Davies A, Cost G J, Caputo E, Li J, Hieter P, Boeke J D. Yeast. 1998;14:115–132. doi: 10.1002/(SICI)1097-0061(19980130)14:2<115::AID-YEA204>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 12.Winzeler E A, Shoemaker D D, Astromoff A, Liang H, Anderson K, Andre B, Bangham R, Benito R, Boeke J D, Bussey H, et al. Science. 1999;285:901–906. doi: 10.1126/science.285.5429.901. [DOI] [PubMed] [Google Scholar]
- 13.Paques F, Haber J E. Mol Cell Biol. 1997;17:6765–6771. doi: 10.1128/mcb.17.11.6765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Enzlin J H, Scharer O D. EMBO J. 2002;21:2045–2053. doi: 10.1093/emboj/21.8.2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wilson T E, Grawunder U, Lieber M R. Nature. 1997;388:495–498. doi: 10.1038/41365. [DOI] [PubMed] [Google Scholar]
- 16.Prakash S, Prakash L. Mutat Res. 2000;451:13–24. doi: 10.1016/s0027-5107(00)00037-3. [DOI] [PubMed] [Google Scholar]
- 17.Sastry S, Ross B M. J Biol Chem. 1998;273:9942–9950. doi: 10.1074/jbc.273.16.9942. [DOI] [PubMed] [Google Scholar]
- 18.Foiani M, Pellicioli A, Lopes M, Lucca C, Ferrari M, Liberi G, Muzi Falconi M, Plevani P. Mutat Res. 2000;451:187–196. doi: 10.1016/s0027-5107(00)00049-x. [DOI] [PubMed] [Google Scholar]
- 19.Le S, Moore J K, Haber J E, Greider C W. Genetics. 1999;152:143–152. doi: 10.1093/genetics/152.1.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Signon L, Malkova A, Naylor M L, Klein H, Haber J E. Mol Cell Biol. 2001;21:2048–2056. doi: 10.1128/MCB.21.6.2048-2056.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Malkova A, Signon L, Schaefer C B, Naylor M L, Theis J F, Newlon C S, Haber J E. Genes Dev. 2001;15:1055–1060. doi: 10.1101/gad.875901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pichierri P, Franchitto A, Piergentili R, Colussi C, Palitti F. Carcinogenesis. 2001;22:1781–1787. doi: 10.1093/carcin/22.11.1781. [DOI] [PubMed] [Google Scholar]
- 23.Bardwell A J, Bardwell L, Tomkinson A E, Friedberg E C. Science. 1994;265:2082–2085. doi: 10.1126/science.8091230. [DOI] [PubMed] [Google Scholar]
- 24.Cox M M, Goodman M F, Kreuzer K N, Sherratt D J, Sandler S J, Marians K J. Nature. 2000;404:37–41. doi: 10.1038/35003501. [DOI] [PubMed] [Google Scholar]
- 25.Tsao Y P, Russo A, Nyamuswa G, Silber R, Liu L F. Cancer Res. 1993;53:5908–5914. [PubMed] [Google Scholar]
- 26.Redinbo M R, Stewart L, Kuhn P, Champoux J J, Hol W G. Science. 1998;279:1504–1513. doi: 10.1126/science.279.5356.1504. [DOI] [PubMed] [Google Scholar]
- 27.Strumberg D, Pilon A A, Smith M, Hickey R, Malkas L, Pommier Y. Mol Cell Biol. 2000;20:3977–3987. doi: 10.1128/mcb.20.11.3977-3987.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kaliraman V, Mullen J R, Fricke W M, Bastin-Shanower S A, Brill S J. Genes Dev. 2001;15:2730–2740. doi: 10.1101/gad.932201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen X B, Melchionna R, Denis C M, Gaillard P H, Blasina A, Van de Weyer I, Boddy M N, Russell P, Vialard J, McGowan C H. Mol Cell. 2001;8:1117–1127. doi: 10.1016/s1097-2765(01)00375-6. [DOI] [PubMed] [Google Scholar]
- 30.Doe C L, Ahn J S, Dixon J, Whitby M C. J Biol Chem. 2002;277:32753–32759. doi: 10.1074/jbc.M202120200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.