Abstract
Some members of the major histocompatibility complex (MHC) class I gene family are encoded outside the MHC. Here we describe a family of mouse class I-like genes mapping to the vicinity of the leukocyte receptor complex (LRC) on chromosome 7. This family, which we call Mill (MHC class I-like located near the LRC), has two members designated Mill1 and Mill2. Both genes are predicted to encode membrane glycoproteins with domain organization essentially similar to that of MHC class I heavy chains. The following features of Mill are noteworthy. (i) The deduced MILL proteins lack most of the residues known to be involved in the docking of peptides in classical MHC class I molecules. (ii) Among the known members of the class I gene family, MILL1 and MILL2 are related most closely to MICA/MICB encoded in the human MHC. (iii) Unlike all other known members of the class I gene family, Mill1 and Mill2 have an exon between those coding for the signal peptide and the α1 domain. (iv) Mill1 has a more restricted expression profile than Mill2. (v) The gene orthologous to Mill1 or Mill2 apparently is absent in the human. (vi) Mill1 and Mill2 show a limited degree of polymorphism in laboratory mice. The observation that the Mill family is related most closely to the MIC family, together with its apparent absence in the human, suggests its involvement in innate immunity.
All jawed vertebrates have two classes of structurally and functionally distinct major histocompatibility complex (MHC) molecules that present peptides to T cell receptors, thereby initiating an adaptive immune response (1). Class I molecules are integral membrane glycoproteins made up of a heavy chain and β2-microglobulin that present peptides derived from intracellularly synthesized proteins to CD8+ T cells (2). On the other hand, class II molecules, consisting of two integral membrane glycoproteins designated α and β chains, present peptides produced in or directed to the endosomal/lysosomal compartment to CD4+ T cells (3). In most animals including human and mouse, class I genes (the genes for the heavy chain of class I molecules) are tightly linked to class II genes (the genes for the α and β chains of class II molecules), collectively forming a gene cluster (1, 4, 5).
Although class I genes that play a major role in antigen presentation map to the MHC, several class I-like genes located outside the MHC have been identified in human and mouse. Examples of such class I-like genes in mice include Cd1 (Cd1d1 and Cd1d2) (6, 7), Azgp1 (the gene for zinc-α2-glycoprotein) (8, 9), Fcgrt (the gene for the heavy chain of the neonatal IgG Fc receptor) (10, 11), Procr (the gene for the endothelial protein C receptor) (12), H2ls (the gene for the H2 complex class 1-like polypeptide) (13, 14), Hfe (the hemochromatosis gene) (15, 16), Raet1a–Raet1e (the genes for retinoic acid early transcripts 1α–ɛ) (17, 18), and H60 (histocompatibility 60) (19). Although these genes have diverse functions, some unrelated to immunity, they all have structural similarities to MHC class I genes and thus qualify as members of the class I gene family.
We describe here a family of mouse class I-like genes located outside the MHC. This gene family, which maps to the vicinity of the leukocyte receptor complex (LRC) on chromosome 7, has two members. Interestingly, the proteins encoded by this gene family are related most closely to the MICA/MICB molecules encoded in the human MHC. After consultation with the International Committee on Standardized Genetic Nomenclature for Mice, we propose to call this newly discovered gene family Mill (MHC class I-like located near the LRC) and its two members Mill1 and Mill2.
Materials and Methods
Isolation of Mill1 and Mill2 cDNA by RT-PCRs.
A computer search of the mouse EST section of the GenBank database resulted in the identification of three clones predicted to encode two distinct, class I heavy chain-like molecules. Based on the sequences of these clones, we designed primers that enabled the isolation of full coding sequences of Mill1 and Mill2 cDNA. The primer sequences were 5′-TGGCTTGGGATCTTCAAAGT-3′ (sense) and 5′-TCCTCTGTCTTGTTTGGCTGTT-3′ (antisense) for Mill1 and 5′-TTTGCACAAAACTCCATTTGA-3′ (sense) and 5′-AGAAACAGCCAAGCCTCAGTC-3′ (antisense) for Mill2. The cDNA templates for PCR were synthesized as described (20) by using total cellular RNAs isolated from the skin of 1-day-old BALB/cCrSlc mice (Mill1) and the lung of adult BALB/cCrSlc mice (Mill2). PCR products were cloned into the pGEM-T Easy vector (Promega), and their sequences were determined by using an automated sequencer. Potential PCR errors were eliminated by setting up PCRs in triplicate and sequencing multiple clones for each reaction.
Phylogenetic Analysis.
Amino acid sequences of representative class I heavy chains and heavy chain-like molecules (hereafter referred to as class I and class I-like molecules, respectively) derived from human and mouse were aligned with the CLUSTAL W program (21). The alignment then was adjusted by eye to maximize sequence similarity. Reliable alignment was possible only in the α1–α3 domains (or the α1 and α2 domains for class I-like molecules lacking the α3 domain). We therefore excluded the remaining regions from the analysis. The distance matrix was obtained by calculating p distances for all pairs of sequences. Sites containing gaps were excluded from the analysis by using the pairwise-deletion option. Neighbor-joining trees were constructed by using MEGA version 2.1 (22). The reliability of branching patterns was assessed by bootstrap analysis (1,000 replications). Maximum-likelihood trees were constructed by using the TREE-PUZZLE 5.0 program (23). We used the JTT matrix (24) as a model of substitution, a uniform rate as a model of rate heterogeneity, and default options in all other settings. We searched for tree topologies with the largest log likelihood by repeating quartet-puzzling steps 10,000 times.
Interspecific Backcross Mapping.
We mapped the two Mill genes as described (25) by PCR using the [(C57BL/6Ei × Mus spretus)F1 × M. spretus] backcross DNA panel purchased from The Jackson Laboratory. The primer pairs used for mapping were 5′-TTCGATGATGAGCCCTTCC-3′ (sense, located in exon 3) and 5′-GATCTGTATGCACTGAGCACCA-3′ (antisense, located in intron 3) for Mill1 and 5′-GAGAGACTAACAATCTGTTGAAGGT-3′ (sense, located in exon 3) and 5′-TCCCATTACTCAGAAGCTCACAG-3′ (antisense, located in exon 4) for Mill2. These primer pairs amplified the bands of 215 bp (Mill1) and 337 bp (Mill2) only in C57BL/6Ei mice. We designed these primers by comparing the partial genomic sequences of Mill1 and Mill2 in C57BL/6J and M. spretus (GenBank accession nos. AB086329, AB086337, AB086344, and AB086352). Mill1 and Mill2 were mapped by using the same cycling conditions (35 cycles of 30 sec at 94°C, 1 min at 56°C, and 40 sec at 72°C).
Analysis of the Gene Structure.
Organizations of the Mill genes were deduced by comparing the cDNA sequences determined above and the GenBank EST sequences with the genomic sequences in the Mouse Genome Sequencing Consortium (MGSC) version 3 assembly (www.ensembl.org/Mus_musculus/) and in the Celera database (Celera Genomics, Rockville, MD). For Mill1, a high-throughput genomic sequence was available in the GenBank database.
Analysis of Polymorphism in Laboratory Mice.
Genomic fragments encompassing exon 3 (coding for the α1 domain), intron 3, and exon 4 (coding for the α2 domain) of Mill1 and Mill2 were amplified by PCR from the genomic DNA isolated from a total of 14 inbred strains listed in Table 1. The primer sequences were 5′-CCCATGAACCTGTCTCCTCA-3′ (sense, located in intron 2) and 5′-GGTTCTGGAAACCAACTCTGG-3′ (antisense, located in intron 4) for Mill1 and 5′-CAATCATCTGGGTTCAGTCCC-3′ (sense, located in intron 2) and 5′-CATCTTACCATCCAAGCCCAA-3′ (antisense, located in intron 4) for Mill2. PCRs were set up in triplicate, and multiple clones were sequenced for each reaction.
Table 1.
Polymorphism of Mill1 and Mill2 in laboratory mice
| Mice | Codon
no.*
|
||||||
|---|---|---|---|---|---|---|---|
|
Mill1
|
Mill2
|
||||||
| 87 | 206 | 40 | 83 | 94 | 166 | 190 | |
| AKR/J, A/WySnJ, BALB/c, C57BL/6J, C57BL/10J, C57L/J, RIIIS/J, and SJL/J | GGA | AAT | CAG | AAC | GGG | CAT | GCT |
| Gly | Asn | Gln | Asn | Gly | His | Ala | |
| P/J and DBA/2J | GCA | GAT | CAG | AAC | GGG | CAT | GCT |
| Ala | Asp | Gln | Asn | Gly | His | Ala | |
| 129X1/SvJ, C3H/HeJ, CBA/J, and NZB/B1NJ | GCA | GAT | GGG | AAG | GAG | CGT | ACT |
| Ala | Asp | Gly | Lys | Glu | Arg | Thr | |
Codon numbers refer to those of mature MILL proteins.
Expression Analysis.
Expression patterns of Mill1 and Mill2 were analyzed by RT-PCR as described (26). Briefly, total cellular RNAs isolated from various organs of BALB/cCrSlc mice or cell lines were converted to cDNA with the Superscript II kit (GIBCO/BRL). By using the cDNA and gene-specific primers, 30–35 cycles of PCR (35 cycles for Mill1 and 30 cycles for Mill2 and β-actin) were conducted under the following conditions: denaturation at 94°C for 30 sec, annealing at 55–62°C for 1 min (62°C for Mill1, 60.5°C for Mill2, and 55°C for β-actin), and extension at 72°C for 1 min. The primer sequences were 5′-CACACTCTGCGCTATGACCT-3′ (sense) and 5′-ATATTGTGGTTGCCGTGCTT-3′ (antisense) for Mill1, 5′-TTTGGGCTGTGAGCTTCTGAG-3′ (sense) and 5′-AGTCCTGGTCCTGTCCTTTGT-3′ (antisense) for Mill2, and 5′-TGTTACCAACTGGGACGACA-3′ (sense) and 5′-CTTTTCACGGTTGGCCTTAG-3′ (antisense) for β-actin. Neonatal tissues were obtained from 1-day-old BALB/cCrSlc mice. Mouse cell lines were purchased from RIKEN BioResource Center (Tsukuba, Japan).
“Zoo-Blot” Analysis.
BamHI digests of genomic DNA were electrophoresed on a 0.8% agarose gel, blotted to a nylon membrane, and hybridized with a digoxigenin-labeled Mill1 cDNA probe spanning the α1–α3 domains (residues 30–294 of MILL1 in Fig. 1) according to the instructions of the manufacturer (Roche Diagnostics). Hybridization was performed at 42°C as described (27) under reduced stringency in a solution containing 30% formamide, 0.1% N-lauroylsarcosine, 0.02% SDS (NaDodSO4), and 2% blocking solution. Final washing conditions were 2× standard saline citrate (SSC)/0.1% NaDodSO4 at 50°C.
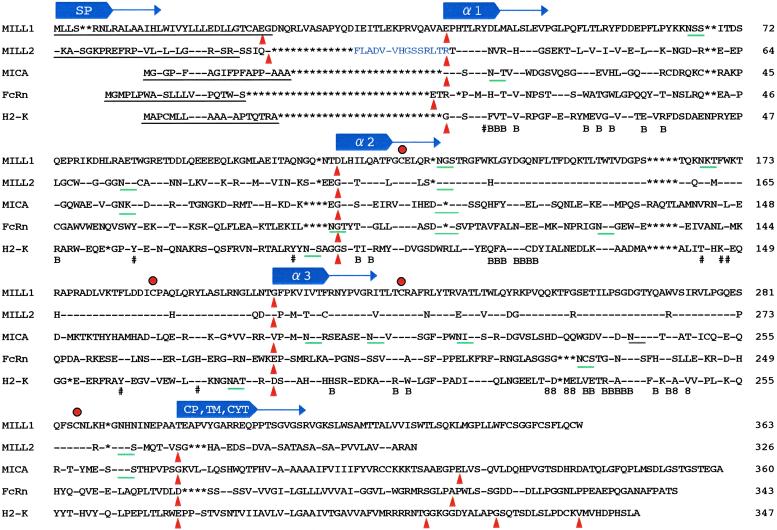
Figure 1.
Deduced amino acid sequences of mouse MILL1 and MILL2. N-terminal sequences predicted to function as signal peptides are underlined. Red dots and green bars indicate conserved cysteine residues and potential N-linked glycosylation sites, respectively. Filled triangles in red indicate exon–intron boundaries. The signs − and * indicate identity with the top sequence and absence of residues, respectively. The sign # below the H2-K sequence indicates residues important in anchoring the N and C termini of peptides. 8 and B refer to those residues of the H2-K molecule proposed to interact with CD8 and β2-microglobulin, respectively. In a shorter version of Mill2 cDNA, the 15 amino acids immediately preceding the α1 domain (indicated in blue) are absent. CP/TM/CYT stands for the connecting peptide/transmembrane region/cytoplasmic region. The GenBank accession numbers for human MICA, mouse FcRn, and H2-K are A55739, I56197, and I49713, respectively.
Results
Cloning of Mill1 and Mill2 cDNA.
To search for class I-like genes not described previously, we performed a BLAST search of the GenBank database by using various class I amino acid sequences as queries. This search resulted in the identification of one high-throughput sequence (accession no. AC079493.1) containing a gene potentially capable of encoding a new class I-like molecule. Analysis of the mouse EST database at GenBank confirmed the presence of two ESTs (accession nos. BB613427 and BB618324) matching to the gene that we designated Mill1. These ESTs were the 5′-end sequences of clones 4732481C10 and 5530400I18 isolated from the skin and head cDNA libraries of 10-day-old mice, respectively. Independently of this, a BLAST search of the mouse EST database identified one EST (accession no. BB636458) thought to encode yet another new class I-like molecule. This EST, derived from the gene that we designated Mill2, was the 5′-end sequence of clone A530001N15 isolated from an adult male aorta/vein cDNA library. The 3′-end sequences of clones 5530400I18 and A530001N15 were available at GenBank under accession nos. AV293819 and BB212568, respectively. Based on the 5′- and 3′-end sequences of these clones, we designed PCR primers that would allow for the amplification of the complete coding regions of Mill1 and Mill2 cDNA. The cDNA sequences thus obtained were deposited in the GenBank database under accession nos. AB086265 (Mill1) and AB086267 (Mill2).
MILL1 and MILL2 Have Domain Organizations Characteristic of MHC Class I Molecules.
The deduced MILL1 molecule is made up of 395 amino acids, of which the N-terminal 32 residues were predicted to comprise the signal peptide (Fig. 1). The mature form of the MILL1 polypeptide has 363 amino acids, with a calculated molecular mass of 41,180.8. The C-terminal end of MILL1 contains a stretch of hydrophobic amino acids that presumably constitutes the transmembrane region. On the other hand, MILL2 has 355 amino acids, of which the N-terminal 29 residues were predicted to comprise the signal peptide (Fig. 1). Hence, the mature MILL2 polypeptide has 326 amino acids, with a calculated molecular mass of 36,047.6. Similar to MILL1, the C-terminal end of MILL2 has a stretch of hydrophobic amino acids that presumably constitutes the transmembrane region. Both MILL1 and MILL2 have three potential N-linked glycosylation sites in the predicted extracellular regions.
Comparison of MILL1 and MILL2 with three representative members of the MHC class I family provides convincing evidence that both MILL molecules have three extracellular domains characteristic of the class I gene family (Fig. 1). The SMART program (28) predicted that residues 28–204 of MILL1 and 20–196 of MILL2 match to the Pfam (Protein families database maintained by The Sanger Institute, Hinxton, U.K.) entry PF00129 (class I histocompatibility antigen, domains α1 and α2), whereas residues 223–295 of MILL1 and 215–287 of MILL2 have features typical of IgC1 domains. The overall amino acid sequence identities of the three extracellular domains of MILL1 to the corresponding domains of MICA, H2-K, and mouse FcRn (the heavy chain of the neonatal IgG Fc receptor) were 33%, 27%, and 24%, respectively, and the corresponding figures for MILL2 were 35%, 29%, and 24%, respectively. MILL1 and MILL2 show little sequence similarity to each other in the regions other than the α1–α3 domains. However, in these domains their sequences are well conserved (70% amino acid sequence identity). Interestingly, the extent of sequence identity differs markedly between the α1 and α2/α3 domains. The amino acid sequences of MILL1 and MILL2 are only 43% identical in the α1 domain, whereas they are 87% and 78% identical in the α2 and α3 domains, respectively. Another notable feature of MILL1 and MILL2 is that they have insertions of amino acids between the signal peptide and the α1 domain. As described later, these residues are encoded by an exon not found in other members of the class I gene family. For Mill2, we also isolated cDNA clones that spliced out this exon. This shorter form of cDNA sequence was deposited in the GenBank database under accession no. AB086266.
Classical class I molecules such as H2-K have eight highly conserved residues important in anchoring the N and C termini of peptides (29): Tyr-7, Tyr-59, Tyr-159, Tyr-171, and Tyr-84 or Arg-84, Thr-143, Lys-146, and Trp-147 (numbering based on H2-K). MILL1 and MILL2 only have three such residues, suggesting that they are unlikely to bind peptides at least in a manner similar to classical class I molecules. In H2-K, six residues in the α3 domain are thought to interact with CD8 (30), of which only one is shared between MILL and H2-K. Furthermore, most of the residues known to interact with β2-microglobulin in classical class I molecules (31) are substituted to other amino acids in MILL. Thus, neither MILL1 nor MILL2 is likely to interact with β2-microglobulin or CD8.
Phylogenetic Analysis Indicates That MILL Molecules Are Related Most Closely to MICA/MICB.
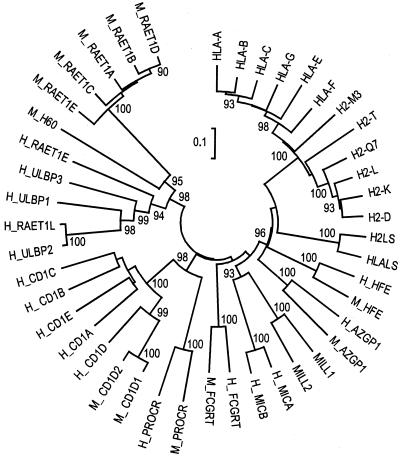
To address the relationship of MILL to other known members of the class I gene family more precisely, we constructed a neighbor-joining tree (Fig. 2). MILL1 clearly was related most closely to MILL2, justifying our proposal that Mill1 and Mill2 constitute a previously uncharacterized family of MHC class I-like genes. Among more distantly related members of the class I gene family, MICA and MICB were related most closely to MILL1 and MILL2. The node connecting MICA/MICB and MILL1/MILL2 was supported by a high bootstrap value. To test whether this branching pattern can be reproduced with a different tree-building algorithm, we also made a tree by using the maximum-likelihood method. Here again, MICA/MICB and MILL1/MILL2 formed a cluster connected by a single node (see Fig. 6, which is published as supporting information on the PNAS web site, www.pnas.org). We also made domain-by-domain comparison by constructing neighbor-joining trees by using only the α1, α2, or α3 domain sequences of the same data set. In each domain, MICA/MICB and MILL1/MILL2 formed a cluster connected by a single node (data not shown).
Figure 2.
Neighbor-joining tree of mammalian MHC class I and class I-like molecules. Numbers at the nodes represent the bootstrap confidence level in percentage. Only bootstrap values over 90 are indicated. M_ and H_ stand for mouse and human, respectively. Database accession nos.: HLA-A, P18462; HLA-B, HLHUB7; HLA-C, NP_002108.1; HLA-E, NP_005507.1; HLA-F, NP_061823.1; HLA-G, A39953; H2-K, I49713; H2-D, NP_034510.1; H2-L, I54069; H2-T, P14433; H2-Q7, P14429; H2-M3, NP_038847.1; H2LS, JC5663; HLALS, A57136; H_HFE, Q30201; M_HFE, P70387; H_AZGP1, NP_001176.1; M_AZGP1, Q64726; MILL1 and MILL2, this study; H_MICA, A55739; H_MICB, NP_005922.1; H_FCGRT, I38720; M_FCGRT, I56197; H_PROCR, Q9UNN8; M_PROCR, A55945; H_CD1A, AAA51931.1; H_CD1B, P29016; H_CD1C, P29017; H_CD1D, P15813; H_CD1E, NP_112155.1; M_CD1D1, I49581; M_CD1D2, NP_031666.1; H_ULBP1, NP_079494.1; H_ULBP2, NP_079493.1; H_ULBP3, NP_078794.1; H_RAET1L, AAK91503.1; H_RAET1E, AAL76417.1; M_RAET1A, 2206404A; M_RAET1B, NP_033043.1; M_RAET1C, NP_033044.1; M_RAET1D, NP_064414.1; M_RAET1E, AAL11004.1; and M_H60, AAC63305.1.
Consistent with the results of phylogenetic-tree analysis, simple amino acid sequence comparison also indicated that the Mill family was related most closely to the MIC family. Of all the molecules shown in Fig. 2 except MILL2, the α1–α3 domains of MILL1 showed the highest amino acid sequence identity to those of MICB (36%), followed by those of MICA (33%). Likewise, the α1–α3 domains of MILL2 were most similar to those of MICA/MICB (35% amino acid identity). The next most similar class I-like molecules to MILL1/MILL2 were HFE (31–32%), which was followed by H2LS/HLALS (30–31%), MHC-encoded class I molecules other than MICA/MICB (25–30%), FcRn (24–27%), zinc-α2-glycoproteins (26%), and CD1 (15–23%). Class I-like molecules with two extracellular domains (ULBP, RAET1, and endothelial protein C receptors) showed amino acid sequence identities of 14–27% to the corresponding domains of MILL1/MILL2.
Structures of the Mill Genes.
The high-throughput sequence deposited in the GenBank database (accession no. AC079493.1) contained the entire Mill1 gene. Comparison of this genomic sequence, our cDNA sequence, and the ESTs enabled us to decipher the locations of all exon–intron boundaries in the Mill1 gene (Fig. 3A). Similar comparison with the genomic sequence in the MGSC version 3 assembly enabled us to estimate the size of all introns. For Mill2, we deduced its exon–intron organization by comparing our cDNA and the EST sequences with the genomic sequence in the MGSC version 3 assembly. We confirmed the accuracy of the maps shown in Fig. 3A by inspecting the Celera database and also by determining all exon–intron boundaries experimentally with PCR (data not shown).
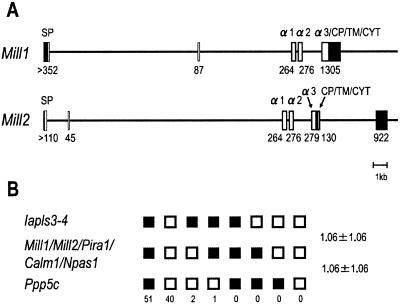
Figure 3.
(A) Exon–intron organization of mouse Mill1 and Mill2. Coding regions are shown as open boxes. Filled boxes indicate 5′- and 3′-untranslated regions. The nucleotide sequences surrounding the exon–intron boundaries conformed to the GT/AG rule without any exception. In all boundaries, splicing occurs between the first and second bases of the codon. Exon size in bp (shown below each box) was deduced on the basis of our cDNA sequences as well as the ESTs. SP, signal peptide. (B) Segregation patterns of Mill1, Mill2, and neighboring loci in [(C57BL/6Ei × M. spretus)F1 × M. spretus] interspecific backcross mice. Each column represents the chromosome identified in the backcross progeny that was inherited from the (C57BL/6Ei × M. spretus)F1 parent. Black boxes indicate the inheritance of the C57BL/6Ei allele, and white boxes indicate the inheritance of the M. spretus allele. The number of offspring inheriting each type of chromosome is shown at the bottom of each column. Recombination frequencies between pairs of loci (percentage recombination with standard errors) are given on the right. Gene symbols not explained in the text: Iapls3-4, intracisternal A particle, lymphocyte specific 3-4; Calm1, calmodulin 1; Npas1, neuronal PAS domain protein 1; and Ppp5c, protein phosphatase 5, catalytic subunit.
The structures of Mill1 and Mill2 differ from those of all other known members of the class I gene family in that they have an exon between the exons coding for the signal peptide and the α1 domain (Fig. 3A). This extra exon accounts for the MILL-specific insertion of amino acids between the signal peptide and the α1 domain (Fig. 1). Interestingly, the organization of Mill1 differs from that of Mill2 at its 3′ end. Although Mill2 has an intron at the end of the α3 domain as seen in most other members of the class I gene family, Mill1 has no such intron. Instead, the 3′-untranslated region and the α3 domain of Mill1 are encoded by a single exon.
Mill1 and Mill2 Are Located in the Vicinity of the LRC.
We mapped Mill1 and Mill2 by using progeny derived from the interspecific backcross. Analysis of 94 progeny showed that both Mill1 and Mill2 map to the paracentromeric region of chromosome 7 (Fig. 3B). No recombination was observed between Mill1 and Mill2. Furthermore, there was no recombination between Mill1/Mill2 and Pira1 (paired Ig-like receptor A1), one of the genes located in the LRC, indicating that they lie within 3.8 centimorgans at the 95% confidence level. Inspection of the MGSC version 3 assembly indicates that Mill1 and Mill2 are ≈500 kb apart on chromosome 7, consistent with our mapping results.
Mill1 and Mill2 Have at Least Two Alleles in Laboratory Mice.
Genomic fragments containing the α1 domain, intron 3, and the α2 domain of Mill1 and Mill2 were cloned by PCR from 14 laboratory mouse strains, and their sequences were determined (Table 1). Mill1 had two alleles distinguished by two conservative amino acid substitutions. Mill2 was also diallelic. The two alleles of Mill2 differed by five amino acids, of which the three substitutions in the α1 domain were nonconservative. P/J and DBA/2J carried the Mill1 allele of the 129 X 1/SvJ type and the Mill2 allele of the AKR/J type, suggesting that these strains carry a recombinant Mill haplotype.
Expression Patterns of Mill1 and Mill2 Are Markedly Different.
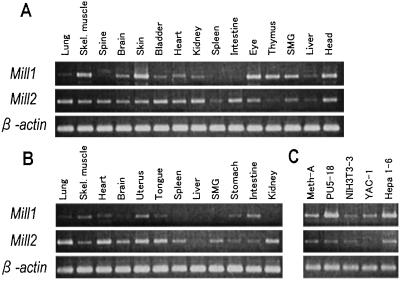
Expression patterns of Mill1 and Mill2 were examined by RT-PCR (Fig. 4). Mill1 was transcribed abundantly in the skin and head of neonatal mice, consistent with the existence of the ESTs derived from these tissues. Other neonatal tissues in which Mill1 transcripts were abundant included skeletal muscle, eyes, thymus, and submandibular glands. Interestingly, Mill1 was expressed less abundantly in most adult organs. In contrast, Mill2 was expressed more ubiquitously in both adult and neonatal organs. Mill1 and Mill2 transcripts were detectable in almost all cell lines. Mill1 was transcribed abundantly in PU5-18 and Hepa 1-6.
Figure 4.
Expression profiles of mouse Mill1 and Mill2. Expression patterns were analyzed by RT-PCR using a panel of cDNA prepared from various organs of 1-day-old neonatal mice (A), those of adult mice (B), or cell lines (C). Primers for Mill1 and Mill2 were designed in such a way that they span at least one intron so as to avoid the amplification of contaminated genomic DNA. Meth-A, methylcholanthrene-induced sarcoma; PU5-18, lymphoid tumor; NIH 3T3-3, fibroblast-like; YAC-1, lymphoma; and Hepa 1-6, hepatoma. SMG, submandibular glands.
Mill Orthologs Are Most Likely Absent in Human.
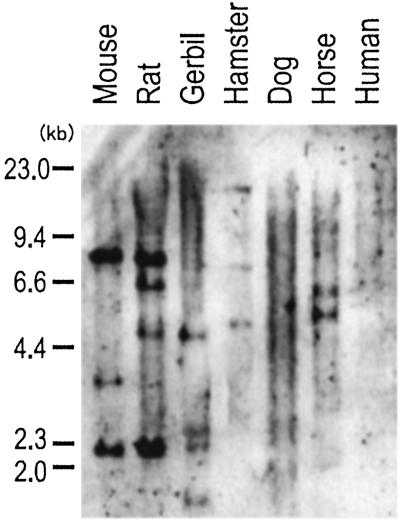
We performed Zoo-blot analysis by using the Mill1 cDNA spanning the α1–α3 domains as a probe (Fig. 5). Because the α2 and α3 domain sequences of MILL1 and MILL2 are sufficiently similar, this probe cross-hybridizes to Mill2 under reduced stringency conditions. In rodents such as rats, gerbils, and Syrian hamsters, we detected hybridizing bands. Bands were detected also in horse but not in human or dog. Consistent with this result, a computer search of the GenBank and Celera databases failed to identify human class I-like sequences corresponding to Mill1 or Mill2. Thus, Mill orthologs are most likely absent in the human. In C57BL/6J mice, the probe detected three BamHI fragments of 8.2, 3.6, and 2.2 kb. Inspection of the Celera database indicates that the 8.2-kb band is derived from Mill1 and the remaining two bands from Mill2. Thus, the mouse Mill gene family is unlikely to have members other than Mill1 and Mill2.
Figure 5.
Zoo-blot analysis of the Mill genes. Mouse and rat DNA were from C57BL/6J and WKA, respectively.
Discussion
In the present study, we described the identification and characterization of two class I-like genes located in the vicinity of the LRC on mouse chromosome 7. Although the class I-like IgG Fc receptor gene, Fcgrt, is on the same chromosome (11), it is located in a more telomeric region and clearly is a distinct entity. Indeed, MILL1 and MILL2 are related only distantly to any known members of the class I family (Figs. 1 and 2), supporting our proposal that Mill1 and Mill2 are the members of a previously uncharacterized family of MHC class I-like genes.
Among the known members of the class I family, MILL molecules seem related most closely to the MIC family encoded in the HLA complex (Fig. 2). This observation might give us a hint as to the function of MILL. MICA/MICB are the stress-inducible class I-like molecules that serve as ligands to an activating natural killer (NK) receptor NKG2D (32–34). Engagement of NKG2D by MICA stimulates NK cells and some γ/δ T cells and costimulates CD8+ α/β T cells (34–36). Interestingly, the mouse MHC does not contain class I-like genes thought to be orthologous to MICA or MICB (37). Indeed, among many members of the class I gene family, MICA and MICB are the only genes the rodent counterparts of which have not been identified (Fig. 2). Conversely, Mill1 and Mill2 appear to lack the human counterparts (Fig. 5) and are the only genes the human counterparts of which have not been identified. Hence, MILL might be the elusive rodent counterpart of human MIC. The observation that, similar to MIC, Mill1 has a restricted tissue distribution (Fig. 4) is consistent with this hypothesis. However, there also are observations that are in apparent conflict with the hypothesis or suggest functional differences between MIC and MILL. First, the promoter regions of MICA and MICB contain a heat-shock element, and the transcription of these genes is induced with heat shock (33). Inspection of the MGSC version 3 assembly and the Celera database indicates that the promoter region of neither Mill1 nor Mill2 contains a sequence that perfectly matches a consensus heat-shock element. Consistent with this, transcription of neither Mill1 nor Mill2 was induced appreciably by heat-shock treatment in the five cell lines in Fig. 4 (M.S. and M.K., unpublished observations). Second, MICA and MICB have many alleles (38), whereas we identified only two alleles each for Mill1 and Mill2 in a limited survey of laboratory mice (Table 1). Third, previous Zoo-blot experiments suggested that most mammalian species other than mice have MIC-like genes (32). Our Zoo-blot experiments indicate that some nonrodent species such as horse presumably have Mill-like genes (Fig. 5), suggesting that both gene families might coexist in some species. Clearly, the idea that MILL might perform a function equivalent or similar to that of MIC is only one of many plausible hypotheses at the present time. Biochemical identification of interacting molecules, if any, may provide an insight into the function of the MILL molecules.
The human genome contains two major clusters of NK receptor genes: the NK complex of lectin-like genes on 12p13.1 and the LRC of Ig-like genes on 19q13.4 (4). In the mouse, the NK complex is on chromosome 6, and the LRC is on the paracentromeric region of chromosome 7. Of particular interest is the proposal that the regions containing the two major clusters of NK receptor genes are paralogous to the MHC and that they may have originated from a single ancestral region (4, 39, 40). The identification of class I-like genes tightly linked to the LRC seems consistent with this proposal.
Supplementary Material
Acknowledgments
We thank Drs. Katsumi Maenaka, Martin F. Flajnik, Tatsuya Ota, and the members of our laboratory for discussion. This work was supported in part by Grants-in-Aid for Scientific Research from the Ministry of Education, Culture, Sports, Science, and Technology of Japan and by the Joint Research Project (Soken/K01-4) of Sokendai.
Abbreviations
- LRC
leukocyte receptor complex
- MGSC
Mouse Genome Sequencing Consortium
- NK
natural killer
Footnotes
References
- 1.Flajnik M F, Kasahara M. Immunity. 2001;15:351–362. doi: 10.1016/s1074-7613(01)00198-4. [DOI] [PubMed] [Google Scholar]
- 2.Rock K L, Goldberg A L. Annu Rev Immunol. 1999;17:739–779. doi: 10.1146/annurev.immunol.17.1.739. [DOI] [PubMed] [Google Scholar]
- 3.Cresswell P. Annu Rev Immunol. 1994;12:259–293. doi: 10.1146/annurev.iy.12.040194.001355. [DOI] [PubMed] [Google Scholar]
- 4.Trowsdale J. Immunity. 2001;15:363–374. doi: 10.1016/s1074-7613(01)00197-2. [DOI] [PubMed] [Google Scholar]
- 5.Klein J. Natural History of the Major Histocompatibility Complex. New York: Wiley; 1986. [Google Scholar]
- 6.Calabi F, Milstein C. Nature. 1986;323:540–543. doi: 10.1038/323540a0. [DOI] [PubMed] [Google Scholar]
- 7.Balk S P, Bleicher P A, Terhorst C. J Immunol. 1991;146:768–774. [PubMed] [Google Scholar]
- 8.Araki T, Gejyo F, Takagaki K, Haupt H, Schwick H G, Bürgi W, Marti T, Schaller J, Rickli E, Brossmer R, et al. Proc Natl Acad Sci USA. 1988;85:679–683. doi: 10.1073/pnas.85.3.679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ueyama H, Naitoh H, Ohkubo I. J Biochem (Tokyo) 1994;116:677–681. doi: 10.1093/oxfordjournals.jbchem.a124579. [DOI] [PubMed] [Google Scholar]
- 10.Simister N E, Mostov K E. Nature. 1989;337:184–187. doi: 10.1038/337184a0. [DOI] [PubMed] [Google Scholar]
- 11.Ahouse J J, Hagerman C L, Mittal P, Gilbert D J, Copeland N G, Jenkins N A, Simister N E. J Immunol. 1993;151:6076–6088. [PubMed] [Google Scholar]
- 12.Fukudome K, Esmon C T. J Biol Chem. 1995;270:5571–5577. doi: 10.1074/jbc.270.10.5571. [DOI] [PubMed] [Google Scholar]
- 13.Hashimoto K, Hirai M, Kurosawa Y. Science. 1995;269:693–695. doi: 10.1126/science.7624800. [DOI] [PubMed] [Google Scholar]
- 14.Yamaguchi H, Hirai M, Kurosawa Y, Hashimoto K. Biochem Biophys Res Commun. 1997;238:697–702. doi: 10.1006/bbrc.1997.7379. [DOI] [PubMed] [Google Scholar]
- 15.Feder J N, Gnirke A, Thomas W, Tsuchihashi Z, Ruddy D A, Basava A, Dormishian F, Domingo R, Jr, Ellis M C, Fullan A, et al. Nat Genet. 1996;13:399–408. doi: 10.1038/ng0896-399. [DOI] [PubMed] [Google Scholar]
- 16.Hashimoto K, Hirai M, Kurosawa Y. Biochem Biophys Res Commun. 1997;230:35–39. doi: 10.1006/bbrc.1996.5889. [DOI] [PubMed] [Google Scholar]
- 17.Zou Z, Nomura M, Takihara Y, Yasunaga T, Shimada K. J Biochem (Tokyo) 1996;119:319–328. doi: 10.1093/oxfordjournals.jbchem.a021242. [DOI] [PubMed] [Google Scholar]
- 18.Cerwenka A, Bakker A B, McClanahan T, Wagner J, Wu J, Phillips J H, Lanier L L. Immunity. 2000;12:721–727. doi: 10.1016/s1074-7613(00)80222-8. [DOI] [PubMed] [Google Scholar]
- 19.Malarkannan S, Shih P P, Eden P A, Horng T, Zuberi A R, Christianson G, Roopenian D, Shastri N. J Immunol. 1998;161:3501–3509. [PubMed] [Google Scholar]
- 20.Kasahara M, Vazquez M, Sato K, McKinney E C, Flajnik M F. Proc Natl Acad Sci USA. 1992;89:6688–6692. doi: 10.1073/pnas.89.15.6688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Thompson J D, Higgins D G, Gibson T J. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kumar S, Tamura K, Jakobsen I B, Nei M. Bioinformatics. 2001;17:1244–1245. doi: 10.1093/bioinformatics/17.12.1244. [DOI] [PubMed] [Google Scholar]
- 23.Schmidt H A, Strimmer K, Vingron M, von Haeseler A. Bioinformatics. 2002;18:502–504. doi: 10.1093/bioinformatics/18.3.502. [DOI] [PubMed] [Google Scholar]
- 24.Jones D T, Taylor W R, Thornton J M. Comput Appl Biosci. 1992;8:275–282. doi: 10.1093/bioinformatics/8.3.275. [DOI] [PubMed] [Google Scholar]
- 25.Kasahara M, Hayashi M, Tanaka K, Inoko H, Sugaya K, Ikemura T, Ishibashi T. Proc Natl Acad Sci USA. 1996;93:9096–9101. doi: 10.1073/pnas.93.17.9096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yawata M, Murata S, Tanaka K, Ishigatsubo Y, Kasahara M. Immunogenetics. 2001;53:119–129. doi: 10.1007/s002510100308. [DOI] [PubMed] [Google Scholar]
- 27.Kandil E, Noguchi M, Ishibashi T, Kasahara M. J Immunol. 1995;154:5907–5918. [PubMed] [Google Scholar]
- 28.Schultz J, Copley R R, Doerks T, Ponting C P, Bork P. Nucleic Acids Res. 2000;28:231–234. doi: 10.1093/nar/28.1.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Madden D R, Gorga J C, Strominger J L, Wiley D C. Cell. 1992;70:1035–1048. doi: 10.1016/0092-8674(92)90252-8. [DOI] [PubMed] [Google Scholar]
- 30.Salter R D, Benjamin R J, Wesley P K, Buxton S E, Garrett T P, Clayberger C, Krensky A M, Norment A M, Littman D R, Parham P. Nature. 1990;345:41–46. doi: 10.1038/345041a0. [DOI] [PubMed] [Google Scholar]
- 31.Saper M A, Bjorkman P J, Wiley D C. J Mol Biol. 1991;219:277–319. doi: 10.1016/0022-2836(91)90567-p. [DOI] [PubMed] [Google Scholar]
- 32.Bahram S, Bresnahan M, Geraghty D E, Spies T. Proc Natl Acad Sci USA. 1994;91:6259–6263. doi: 10.1073/pnas.91.14.6259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Groh V, Bahram S, Bauer S, Herman A, Beauchamp M, Spies T. Proc Natl Acad Sci USA. 1996;93:12445–12450. doi: 10.1073/pnas.93.22.12445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bauer S, Groh V, Wu J, Steinle A, Phillips J H, Lanier L L, Spies T. Science. 1999;285:727–729. [PubMed] [Google Scholar]
- 35.Groh V, Steinle A, Bauer S, Spies T. Science. 1998;279:1737–1740. doi: 10.1126/science.279.5357.1737. [DOI] [PubMed] [Google Scholar]
- 36.Groh V, Rhinehart R, Randolph-Habecker J, Topp M S, Riddell S R, Spies T. Nat Immunol. 2001;2:255–260. doi: 10.1038/85321. [DOI] [PubMed] [Google Scholar]
- 37.Amadou C, Kumánovics A, Jones E P, Lambracht-Washington D, Yoshino M, Fischer Lindahl K. Immunol Rev. 1999;167:211–221. doi: 10.1111/j.1600-065x.1999.tb01394.x. [DOI] [PubMed] [Google Scholar]
- 38.Bahram S. Adv Immunol. 2000;76:1–60. doi: 10.1016/s0065-2776(01)76018-x. [DOI] [PubMed] [Google Scholar]
- 39.Kasahara M. Immunol Rev. 1999;167:17–32. doi: 10.1111/j.1600-065x.1999.tb01379.x. [DOI] [PubMed] [Google Scholar]
- 40.Du Pasquier L. Curr Top Microbiol Immunol. 2000;248:159–185. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.