Abstract
The identification of small molecules that fall within the biologically relevant subfraction of vast chemical space is of utmost importance to chemical biology and medicinal chemistry research. The prerequirement of biological relevance to be met by such molecules is fulfilled by natural product-derived compound collections. We report a structural classification of natural products (SCONP) as organizing principle for charting the known chemical space explored by nature. SCONP arranges the scaffolds of the natural products in a tree-like fashion and provides a viable analysis- and hypothesis-generating tool for the design of natural product-derived compound collections. The validity of the approach is demonstrated in the development of a previously undescribed class of selective and potent inhibitors of 11β-hydroxysteroid dehydrogenase type 1 with activity in cells guided by SCONP and protein structure similarity clustering. 11β-hydroxysteroid dehydrogenase type 1 is a target in the development of new therapies for the treatment of diabetes, the metabolic syndrome, and obesity.
Keywords: chemical biology, compound libraries, hydroxysteroid dehydrogenase, cheminformatics
The efficient identification of small molecules that modulate protein function in vitro and in vivo is at the heart of chemical biology and medicinal chemistry research and the development of new therapies and diagnostics for disease. Key to their discovery is the identification and charting of biologically relevant space, i.e., the regions of complete chemical space that are relevant to biology (1–5). The underlying structures of evolutionary selected natural products (NPs) define structural prerequisites for binding to proteins (4, 6). Their structural scaffolds represent the biologically relevant and prevalidated fractions of chemical structure space explored by nature so far. Consequently, the probability that compound libraries designed to mimic the structures and properties of NP classes will be biologically relevant is high, and it is also to be expected that “NP-guided compound library development” (1, 4) will prove to be a viable guiding principle for the identification of small molecules for chemical biology and medicinal chemistry research (1–6).
A systematic structure-orientated organizing principle of the known NPs combined with annotations of biological origin and pharmacological activity would chart the regions of chemical space explored by nature, provide a structural rationalization and categorization of NP diversity, and also provide guidance for the development of NP-like compound libraries.
Statistical analyses of different NP databases have been performed in a few cases (7–10); however, a systematic and annotated structural categorization of NPs leading to development principles for compound library design is missing.
Here, we introduce a structural classification of NPs (SCONP) as a idea- and hypothesis-generating tool to define structural relationships between different NP classes in a tree-like arrangement and for the design of NP-derived compound libraries.
Materials and Methods
Cheminformatics. The CRC Dictionary of Natural Products (DNP) (11), which lists 190,939 records, was used as the basis for the analysis of NP structure. The molecular structures of the MDL structure data file (SD file) version of the DNP were standardized and subjected to in silico deglycosylation based on substructural patterns. Subsequently, all terminal side chains were pruned to obtain the scaffold. The scaffolds obtained in this way were grouped hierarchically by establishing parent–child relationships between the scaffolds whereby the parent scaffold represents a substructure of the child scaffold. In case of several possible parent scaffolds, the prioritization rules given in Supporting Materials and Methods, which is published as supporting information on the PNAS web site, were applied. The parent–child relationships then were assembled to the classification tree. These operations were performed by using java (Sun Microsystems, Mountain View, CA) routines written in house and based on the molinspiration toolkit (Molinspiration Cheminformatics, Bratislava, Slovak Republic, www.molinspiration.com). A representation of the tree is available from H.W. upon request.
From the biological source field (BSRC) field in the DNP, the genus of the source organism was extracted, and its taxonomic classification was identified in the Integrated Taxonomic Information System (ITIS) database (www.itis.usda.gov). For species not listed in ITIS, information was amended by using the National Center for Biotechnology Information taxonomy browser (www.ncbi.nlm.nih.gov/Taxonomy/taxonomyhome.html) and joined to the ITIS taxonomic tree at the lowest possible taxonomic level. The canonical Simplified Molecular Input Line Entry Specification (SMILES) (12) without stereochemistry of the structures in the DNP and the mdl Drug Data Report (MDDR) database (www.mdl.com) were matched against each other to join biological activity information contained in the MDDR to the DNP data set. Both database-joining operations were performed by using pipelinepilot software (www.scitegic.com).
Inhibition of 11β-Hydroxysteroid Dehydrogenases (11βHSDs). The 11βHSD type 1 (11βHSD1)-dependent oxoreduction of cortisone and 11βHSD type 2 (11βHSD2)-dependent oxidation of cortisol were measured in lysates of stably transfected HEK-293 cells as described in ref. 13. The rate of conversion of cortisone to cortisol or the reverse reaction was determined by using [1,2,6,7-3H]-labeled substrate at a final concentration of 200 nM cortisone or 25 nM cortisol, respectively, in the presence of inhibitor (0–200 μM). Data represent mean ± SD of at least four independent experiments. To exclude the possibility that the observed reduction in 11βHSD1 activity is due to promiscuous inhibition of the enzyme, the inhibition of the oxoreduction of cortisone by 3, 4, and 5 was measured in the presence of 0.1% Triton X-100 (14). The calculated IC50 values were comparable with those obtained in the absence of the detergent.
Nuclear Translocation Assay. HEK-293 cells (300,000 cells per well) were grown on poly(l-lysine)-coated glass slides in six-well plates containing 2 ml of DMEM supplemented with 10% FCS. Cells were transfected according to the calcium phosphate precipitation method with 1 μg of GFP-human glucocorticoid receptor (GR) expression plasmid (15) and either 0.5 μg of human 11βHSD1 plasmid or empty pcDNA3 vector (Invitrogen) per well. After 6 h, cells were washed twice with medium that was charcoal-stripped twice to remove steroids, followed by incubation in this medium for another 18 h. Cells then were preincubated for 30 min with inhibitor or GR antagonist as indicated, followed by the addition of 500 nM cortisone and further incubation for 40 min. Cells were washed and fixed with 4% paraformaldehyde for 10 min, and localization of GFP-GR was determined by fluorescence microscopy. Three independent transfection experiments were carried out whereby 300 fluorescent cells were determined per sample by an observer who was blinded to the cell treatment.
For further descriptions of cheminformatics methods, a summary of the library synthesis, and the transactivation assay, see Supporting Materials and Methods and Scheme 1, which is published as supporting information on the PNAS web site.
Results
The DNP, which lists data on 190,939 NPs, was used as the basis for the analysis of NP structure. For primary molecular processing of the database data were converted from mdl structure data file molecular format to Daylight SMILES (Simplified Molecular Input Line Entry Specification) (12) strings. In this process, also records without structural data and records with errors were removed. Further standardization was performed by normalizing charges and removing counterions and smaller parts (e.g., water, salts associated with compound structures). Stereochemistry could not be considered in the course of this basic cheminformatics analysis because for many NPs the absolute and relative configuration has not been determined. Instead, the different possible configurations of the NP scaffolds had to be treated as being equivalent.
However, in general, a possible subsequent synthesis effort planned on the basis of our analysis (see below), must take this self-limitation into account and possibly compensate it by the synthesis of different stereoisomers with the same underlying structural scaffold. The biological activity of NPs and compounds derived thereof is certainly determined to a major extent by their chirality. For a useful structure-based organizing principle of NPs, however, abstraction of structural information by focusing on 2D structures seemed to be acceptable. This finding is supported by several previous studies that showed that cheminformatics analyses employing 3D molecular descriptors do not perform better than analyses based on 2D molecular structures (16–18).
Duplicates were not eliminated from the data set because these may represent stereoisomers or may be differently annotated (e.g., biological origin or function). The resulting set of 171,045 structures was processed. We found that 154,428 of the structures contain rings (90%). Because the overwhelming majority of the compounds used in medicinal chemistry and chemical biology research also contains rings (19), subsequent analysis focused on the ring-containing NPs.
Cheminformatics analysis of the ring-containing NPs started with the removal of acyclic substituents. Cyclic substituents were regarded as being part of the scaffold. We extracted 31,011 unique scaffolds [i.e., frameworks as unions of ring systems (20) including also exocyclic double bonds and possible linking chains between rings] from the ring-containing NPs. After careful visual inspection of this initial result, it was found that redundancies occurred because of different glycosylation patterns of a given aglycon (scaffold) (for an example, see Fig. 4, which is published as supporting information on the PNAS web site).
Glycosidic moieties, unless they contain complex sugars (for example, in glycosidic antibiotics, such as aminoglycosides, vancomycin, and others), usually exert a modulating effect on the aglycon's biological activity, e.g., modulation of solubility or release from an inactive precursor by a glycosidase (21–23). Therefore, we introduced an additional step of in silico deglycosylation of standard O-glycosides before extracting scaffolds. The analysis was performed with a special computational algorithm written in java (Sun Microsystems) by using recursive substructure matching. Altogether 25,337 NP molecules were deglycosylated (14.8% of the whole NP data set) by removing 1–12 sugar moieties from the parent glycosides, yielding at the end 149,513 NP structures with rings, which in turn provided 24,891 unique deglycosylated scaffolds (reduction of the number of distinct scaffolds by 20%). In an initial attempt to develop a structural organizing principle for NPs, cluster analysis was performed on the NP structures. Jarvis–Patrick clustering (24) of the NPs' molecular fingerprints using the Tanimoto coefficient (25) as quantitative similarity measure (Tanimoto index ≥ 0.9) led to a grouping of NPs into 42 large clusters containing ≥100 NP molecules, 1,507 medium clusters containing ≥10 NP molecules, 2,851 small clusters (containing ≥5 NP molecules), and ≈50,000 singletons. Thus, this clustering procedure did not provide a useful principle of structural classification.
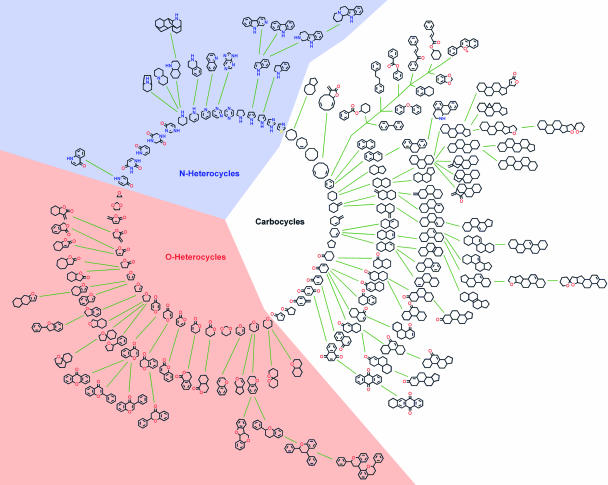
For an alternative hierarchical tree-like classification of NP scaffolds, a computational algorithm was developed that identifies the parent scaffold of each individual scaffold. It searches for substructures in each scaffold that represent NP scaffolds and groups them hierarchically according to decreasing number of rings, i.e., scaffold size. Thus, a parent scaffold represents a substructure of a respective query scaffold. This hierarchical analysis generated a scaffold tree with several hierarchical levels and with single rings being the roots that were grouped into carbocycles, N- and O-heterocycles. Each scaffold on a given hierarchical level can be annotated with several individual NPs and represents a node from which further arborization may lead to more complex scaffolds. For the identification of “parent scaffolds,” a set of rules was used that provides results close to the way of thinking of synthetic chemists. First, the parent had to be a substructure of the child scaffold. Second, no breaking of ring bonds in a child was allowed. In case several candidates were available, the parent was selected such as to get the maximum number of heteroatoms in the parent and then by choosing the parent of maximum size. If it was not possible to make a decision based on these rules simply the most frequent scaffold was chosen as parent scaffold (see Supporting Materials and Methods for a detailed description of the algorithm).
Such a modular tree-like arrangement of NPs (see Fig. 1) allows for the representation of very rare scaffolds representing only a few or only one NP as a branch in the tree and thereby for their highly dynamic and readily extendable structural classification and correlation with other scaffolds. Regular clustering approaches would define them as singletons, and structural similarities with other NPs would not be detected.
Fig. 1.
Graphical star-like representation of the NP scaffold tree. For clarity of the graphic illustration, only scaffolds are shown that represent cumulatively at least 0.2% of the NP population in the DNP.
Quantitative analysis of the NP scaffold tree revealed that scaffolds with three rings are most often found in NPs (see Fig. 5, which is published as supporting information on the PNAS web site) with scaffolds incorporating two or four rings lying within the range of 1 SD of the calculation. We calculated the van der Waals volumes of the deglycosylated NPs by using a java program (Novartis, Basel) from 3D structures generated by the program corina 3.1 (Molecular Networks, Erlangen, Germany) from molecule SMILES (Simplified Molecular Input Line Entry Specification). We determined that the volumes of the majority of the NPs containing two to four rings range from 100 to 500 Å3 with the maximum at ≈250 Å3 (see Fig. 6A, which is published as supporting information on the PNAS web site). A statistical evaluation of the volumes found in a data sample of 18,402 cavities extracted from the Protein Data Bank (www.pdb.org) revealed that most cavities fall into a volume range between 300 and 800 Å3 (26). Thus, the average volume of the two- to four-ring NPs correlates with and can be mapped to the average dimensions of protein cavities, taking into account that ligands of proteins often do not fill the whole volume of a given cavity. A similar analysis of the volume distribution of ≈30,000 drugs from the World Drug Index (Thomson Derwent, Philadelphia, www.thomsonderwent.com) revealed furthermore that the volumes of the two- to four-ring -containing NPs are also comparable with those found in drugs (see Fig. 6B).
The NP scaffold tree can be used as a strategic and guiding tool for the selection of underlying frameworks for NP-inspired compound library development in different ways. The most logical and apparent approach is to select the scaffold of a given NP and close structural neighbors to guide the synthesis of compound libraries. These NP-derived compound collections should yield relatively high hit rates at comparably small library size. Such an approach should be of general value and applicability, and current experience with several NP-derived compound collections prepared by others and by us confirms this expectation (refs. 27–31; see also references within ref. 29 for recent reviews of NP guided compound library synthesis).
In addition to the most logical use of the NP scaffold tree, we envisioned that for compound library development possible structurally simplified analogs with prevailing biological relevance (not necessarily identical activity) might be found by brachiation through the branches of the tree. Structural simplification would then be sought by identification of scaffolds according to the classification principle used for the construction of the tree, i.e., core structures of less complex NPs, which form a substructure of the scaffold characteristic for the query structure. Notably, these scaffolds still encode for the property to bind to proteins because they occur in NPs. We also note that this brachiation does not imply an evolutionary or biosynthetic relationship between the scaffolds involved; rather, the argument is exclusively based on structural, i.e., chemical relationships. We also stress that if structural simplification extends too far, most probably the guiding biological activity may be lost.
Because on each level of lower complexity several scaffolds may be identified and because a priori it is not obvious on which level the analysis should halt (i.e., down to which level the desired biological activity will remain preserved), for the final choice of the scaffold of the compound library to be synthesized, a second criterion needs to be introduced.
Such criteria could, for instance, be biological activity of compound classes represented by individual scaffolds or biological origin. Such information can be encoded for the different hierarchical levels and scaffolds of the tree (for an annotation example with biological origin, see Fig. 7, which is published as supporting information on the PNAS web site). Biological activity might be a very relevant second criterion, but currently it can only be applied for a relatively small subfraction of the NPs analyzed. Comparison of the NPs in the DNP with the mdl Drug Data Report (MDDR) database (www.mdl.com), which lists biological activity of 153,366 compounds, revealed that only for 2,110 of the NPs listed in the DNP biological activity was reported in the MDDR. Furthermore, biological activity is very ill-defined, ranging from the description of precise targets to very general annotations, e.g., “cytotoxic.” It also is not general and may change frequently because it depends on the type and number of assays to which the NPs were and will be subjected.
The ability to bind to proteins is a hierarchically higher consideration than bioactivity because it also addresses the fact that NPs are biosynthesized by and therefore bind to proteins irrespective of activity in an assay. Thus, functional, mechanistic, evolutionary, and structural analogy between possible protein targets appear to be particularly suitable second criteria for selection of a structurally simpler scaffold for library development.
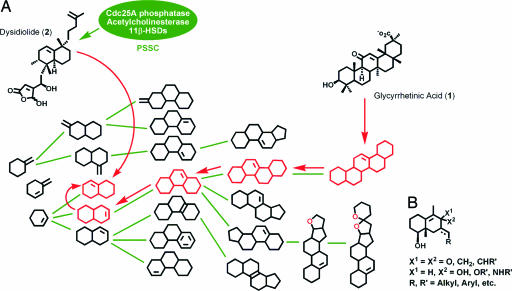
To investigate whether this approach may indeed be viable, we used the naturally occurring 11βHSD1 and 11βHSD2 ligand glycyrrhetinic acid (GA, 1; Fig. 2A) as query compound. 11βHSD1, which catalyzes the conversion of inactive cortisone into active cortisol, is a promising target for the development of new drugs (13, 32), and isoenzyme-selective 11βHSD1 inhibitors are very actively being sought.
Fig. 2.
Strategic use of the NP tree. (A) The frequency distribution pattern first guided brachiation from the complex pentacyclic starting scaffold of GA (1) in the direction of reduced complexity to two- to four-ring-containing NPs, which are the most abundant ones (see Fig. 5). A second independent criterion (PSSC) finally leads to the selection of the 1,2,3,4,4a,5,6,7-octahydronaphthalene scaffold as biologically relevant starting point for compound library generation. (B) General structure of the library synthesized.
Analysis of the pentacyclic core structure of the NP GA leads to an assignment of the NP scaffold to a branch within the NP tree (see Fig. 2 A). Brachiation in the direction of reduced complexity leads to a subset of two- and three-ring systems that were chosen because they occur most frequently (see above) and because of their accessibility by chemical synthesis.
For the choice of the precise scaffold of a compound library, we used as the second decisive criterion similarity between possible target proteins (see above). For this purpose a priori the established grouping of proteins into protein families according to evolutionary relationship and according to their function is certainly most appropriate and logical. As an alternative to these groupings, we have recently introduced protein structure similarity clustering (PSSC) (4) as an abstracting guiding principle for compound library development. PSSC focuses on sheer structural similarity in the ligand-sensing cores of proteins to group them into a structure similarity cluster. The structure of a known ligand of a cluster member protein, e.g., a NP, is used for the development of ligands for other cluster member proteins. Following this logic, we previously identified a protein structure similarity cluster (PSSC) containing the 11βHSDs, acetylcholinesterase, and Cdc25A phosphatase (4). Dysidiolide (2) is a known naturally occurring inhibitor of Cdc25A (33). This NP embodies the 1,2,3,4,4a,5,6,7-octahydronaphthalene scaffold, which also was identified as possible library scaffold by the brachiation approach (see Fig. 2 A). Thus, we investigated whether a compound library based on this scaffold would yield inhibitors of 11βHSD. 11βHSD inhibitors with this scaffold have not been described before.
The substitution pattern of the octahydronaphthalene system in dysidiolide and of the unsaturated C/D ring system in GA only partly match. Therefore, and to increase the diversity of the library, substitution patterns with different degrees of overlap between the GA and the Dysidiolide core structures were introduced into the basic core scaffold during library synthesis (see Fig. 2B for a general structural description of the library). The scaffolds themselves were synthesized employing the Robinson annulation as key step. For library synthesis on solid support, the scaffolds were equipped with a secondary alcohol for attachment to the polymeric carrier. Diversification of the basic scaffolds was achieved by means of aldol and Wittig reactions and Pd(0)-catalyzed coupling reactions (see Supporting Materials and Methods and Table 2, which is published as supporting information on the PNAS web site, for a description of the synthesis route and characterization of compounds 3–5).
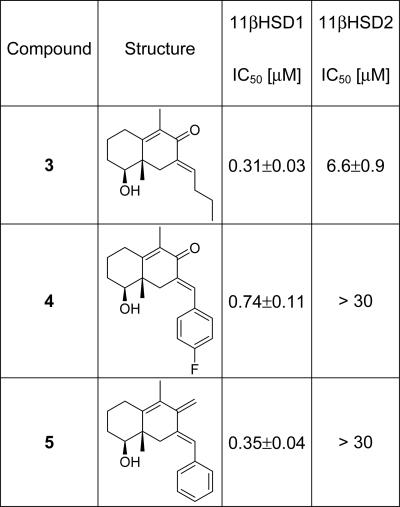
A collection of 162 compounds was synthesized and investigated biochemically for inhibition of 11βHSD1 and 11βHSD2 (see Supporting Materials and Methods) (13). Compounds displaying IC50 values of ≤10 μM were considered as hits.
The collection contained 30 11βHSD1 inhibitors with IC50 values of 0.31–9.1 μM. Four 11βHSD1 inhibitors were in the nanomolar range (IC50 values of 0.31–0.74 μM). Three compounds inhibited 11βHSD2 with IC50 values of 2.0–6.6 μM. The results for the most relevant compounds are given in Table 1. Most remarkably, even at this comparably small library size, the hits indicated a pronounced degree of selectivity for the isoenzymes 11βHSD1 and 11βHSD2. Twenty-eight compounds selectively inhibited 11βHSD1. To demonstrate cellular activity of the previously undescribed inhibitor class, translocation and transactivation assays (see Supporting Materials and Methods for the description of the assays) were performed for compound 5 (see Table 1), one of the most potent and selective 11βHSD1 inhibitors (IC50 = 0.35 μM).
Table 1.
Nanomolar inhibitors of 11βHSD1 identified by means of combined application of SCONP and PSSC
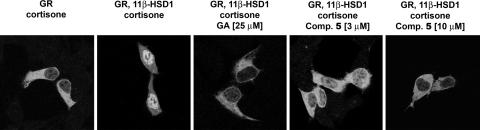
To this end, HEK-293 cells that do not express 11βHSDs (34) and in which the GR is present at only very low levels were transfected with a GFP-human GR expression plasmid (15, 35). In the absence of cortisol, GR is cytosolic. If the potent GR agonist cortisol is produced from externally added cortisone through reduction by 11βHSD1, it binds GR and induces the translocation of the receptor to the nucleus and stimulation of transactivation.
As shown in Fig. 3, upon addition of cortisone to cells expressing 11βHSD1, a dose-dependent induction of translocation of GR into the nucleus was observed, and also stimulation of transactivation was recorded. Glucocorticoid-dependent nuclear translocation of GR and transactivation were blocked in the presence of the unspecific 11βHSD inhibitor GA (1) (Fig. 3). Both nuclear translocation and GR-dependent transactivation were abolished upon coincubation of cells with cortisone and the selective 11βHSD1 inhibitor 5, indicating the capability of 5 to efficiently inhibit the conversion of cortisone to cortisol in intact cells. Less than 50% of GR molecules localized to the nucleus in the presence of 3 μM compound 5 (see Fig. 8, which is published as supporting information on the PNAS web site), and transactivation by GR diminished to ≈40% (see Fig. 9, which is published as supporting information on the PNAS web site). Nuclear translocation and transactivation were completely blocked at 10 μM 5.
Fig. 3.
Inhibition of 11βHSD1-dependent translocation of GR by compound 5. HEK-293 cells were transfected with GFP-GR alone or together with 11βHSD1. Cells were incubated 6 h posttransfection in steroid-free medium for 18 h. Where indicated, cells were preincubated for 30 min with 25 μMGA, 3 or 10 μM of inhibitor 5, followed by incubation for another 40 min with 500 nM cortisone. GFP-GR was detected by immunofluorescence microscopy. By converting inactive cortisone to cortisol, 11βHSD1 mediated nuclear translocation of GR. Inhibition of 11βHSD1 prevented the formation of cortisol and nuclear translocation of GR.
Discussion
We have developed a chemoinformatics approach to the SCONP according to their underlying scaffolds. The resulting NP scaffold tree should be regarded as a guiding and hypothesis-generating tool for charting the chemical space explored by nature in the context of NP-inspired compound library design and development for chemical biology and medicinal chemistry research.
Its most logical application for compound library development is the selection of library scaffolds based on relevance to nature and leading to compound collections that can be regarded as biologically prevalidated. Because NPs emerge by means of biosynthesis by proteins and often fulfill various biological functions through interaction with multiple proteins, the investigation of such compound collections in biochemical and biological screens should yield high hit rates at comparably small library size. Current experience with NP-inspired compound libraries synthesized and evaluated biochemically and biologically by others and by us supports this notion (see above), and the compound collection investigated here provides further proof-of-principle. Although the choice of NP-derived scaffolds currently is based on individual insight and knowledge about particular NP classes, the SCONP tree provides a systematization based on NP structure and allows for a statistically founded choice.
The systematic application of the NP scaffold tree for the establishment of a larger compound library from several smaller compound collections should allow for the assembly of an elaborate, yet still comparatively small, screening library that is diverse because it builds on the diversity found in nature and enriched in biologically relevant members because the scaffolds of the individual sublibraries are biologically prevalidated. Such a library as a whole also should yield comparably high hit rates in different biochemical and biological screens at comparably small size.
In addition to this most logical application of SCONP, a less obvious and probably less general, however, if successful, very valuable, application of the scaffold tree may be viable. Brachiation within the tree from an outer branch to structurally less complex scaffolds on an inner branch by means of intermediate naturally occurring parent scaffolds and leading to structurally simplified compound classes with similar biological activity was demonstrated to be possible for the pentacyclic unspecific 11βHSD inhibitor GA and the octahydronaphtalene scaffold identified in this way. If such a brachiation is performed, a second criterion is required for the choice of the smaller scaffold because typically on inner branches several scaffolds will be identified and because it is not obvious how far the brachiation can be allowed to proceed without loss of the desired biological activity. In the example pursued here, PSSC was chosen as second criterion, but other arguments like evolutionary and mechanistic relationships between possible target proteins and biological activity are very likely to be equally applicable or superior.
The successful experimental verification of this less-obvious approach demonstrates that brachiation may indeed be a viable procedure for the identification of structurally simpler compound classes with retained ability to bind to proteins and biological activity. However, we stress that generality cannot be claimed based on the example detailed above alone and that we do not intend to do so. Rather, reduction from larger to smaller scaffolds probably may be possible in only a limited number of cases and would depend on the complexity of the scaffold. Future investigations of further examples will be required to determine whether or not this far-going and more radical instrumentalization of the NP scaffold tree is generally applicable.
Supplementary Material
Acknowledgments
We thank Heidi Jamin for excellent technical support. This work was supported by the Max-Planck-Gesellschaft, the Deutsche Forschungsgemeinschaft, and the Fonds der Chemischen Industrie. M.A.K. is a scholar of the Studienstiftung des deutschen Volkes. A.O. is a fellow of the Cloëtta Research Foundation and was supported by the Swiss National Science Foundation and the Swiss Cancer League.
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: NP, natural product; SCONP, structural classification of NPs; DNP, Dictionary of Natural Products; PSSC, protein structure similarity clustering; 11βHSD, 11β-hydroxysteroid dehydrogenase; 11βHSD1, 11βHSD type 1; 11βHSD2, 11βHSD type 2; GR, glucocorticoid receptor; GA, glycyrrhetinic acid.
References
- 1.Breinbauer, R., Vetter, I. R. & Waldmann, H. (2002) Angew. Chem. Int. Ed. 41, 2879–2890. [DOI] [PubMed] [Google Scholar]
- 2.Koch, M. A., Breinbauer, R. & Waldmann, H. (2003) Biol. Chem. 384, 1265–1272. [DOI] [PubMed] [Google Scholar]
- 3.Koch, M. A. & Waldmann, H. (2004) in Chemogenomics in Drug Discovery: A Medicinal Chemistry Perspective, eds. Kubinyi, H. & Müller, G. (Wiley–VCH, Weinheim, Germany), pp. 377–403.
- 4.Koch, M. A., Wittenberg, L.-O., Basu, S., Jeyaraj, D. A., Gourzoulidou, E., Reinecke, K., Odermatt, A. & Waldmann, H. (2004) Proc. Natl. Acad. Sci. USA 101, 16721–16726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dobson, C. M. (2004) Nature 432, 824–828. [DOI] [PubMed] [Google Scholar]
- 6.Clardy, J. & Walsh, C. (2004) Nature 432, 829–837. [DOI] [PubMed] [Google Scholar]
- 7.Feher, M. & Schmidt, J. M. (2003) J. Chem. Inf. Comput. Sci. 43, 218–227. [DOI] [PubMed] [Google Scholar]
- 8.Henkel, T., Brunne, R. M., Muller, H. & Reichel, F. (1999) Angew. Chem. Int. Ed. 38, 643–647. [DOI] [PubMed] [Google Scholar]
- 9.Lee, M. L. & Schneider, G. (2001) J. Comb. Chem. 3, 284–289. [DOI] [PubMed] [Google Scholar]
- 10.Wessjohann, L. A., Ruijter, E., Garcia-Rivera, D. & Brandt, W. (2005) Mol. Diversity 9, 171–186. [DOI] [PubMed] [Google Scholar]
- 11.Dictionary of Natural Products (Chapman & Hall/CRC Informa, London), Version 14.1, 2005.
- 12.Weininger, D. (1988) J. Chem. Inf. Comput. Sci. 28, 31–36. [DOI] [PubMed] [Google Scholar]
- 13.Schweizer, R. A., Atanasov, A. G., Frey, B. M. & Odermatt, A. (2003) Mol. Cell. Endocrinol. 212, 41–49. [DOI] [PubMed] [Google Scholar]
- 14.McGovern, S. L., Helfand, B. T., Feng, B. & Shoichet, B. K. (2003) J. Med. Chem. 46, 4265–4272. [DOI] [PubMed] [Google Scholar]
- 15.Carey, K. L., Richards, S. A., Lounsbury, K. M. & Macara, I. G. (1996) J. Cell Biol. 133, 985–996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brown, R. D. & Martin, Y. C. (1996) J. Chem. Inf. Comput. Sci. 36, 572–584. [Google Scholar]
- 17.Matter, H. & Potter, T. (1999) J. Chem. Inf. Comput. Sci. 39, 1211–1225. [Google Scholar]
- 18.Sheridan, R. P. & Kearsley, S. K. (2002) Drug Discov. Today 7, 903–911. [DOI] [PubMed] [Google Scholar]
- 19.Fejzo, J., Lepre, C. A., Peng, J. W., Bemis, G. W., Ajay, Murcko, M. A. & Moore, J. M. (1999) Chem. Biol. 6, 755–769. [DOI] [PubMed] [Google Scholar]
- 20.Bemis, G. W. & Murcko, M. A. (1996) J. Med. Chem. 39, 2887–2893. [DOI] [PubMed] [Google Scholar]
- 21.Kren, V. & Martínková, L. (2001) Curr. Med. Chem. 8, 1303–1328. [DOI] [PubMed] [Google Scholar]
- 22.Gleadow, R. M. & Woodrow, I. E. (2002) J. Chem. Ecol. 28, 1301–1313. [DOI] [PubMed] [Google Scholar]
- 23.Wittstock, U., Agerbirk, N., Stauber, E. J., Olsen, C. E., Hippler, M., Mitchell-Olds, T. & Gershenzon, J. (2004) Proc. Natl. Acad. Sci. USA 101, 4859–4864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jarvis, R. A. & Patrick, E. A. (1973) IEEE Trans. Comput. C-22, 1025–1034. [Google Scholar]
- 25.Whittle, M., Gillet, V. J., Willett, P., Alex, A. & Loesel, J. (2004) J. Chem. Inf. Comput. Sci. 44, 1840–1848. [DOI] [PubMed] [Google Scholar]
- 26.Schmitt, S., Kuhn, D. & Klebe, G. (2002) J. Mol. Biol. 323, 387–406. [DOI] [PubMed] [Google Scholar]
- 27.Rosenbaum, C., Baumhof, P., Mazitschek, R., Muller, O., Giannis, A. & Waldmann, H. (2004) Angew. Chem. Int. Ed. 43, 224–228. [DOI] [PubMed] [Google Scholar]
- 28.Rosenbaum, C., Rohrs, S., Muller, O. & Waldmann, H. (2005) J. Med. Chem. 48, 1179–1187. [DOI] [PubMed] [Google Scholar]
- 29.Koch, M. A. & Waldmann, H. (2005) Drug Discov. Today 10, 471–483. [DOI] [PubMed] [Google Scholar]
- 30.Wu, X., Walker, J., Zhang, J., Ding, S. & Schultz, P. G. (2004) Chem. Biol. 11, 1229–1238. [DOI] [PubMed] [Google Scholar]
- 31.Nicolaou, K. C., Pfefferkorn, J. A., Schuler, F., Roecker, A. J., Cao, G. Q. & Casida, J. E. (2000) Chem. Biol. 7, 979–992. [DOI] [PubMed] [Google Scholar]
- 32.Sandeep, T. C., Yau, J. L., MacLullich, A. M., Noble, J., Deary, I. J., Walker, B. R. & Seckl, J. R. (2004) Proc. Natl. Acad. Sci. USA 101, 6734–6739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gunasekera, S. P., McCarthy, P. J., Kelly-Borges, M., Lobkovsky, E. & Clardy, J. (1996) J. Am. Chem. Soc. 118, 8759–8760. [Google Scholar]
- 34.Odermatt, A., Arnold, P., Stauffer, A., Frey, B. M. & Frey, F. J. (1999) J. Biol. Chem. 274, 28762–28770. [DOI] [PubMed] [Google Scholar]
- 35.Odermatt, A., Arnold, P. & Frey, F. J. (2001) J. Biol. Chem. 276, 28484–28492. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.