Abstract
Salmonella enterica relies on a type III secretion system encoded in Salmonella pathogenicity island-2 (SPI-2) to survive and replicate within macrophages at systemic sites during typhoid. SPI-2 virulence is induced upon entry into macrophages, but the mechanisms of SPI-2 gene control in vivo remain unclear, particularly with regard to negative regulators that control the contextual activation of SPI-2. Here, we identified and characterized YdgT as a negative modulator of the SPI-2 pathogenicity island and established that this negative regulation is central to systemic pathogenesis because ydgT mutants overexpressing typhoid virulence genes were ultimately attenuated during infection. ydgT mutants displayed a biphasic virulence phenotype during in vivo competitive infections that consisted of an early “gain-of-virulence” dependent on SPI-2 activation, followed by attenuation later in infection indicating that proper contextual regulation of SPI-2 by YdgT is necessary for full virulence during systemic colonization. These data suggest that overexpression of virulence-associated type III secretion genes can have an adverse effect on bacterial pathogenesis in vivo.
Keywords: in vivo, type III secretion, YdgT, pathogenesis
In Salmonella, two major pathogenicity islands encode type III secretion systems (T3SS) that translocate bacterial virulence proteins into host cells during infection. The T3SS encoded by Salmonella Pathogenicity Island-1 (SPI-1) is required for invasion of epithelial cells (1) and is activated under conditions thought to be present in the intestinal lumen before host cell invasion (2). Gene expression in SPI-1 is activated by HilA, a transcriptional regulator that shares homology in its DNA-binding domain to members of the OmpR/ToxR family (3). Several negative regulators of SPI-1 gene expression have been identified, including HilE (4), Hha (5), Lon protease (6), and others (7). Deletion of some of these genes increases the expression of hilA and confer a “hyperinvasive” phenotype for cultured epithelial cells (5).
SPI-2 (8, 9) encodes a second T3SS, effector proteins, molecular chaperones, and a two-component regulatory system (SsrA/SsrB) that activates SPI-2 promoters (10). This pathogenicity island and related effectors are required for intracellular survival and replication at systemic sites of infection. Other positive regulators of SPI-2-mediated virulence include EnvZ/OmpR (11, 12) and SlyA. SlyA is associated with SPI-2-dependent phenotypes, including survival and replication in macrophages and resistance to oxidative stress (13-15), likely because of its activity on ssrAB promoters (16) and SPI-2 effector promoters (17). The role of PhoP/PhoQ on SPI-2 gene expression has been contentious (10, 18-20), perhaps because of the difficulty in culturing phoP mutants under acidic minimal medium conditions required for SPI-2 activation and the lack of in vivo regulatory data. To date, no SPI-2 repressor has been identified, although the existence of such a modulator is implied by the specific environmental context required for SPI-2 activation.
Here, we establish that negative regulation of SPI-2 is required for full typhoid pathogenesis and identified a protein in Salmonella enterica called YdgT that exerts a negative regulatory activity on SPI-2. YdgT shares similarity to two small proteins, YmoA and Hha, that can negatively modulate bacterial virulence gene expression (21) and which are similar to the N-terminal oligomerization domain of the bacterial nucleoidassociated protein, H-NS (22). This report of a negative regulatory function for YdgT allows us to propose a model whereby Salmonella virulence during typhoid is fine-tuned through a horizontally acquired pathogenicity island-specific activator that integrates with an ancestral negative regulatory circuit.
Materials and Methods
Bacterial Strains and Mutant Construction. S. enterica serovar Typhimurium strain SL1344 was used as the wild-type strain and for mutant construction. An unmarked, in-frame deletion of ydgT was generated by allelic exchange from a counterselectable suicide vector. To generate marked strains for competitive index experiments, a chloramphenicol resistance cassette was integrated into the ushA gene in the Salmonella chromosome. Salmonella reporter strains were generated by introducing a single-copy chromosomal transcriptional fusion of the sseA promoter fused a promoterless lacZ gene as described (23). For a list of strains and plasmids used in this study and detailed methods for the generation of mutants, see Supporting Text and Table 1, which are published as supporting information on the PNAS web site.
Gentamicin Protection Assays. RAW264.7 murine macrophages and HeLa cells were infected with Salmonella as described (24). At the indicated times after infection, gentamicin-treated cells were washed with PBS and then lysed in 1% Triton X-100/0.1% SDS in PBS. Lysates were diluted in PBS and plated onto LB agar followed by incubation at 37°C. Colonies were enumerated and expressed as a percent of intracellular wild-type bacteria.
In Vitro Secretion Assays and Protein Analyses. Bacteria were grown in LB broth, washed in low phosphate, low magnesium-containing medium (LPM) previously shown to induce the expression of SPI-2 (23) and then inoculated into LPM. Cultures were grown at 37°C with shaking for 5 h. Bacteria were collected by centrifugation, and filtered supernatant was precipitated with trichloroacetic acid. Proteins from equivalent numbers of bacteria were analyzed by Western blot using affinity-purified antibodies recognizing SseB, SseD, and SseE (23), or DnaK (Stressgen Biotechnologies, Victoria, Canada) and secondary antibodies conjugated to horseradish peroxidase.
β-Galactosidase Assays. sseA promoter activity was examined by using transcriptional fusions to lacZ and a chemiluminescence assay described in ref. 23. Briefly, a single-copy fusion of lacZ to the sseA promoter (PsseA::lacZ) was constructed in Salmonella by homologous recombination. Reporter strains were inoculated into LPM medium to give A600 ≈ 0.05 and then incubated with shaking at 37°C. Samples were removed for enumeration of colony-forming units (cfu) and for β-galactosidase activity by using a Spectrafluor Plus Luminometer (Tecan, Zurich). Data were expressed as relative light units normalized to cfu. Each condition was performed in quadruplicate and averaged.
Experimental Animals, Single Infections, and Competitive Index Experiments. All protocols were in accordance with the University of British Columbia Animal Care Committee and the Canadian Council on the Use of Laboratory Animals. For single infections, female BALB/c mice (The Jackson Laboratory) were infected by i.p. injection with ≈500 cfu of Salmonella in PBS. For competitive infection experiments, mice were infected i.p. with a mixed inoculum containing a marked strain resistant to chloramphenicol and a second competing strain. At 24 and 55 h after infection, the spleen, liver, and mesenteric lymph nodes were homogenized (Polytron MR-21, Kinematic, Twain Harte, CA) in cold PBS, diluted, and plated on LB medium containing streptomycin for determination of total Salmonella cfu. Colonies were replica-plated under chloramphenicol selection for enumeration of SL1344 ushA::cat or ssrB::aphT ushA::cat. The competitive index (CI) was calculated as [mutant/wild type]output/[mutant/wild type]input.
RNA Isolation, RT-PCR, and Arrays. Detailed methods and protocols for the generation of bacterial RNA, RT-PCR, probe labeling, array hybridization, and scanning can be found in Supporting Text.
Results
Identification of YdgT in Salmonella. YmoA and Hha belong to an emerging family of transcriptional regulators that negatively modulate bacterial virulence gene expression (21). YmoA modulates the expression of Yersinia virulence factors including Yop proteins, the YadA adhesin, and invasin (25, 26). Hha, originally defined as a hemolysin expression modulating protein in Escherichia coli (27), negatively regulates SPI-1 virulence genes in Salmonella (5) and virulence genes in the locus of enterocyte effacement in enterohemorrhagic E. coli (28). We screened the Salmonella genome using blast to identify putative regulatory genes that shared similarity to the YmoA/Hha family of proteins. We identified a Salmonella gene designated ydgT (STM1461), located 38 ORFs downstream of the SPI-2-encoded gene ssaU. ydgT is annotated as a putative cytoplasmic protein in S. enterica and is transcriptionally repressed during early intracellular infection of macrophages (<4 h), but is slightly induced after 8 h of infection (29). YdgT shows 39% identity to Yersinia YmoA and 35% identity to Hha over the entire length of 71 aa (Fig. 1).
Fig. 1.
Alignment of Salmonella YdgT, Salmonella Hha, and Yersinia YmoA. Amino acid sequences for YdgT, Hha, and YmoA were aligned by using clustalw. Amino acid identities are indicated with black shading.
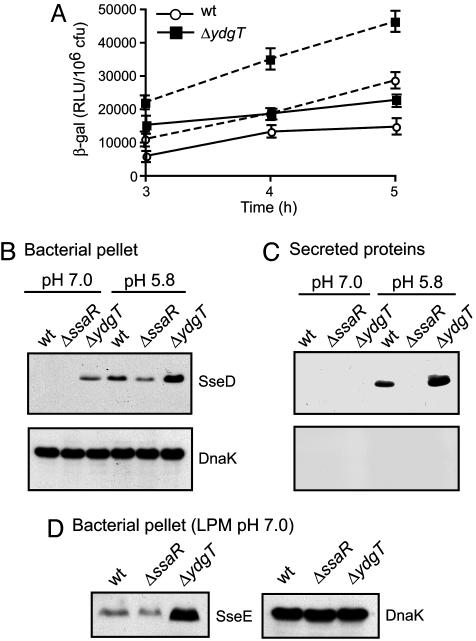
YdgT Is Involved in Repression of SPI-2 Transcription. The presence of a ymoA/hha homologue that is transcriptionally repressed during early intracellular infection when SPI-2 is activated lead us to hypothesize that YdgT might be involved in negatively modulating SPI-2. To test this possibility, we constructed a nonpolar, in-frame deletion of the ydgT gene, which did not affect the phenotype of Salmonella growing on solid media or in various liquid media (data not shown). We then introduced into wild-type and ΔydgT backgrounds a chromosomal transcriptional reporter containing the SPI-2 promoter region upstream of sseA, driving expression of a promoterless lacZ cassette, and measured β-galactosidase activity in whole cell lysates. Loss of ydgT significantly increased sseA promoter activity compared to wild type in LPM, pH 7.0 (Fig. 2A), a condition in which SPI-2 transcriptional activity is low (23). ydgT mutants grown in LPM at pH 5.8, a condition that induces expression and secretion of SPI-2-encoded proteins (18, 23, 30), also exhibited a significant increase in β-galactosidase activity (Fig. 2 A), indicating that the repressive effect of YdgT can occur under both neutral pH and acidic conditions.
Fig. 2.
Characterization of SPI-2 transcription and protein levels. (A) ydgT deletion increases transcription of SPI-2-regulated genes. β-galactosidase activity from a chromosomal PsseA::lacZ reporter in wild-type Salmonella (open circles) and ΔydgT (filled squares). Bacteria were grown in LPM medium at pH 7.0 (solid lines) and pH 5.8 (dashed lines) for the times indicated. Data are means with SD from quadruplicate determinations (pH 7.0 and 5.8, P < 0.05 for ydgT compared to wild type for all time points). Western blot analysis of bacterial cell lysates (pellet) (B and D) and secreted proteins (C) from wild-type, ssaR-, and ydgT- mutants grown in LPM medium. Protein fractions were probed for SseD (B and C, Upper), and SseE (D Left) and the intracellular bacterial protein, DnaK (B and C, Lower and D Right).
Expression and Secretion of SPI-2 Proteins in ydgT Mutants. Next, we examined SPI-2 protein levels in the ydgT mutant compared to wild type. Cell lysates and secreted proteins from bacteria cultured in LPM were analyzed for SseD and SseB, the poreforming and filament components of the SPI-2 type III apparatus, respectively. ydgT mutants showed increased accumulation of SseD (Fig. 2B) and SseB (data not shown) in whole cell lysates compared to wild type. This increase was evident when bacteria were grown under conditions that normally limit early SPI-2 activation, such as growth in LPM medium at neutral pH, but also under SPI-2-inducing conditions when the external medium was acidified after growth at neutral pH. SseD (Fig. 2C) and SseB (not shown) were secreted in a type III-dependent manner in greater abundance from the ydgT mutant compared to the other strain backgrounds, which likely reflected the higher intracellular levels of these SPI-2 substrates. Importantly, enhanced SPI-2 type III secretion occurred only when the external medium was acidified and not at neutral pH, as acidification is necessary for SPI-2 secretion (30, 31). We verified this phenotype by examining another nonsecreted SPI-2 protein, SseE (23), which accumulated in the ydgT mutant but not in wild-type bacteria or the SPI-2 apparatus mutant, ssaR (Fig. 2D). To test whether the loss of ydgT could overcome the requirement for positive regulation of SPI-2, we introduced the ydgT deletion into an ssrB mutant lacking the response regulator of the SsrA/SsrB two-component regulatory system. SPI-2 protein production was eliminated in the ssrB ydgT double mutant (data not shown), indicating that SPI-2 activation requires SsrB even in the absence of YdgT.
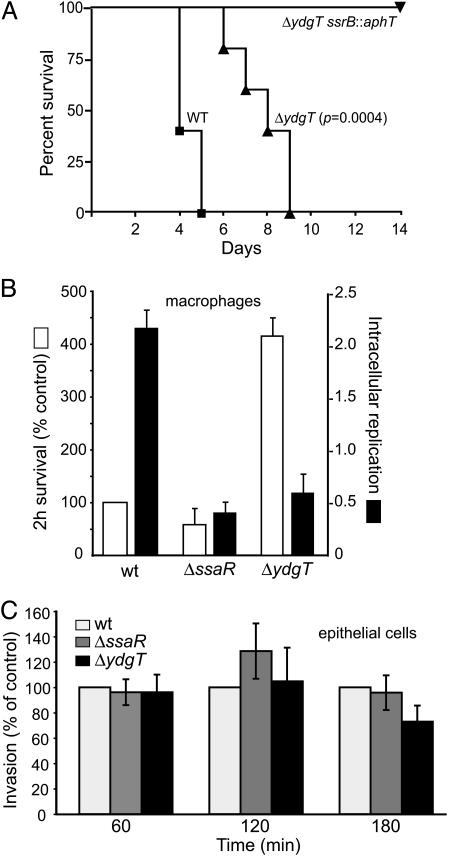
ydgT Mutants Are Attenuated for Virulence in a Mouse Model of Systemic Typhoid. Few studies (5, 32) have examined the in vivo phenotypes associated with loss or overexpression of a molecule that negatively modulates a pathogenicity island containing a T3SS. To assess the role of YdgT in virulence, we infected mice i.p. with wild-type bacteria, ydgT mutants, and ydgT ssrB double mutants and examined survival of the infected animals. The mean survival time was greater for mice infected with the ydgT mutant (7.6 days) compared to wild-type bacteria (4.4 days) (Fig. 3A). Inactivation of ssrB in the ΔydgT background resulted in complete loss of virulence during systemic infection, consistent with the finding that deletion of ydgT does not override the requirement of SsrB for transcriptional activation of the SPI-2 pathogenicity island.
Fig. 3.
In vitro and in vivo phenotypes of the ydgT- strain. (A) Survival plots for mice infected with wild-type Salmonella, ΔydgT, or ΔydgT ssrB::aphT double mutant. Shown is the percentage of mice surviving after infection (P = 0.0004, Log rank for ΔydgT compared to wild type). RAW267.4 cells (B) or HeLa cells (C) were infected with wild-type Salmonella or ΔssaR or ΔydgT mutants, and intracellular bacteria were enumerated at the indicated time after infection. (P > 0.05 for all comparisons in C.)
Deletion of ydgT Enhances Early Survival in Macrophages. SPI-2 activity contributes to intracellular survival and replication in macrophages (33, 34). Because deletion of ydgT seemed to uncouple SPI-2 regulation, we infected RAW264.7 mouse macrophages and HeLa epithelial cells with wild-type Salmonella or ssaR and ydgT mutants and examined invasion and intracellular survival. In macrophages, the numbers of viable intracellular ydgT mutants was significantly higher (≈420%) during the initial 2 h of infection compared to wild type (100%) and the SPI-2 mutant strain, ssaR (≈60%) (Fig. 3B). This increase was not due to differences in phagocytic uptake because the number of intracellular bacteria 15 min after infection was not different between the Salmonella strains (not shown). In longer infection experiments, the ydgT mutant displayed an attenuated intracellular phenotype, whereas wild-type Salmonella replicated to expected levels over a similar time period (Fig. 3B). In contrast, invasion and early survival of the ydgT mutant in epithelial cells, a process that is SPI-2-independent, was not different to wild type or ssaR mutants at various times after infection (Fig. 3C). Together, these data suggest that enhanced SPI-2 activity in the ydgT mutant might promote early intracellular survival in macrophages during a time when wild-type bacteria are still adapting to the intracellular environment. We examined this hypothesis in vivo by using a competitive murine infection model of systemic typhoid.
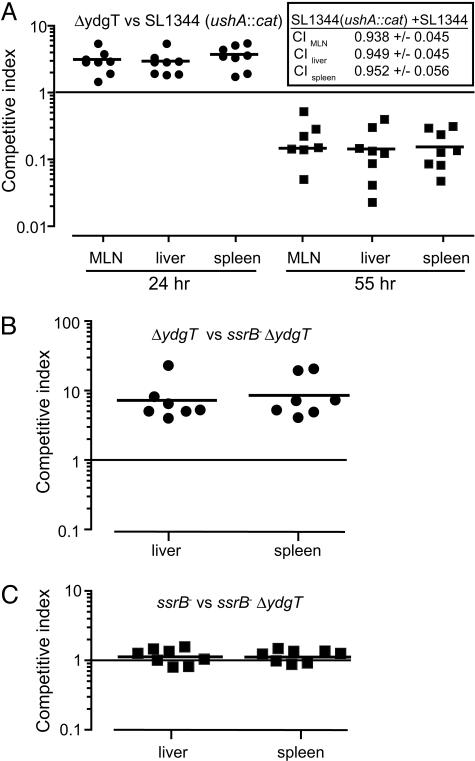
Loss of Negative SPI-2 Regulation Results in Biphasic Virulence in Vivo. Intramacrophage survival and replication allows Salmonella to colonize the mouse spleen and liver (35-37). Because in vitro experiments demonstrated an early survival phenotype in macrophages for ydgT mutants, we examined this at different times during in vivo infection. CI experiments (38) performed with wild-type Salmonella and ydgT mutants in mixed infections of the same animal indicated that the ydgT mutant had an early survival advantage in the reticuloendothelial system compared to wild-type bacteria that was evident in all systemic organs examined (Fig. 4A). However, this advantage was lost during the course of infection, with the ydgT mutant being recovered in significantly lower numbers compared to wild type, which comprised >90% of the output pool in all organs.
Fig. 4.
ydgT mutants have a biphasic systemic virulence phenotype. (A) Groups of mice were infected with a mixed inoculum of ΔydgT and wild-type Salmonella, and the CI for the ydgT- strain were determined in the MLN, liver, and spleen. The CI of the chromosomally marked Cm-resistant SL1344 strain (SL1344 ushA::cat) compared to the unmarked SL1344 parent strain are shown (A Inset). Each data point represents one animal, and horizontal bars indicate means. CI, P < 0.001 (24 and 55 h) for all organs. (B and C) CI of ssrB::aphT ΔydgT double mutants and each of the single mutants at 24 h after infection. Each datum represents one animal, bars indicate means.
The Early ydgT Phenotype in Vivo Depends on SPI-2 Activity. To test whether the enhanced early virulence of ydgT mutants occurred through SPI-2, we performed CI experiments between an ssrB ydgT double mutant and each of the ssrB and ydgT single mutants. The competitive indices of ydgT mutants versus ssrB ydgT double mutants reached as high as 20 (Fig. 4B), indicating that the early enhanced survival phenotype was abolished by removal of the SPI-2 activator, SsrB. In contrast, competitive indices of ssrB mutants versus ssrB ydgT double mutants were near unity (Fig. 4C), consistent with our observation that loss of ydgT does not supersede SsrB for SPI-2 expression and that deletion of ydgT does not confer any additional advantage in the absence of SsrB.
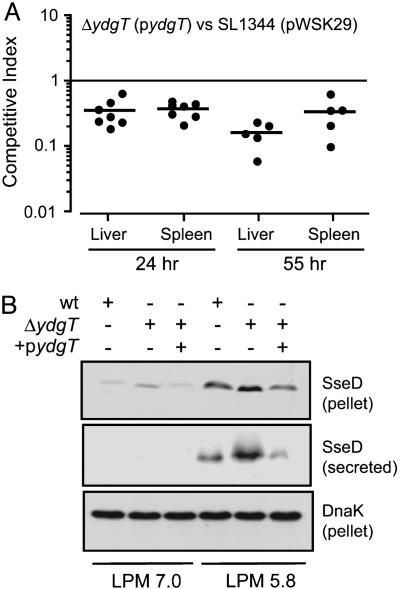
Overexpression of ydgT Attenuates Systemic Virulence. In CI experiments, we tested whether overexpression of ydgT would affect the early SPI-2-dependent “gain-of-virulence” phenotype of ydgT mutants. Overexpression of ydgT eliminated the early phenotype of the ydgT mutant at 24 h, significantly reducing the competitive indices relative to wild type (Fig. 5A). Overexpression of ydgT in wild-type bacteria also reduced the competitive indices relative to the vector alone to 0.15 ± 0.04 and 0.20 ± 0.05 in the liver and spleen, respectively. Furthermore, overexpression of YdgT decreased SseD secretion compared to the ydgT mutant strain (Fig. 5B), suggesting that overexpressing YdgT reduces SPI-2 gene expression below a critical threshold required for efficient systemic infection.
Fig. 5.
Overexpression of YdgT attenuates virulence. (A) CI complementation studies were carried out by using an episomal wild-type allele of ydgT (pydgT) or the plasmid alone (pWSK29). CI was determined at 24 and 55 h after infection in the liver and spleen. (B) SseD in bacterial lysates (pellet) and secreted protein fractions from wild-type Salmonella and ydgT mutants with (+) or without (-) pydgT.
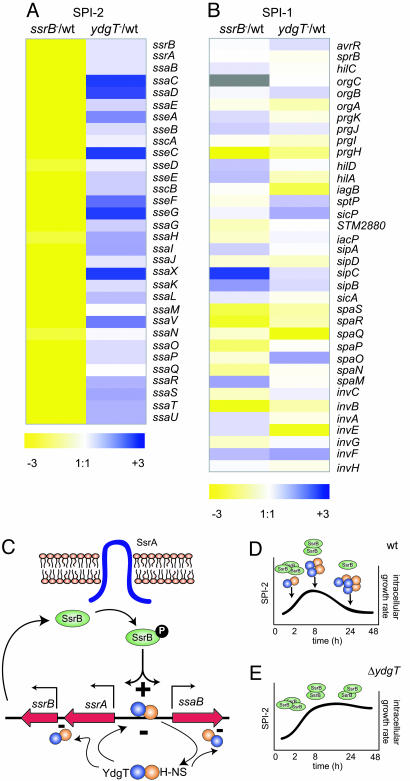
Transcriptional Profiling of SPI-2 in ydgT Mutants. Because deletion of ydgT lead to an early SPI-2-dependent increase in virulence, we used arrays to profile SPI-2 transcripts from wild-type Salmonella, SPI-2-positive regulator mutants (ssrB), and ydgT mutants grown under SPI-2-inducing conditions. Among the 32 structural, regulatory, effector, and chaperone genes encoded in SPI-2, all were down-regulated in the ssrB mutant and robustly expressed in the ydgT mutant. In the ydgT mutant compared to wild type, 7 of the 32 type III secretion-associated genes were up-regulated between 2 and 6-fold, 18 were up-regulated between 1.2 and 1.9-fold, and seven genes were expressed at similar levels between wild-type Salmonella and ydgT mutants (Fig. 6A). Among the more highly abundant transcripts were those from the “structural II” operon including ssaV and STM1410 (named ssaX here), the “structural I” operon (ssaC, ssaD), and the “effector/chaperone” operon (sseC, sseF, sseG). Previously, Cirillo et al. (36) also noted high-level expression in macrophages of transcriptional GFP fusions to targets in the structural I and II operons, suggesting some temporal order to SPI-2 expression. In contrast, transcriptional profiling of T3SS genes in the SPI-1 pathogenicity island did not reveal obvious clustering patterns in the pair-wise comparisons (Fig. 6B), consistent with the SPI-1-dependent phenotypes observed in cell culture.
Fig. 6.
YdgT involvement in repression of the SPI-2 pathogenicity island. Gene expression patterns of SPI-2 (A) and SPI-1 (B) were profiled by using arrays. Shown are the ratios of hybridization intensities of each type III secretion-associated gene in SPI-2 and SPI-1 from the indicated pairs of strains based on merged spots. Colors correspond to the differential gene expression intensity for each gene target. Gray indicates lack of signal in the array output. (C) Proposed model for the negative control of SPI-2 genes in Salmonella. YdgT-H-NS interactions (47) may form repressive complexes on promoters within SPI-2 that interfere with SsrB binding and transcriptional activation. Cycling of YdgT-H-NS complexes between DNAON and DNAOFF states might prevent premature production of SPI-2 proteins and overproduction of SPI-2 proteins inside cells during infection. The presence of YdgT might permit dampening of SPI-2 activation and the intracellular growth rate required for sustained infection and persistence (D), whereas the absence of YdgT ultimately leads to attenuation caused by alterations in intracellular growth and/or fitness (E).
Discussion
In Salmonella, the SPI-2 pathogenicity island and its related type III effectors are required for survival and replication in macrophages in the reticuloendothelial system. Although activators of SPI-2 are well documented, essentially nothing is known about how this pathogenicity island is repressed during stages of infection that are incongruous with SPI-2 activation or how the system is down-regulated once activated in host cells. Because SPI-2 lacks an obvious negative regulator, we hypothesized that the SPI-2 pathogenicity island integrates with ancestral negative regulatory proteins to achieve appropriate genetic control. Here, we identified and characterized a negative modulator of the SPI-2 pathogenicity island and established that negative regulation of SPI-2 is central to typhoid pathogenesis because overexpression of virulence genes required for typhoid reduced the systemic virulence of bacteria during infection in vivo.
Although positive regulation of virulence genes is required for pathogenesis in animals, repression of virulence gene expression until the proper host environment is sensed appears to be similarly crucial for bacterial virulence. This notion has been largely unstudied in in vivo models of bacterial infection, although Salmonella mutants in which the global response regulator PhoP is constitutively active (32) are attenuated for virulence in animals. This attenuation might be linked to the negative effect of PhoP on the SPI-1 invasion locus or, more related to this work, the observed positive effect of PhoP on ssrB expression (39). Derepression of SPI-2 gene expression by loss of ydgT resulted in an early gain-of-virulence suggesting that preformed SPI-2 gene products might enable mutant bacteria to better subvert early antibacterial effector mechanisms of the host. In support of this, others (40) demonstrated that stabilization of virulence gene mRNA through mutation of polynucleotide phosphorylase impacts virulence phenotypes during infection. However, attenuation of the ydgT mutant during infection progression suggests that inappropriate expression of type III virulence traits ultimately attenuates virulence. An evolving concept in Salmonella pathogenesis posits that these bacteria induce responses to temper their intracellular growth rate to establish persistence (41, 42), arguing that SPI-2 should be dampened after establishing an intracellular niche. That moderate ydgT induction is seen in macrophages later in infection, coinciding with a reduction in SPI-2 gene expression (29), suggests that the attenuation of ydgT mutants later in infection could be related to the importance of reducing bacterial intracellular growth rate during infection (Fig. 6D) and the inability of the ydgT mutant to effectively do so (Fig. 6E). The differences in in vitro and in vivo timing of the ydgT phenotype is likely due to the relatively synchronized infection that occurs in cell culture and the lack of this synchronization during infection of animals. Thus, the early virulence phenotype in animals is perceptible over a slightly longer time frame.
One question arising from this work concerns the regulatory mechanisms governing control of virulence genes acquired by horizontal transfer. Although transcription of SPI-2 promoters requires the SPI-2-encoded two-component system of SsrA/SsrB, negative control is achieved, in part, by ancestral YdgT encoded outside of SPI-2. Similar proteins, including YmoA (25, 26) and Hha (5, 28), also perform negative regulatory roles for virulence gene expression in pathogenic bacteria. A common mode of action of these proteins might be through H-NS, a global repressor of gene expression in bacteria (43-45). H-NS-mediated gene repression is thought to occur nonspecifically at regions of DNA curvature (46). However, the observation that E. coli YdgT (47), Hha (22, 48), and Yersinia YmoA (22) can bind to the N-terminal oligomerization domain of H-NS suggests that regulatory specificity might be achieved through interaction with accessory proteins. That the H-NS oligomerization domain also contributes to DNA binding (49) supports the idea that DNA-binding specificity of H-NS could be modulated by heteromeric interactions with such proteins. We (B.K.C. and B.B.F., unpublished data) and others (50) attempted to generate H-NS mutants in S. enterica SL1344, but the resulting strains displayed an aberrant mucoid phenotype, likely because of the adverse effect on chromosome structure. Thus, defining potential DNA consensus sequences for this family of regulators and their heteromeric combinations may aid in the identification of additional virulence genes that require strict regulatory control.
An indirect effect on SPI-2 cannot be ruled out for YdgT and, given the large number of negative regulators described for SPI-1, we predict other repressor molecules to play a role in exerting genetic control over SPI-2-mediated virulence. Other putative regulators might act in conjunction with YdgT, because transcriptional profiling of ydgT mutants indicated modest SPI-2 induction compared to wild type, more consistent with a virulence fine-tuning mechanism rather than an absolute requirement. A fine-tuning mechanism is also suggested because a ydgT-null mutant, although attenuated compared to wild type in single infections, retains some degree of virulence during systemic infection. However, the virulence defect becomes striking when the ydgT mutant is coinfected with wild-type bacteria in the same animal, indicating that selection for proper contextual activation of virulence factors is a strong driving force in the clonal expansion of bacterial pathogens during infection.
Supplementary Material
Acknowledgments
We thank the Finlay laboratory and Jose Puente for critical comments on this work. We also gratefully acknowledge Scott Peterson and the National Institute of Allergy and Infectious Disease (NIAID)-sponsored Pathogen Functional Genomics Resource Center at the Institute for Genomic Research for providing arrays and protocols, and Bob Hancock for the use of array equipment. This work was supported by the Canadian Institutes of Health Research (CIHR) and the Howard Hughes Medical Institute (HHMI). B.K.C. and M.E.W. are postdoctoral fellows of the CIHR and Michael Smith Foundation for Health Research (MSFHR), and N.F.B. is a MSFHR postdoctoral fellow. B.B.F. is a CIHR Distinguished Investigator, an HHMI International Research Scholar, and the University of British Columbia Peter Wall Distinguished Professor.
Conflict of interest statement: No conflicts declared
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: T3SS, type III secretion system; SPI-1, Salmonella pathogenicity island 1; CI, competitive index.
References
- 1.Galan, J. E. (1999) Curr. Opin. Microbiol. 2, 46-50. [DOI] [PubMed] [Google Scholar]
- 2.Galan, J. E. & Curtiss, R., III (1990) Infect. Immun. 58, 1879-1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bajaj, V., Hwang, C. & Lee, C. A. (1995) Mol. Microbiol. 18, 715-727. [DOI] [PubMed] [Google Scholar]
- 4.Baxter, M. A., Fahlen, T. F., Wilson, R. L. & Jones, B. D. (2003) Infect. Immun. 71, 1295-1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fahlen, T. F., Wilson, R. L., Boddicker, J. D. & Jones, B. D. (2001) J. Bacteriol. 183, 6620-6629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boddicker, J. D. & Jones, B. D. (2004) Infect. Immun. 72, 2002-2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fahlen, T. F., Mathur, N. & Jones, B. D. (2000) FEMS Immunol. Med. Microbiol. 28, 25-35. [DOI] [PubMed] [Google Scholar]
- 8.Shea, J. E., Hensel, M., Gleeson, C. & Holden, D. W. (1996) Proc. Natl. Acad. Sci. USA 93, 2593-2597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ochman, H., Soncini, F. C., Solomon, F. & Groisman, E. A. (1996) Proc. Natl. Acad. Sci. USA 93, 7800-7804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Worley, M. J., Ching, K. H. & Heffron, F. (2000) Mol. Microbiol. 36, 749-761. [DOI] [PubMed] [Google Scholar]
- 11.Garmendia, J., Beuzon, C. R., Ruiz-Albert, J. & Holden, D. W. (2003) Microbiology 149, 2385-2396. [DOI] [PubMed] [Google Scholar]
- 12.Lee, A. K., Detweiler, C. S. & Falkow, S. (2000) J. Bacteriol. 182, 771-781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Watson, P. R., Paulin, S. M., Bland, A. P., Libby, S. J., Jones, P. W. & Wallis, T. S. (1999) Infect. Immun. 67, 4950-4954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Buchmeier, N., Bossie, S., Chen, C. Y., Fang, F. C., Guiney, D. G. & Libby, S. J. (1997) Infect. Immun. 65, 3725-3730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Daniels, J. J., Autenrieth, I. B., Ludwig, A. & Goebel, W. (1996) Infect. Immun. 64, 5075-5084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Feng, X., Walthers, D., Oropeza, R. & Kenney, L. J. (2004) Mol. Microbiol. 54, 823-835. [DOI] [PubMed] [Google Scholar]
- 17.Linehan, S. A., Rytkonen, A., Yu, X. J., Liu, M. & Holden, D. W. (2005) Infect. Immun. 73, 4354-4362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Deiwick, J., Nikolaus, T., Erdogan, S. & Hensel, M. (1999) Mol. Microbiol. 31, 1759-1773. [DOI] [PubMed] [Google Scholar]
- 19.Miao, E. A., Freeman, J. A. & Miller, S. I. (2002) J. Bacteriol. 184, 1493-1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Beuzon, C. R., Unsworth, K. E. & Holden, D. W. (2001) Infect. Immun. 69, 7254-7261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Madrid, C., Nieto, J. M. & Juarez, A. (2002) Int. J. Med. Microbiol. 291, 425-432. [DOI] [PubMed] [Google Scholar]
- 22.Nieto, J. M., Madrid, C., Miquelay, E., Parra, J. L., Rodriguez, S. & Juarez, A. (2002) J. Bacteriol. 184, 629-635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Coombes, B. K., Brown, N. F., Valdez, Y., Brumell, J. H. & Finlay, B. B. (2004) J. Biol. Chem. 279, 49804-49815. [DOI] [PubMed] [Google Scholar]
- 24.Coombes, B. K., Brown, N. F., Kujat-Choy, S., Vallance, B. A. & Finlay, B. B. (2003) Microbes Infect. 5, 561-570. [DOI] [PubMed] [Google Scholar]
- 25.Cornelis, G. R., Sluiters, C., Delor, I., Geib, D., Kaniga, K., Lambert de Rouvroit, C., Sory, M. P., Vanooteghem, J. C. & Michiels, T. (1991) Mol. Microbiol. 5, 1023-1034. [DOI] [PubMed] [Google Scholar]
- 26.Ellison, D. W., Young, B., Nelson, K. & Miller, V. L. (2003) J. Bacteriol. 185, 7153-7159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nieto, J. M., Carmona, M., Bolland, S., Jubete, Y., de la Cruz, F. & Juarez, A. (1991) Mol. Microbiol. 5, 1285-1293. [DOI] [PubMed] [Google Scholar]
- 28.Sharma, V. K., Carlson, S. A. & Casey, T. A. (2005) FEMS Microbiol. Lett. 243, 189-196. [DOI] [PubMed] [Google Scholar]
- 29.Eriksson, S., Lucchini, S., Thompson, A., Rhen, M. & Hinton, J. C. (2003) Mol. Microbiol. 47, 103-118. [DOI] [PubMed] [Google Scholar]
- 30.Beuzon, C. R., Banks, G., Deiwick, J., Hensel, M. & Holden, D. W. (1999) Mol. Microbiol. 33, 806-816. [DOI] [PubMed] [Google Scholar]
- 31.Rappl, C., Deiwick, J. & Hensel, M. (2003) FEMS Microbiol. Lett. 226, 363-372. [DOI] [PubMed] [Google Scholar]
- 32.Miller, S. I. & Mekalanos, J. J. (1990) J. Bacteriol. 172, 2485-2490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hensel, M., Shea, J. E., Waterman, S. R., Mundy, R., Nikolaus, T., Banks, G., Vazquez-Torres, A., Gleeson, C., Fang, F. C. & Holden, D. W. (1998) Mol. Microbiol. 30, 163-174. [DOI] [PubMed] [Google Scholar]
- 34.Vazquez-Torres, A., Xu, Y., Jones-Carson, J., Holden, D. W., Lucia, S. M., Dinauer, M. C., Mastroeni, P. & Fang, F. C. (2000) Science 287, 1655-1658. [DOI] [PubMed] [Google Scholar]
- 35.Fields, P. I., Swanson, R. V., Haidaris, C. G. & Heffron, F. (1986) Proc. Natl. Acad. Sci. USA 83, 5189-5193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cirillo, D. M., Valdivia, R. H., Monack, D. M. & Falkow, S. (1998) Mol. Microbiol. 30, 175-188. [DOI] [PubMed] [Google Scholar]
- 37.Shea, J. E., Beuzon, C. R., Gleeson, C., Mundy, R. & Holden, D. W. (1999) Infect. Immun. 67, 213-219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Beuzon, C. R. & Holden, D. W. (2001) Microbes Infect. 3, 1345-1352. [DOI] [PubMed] [Google Scholar]
- 39.Bijlsma, J. J. E. & Groisman, E. A. (2005) Mol. Microbiol. 54, 85-96. [DOI] [PubMed] [Google Scholar]
- 40.Clements, M. O., Eriksson, S., Thompson, A., Lucchini, S., Hinton, J. C., Normark, S. & Rhen, M. (2002) Proc. Natl. Acad. Sci. USA 99, 8784-8789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tierrez, A. & Garcia-Del Portillo, F. (2005) Cell. Microbiol. 7, 901-910. [DOI] [PubMed] [Google Scholar]
- 42.Monack, D. M., Mueller, A. & Falkow, S. (2004) Nat. Rev. Microbiol. 2, 747-765. [DOI] [PubMed] [Google Scholar]
- 43.Dorman, C. J. (2004) Nat. Rev. Microbiol. 2, 391-400. [DOI] [PubMed] [Google Scholar]
- 44.Tupper, A. E., Owen-Hughes, T. A., Ussery, D. W., Santos, D. S., Ferguson, D. J., Sidebotham, J. M., Hinton, J. C. & Higgins, C. F. (1994) EMBO J. 13, 258-268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hulton, C. S., Seirafi, A., Hinton, J. C., Sidebotham, J. M., Waddell, L., Pavitt, G. D., Owen-Hughes, T., Spassky, A., Buc, H. & Higgins, C. F. (1990) Cell 63, 631-642. [DOI] [PubMed] [Google Scholar]
- 46.Owen-Hughes, T. A., Pavitt, G. D., Santos, D. S., Sidebotham, J. M., Hulton, C. S., Hinton, J. C. & Higgins, C. F. (1992) Cell 71, 255-265. [DOI] [PubMed] [Google Scholar]
- 47.Paytubi, S., Madrid, C., Forns, N., Nieto, J. M., Balsalobre, C., Uhlin, B. E. & Juarez, A. (2004) Mol. Microbiol. 54, 251-263. [DOI] [PubMed] [Google Scholar]
- 48.Juarez, A., Nieto, J. M., Prenafeta, A., Miquelay, E., Balsalobre, C., Carrascal, M. & Madrid, C. (2000) Adv. Exp. Med. Biol. 485, 127-131. [DOI] [PubMed] [Google Scholar]
- 49.Bloch, V., Yang, Y., Margeat, E., Chavanieu, A., Auge, M. T., Robert, B., Arold, S., Rimsky, S. & Kochoyan, M. (2003) Nat. Struct. Biol. 10, 212-218. [DOI] [PubMed] [Google Scholar]
- 50.Schechter, L. M., Jain, S., Akbar, S. & Lee, C. A. (2003) Infect. Immun. 71, 5432-5435. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.