Abstract
Breast cancer remains the cancer with the highest mortality among women in the United States. Peptides derived from the oncogenic Tac1 gene (full transcript: βPPT-A) stimulate the proliferation of breast cancer cells (BCCs) via seven-transmembrane G protein-coupled neurokinin 1 (NK1) and NK2 receptors. The NK1 gene could generate full-length (NK1-FL) and truncated (NK1-Tr) transcripts. NK1-Tr lacks 100 residues in their cytoplasmic end, could couple to G proteins, and shows reduced efficiency with respect to internalization and desensitization. This study reports on a role of NK1-Tr in the transformation of nontumorigenic breast cells, and investigates whether Tac1 expression is linked to the generation of NK1-Tr. Western blots and Northern analyses showed coexpressions of NK1-Tr and NK1-FL in BCCs (cell lines and primary cells from patients with different stages of breast cancer). Stable transfections of βPPT-A or NK1-Tr expression vectors in nontumorigenic cells showed each induces the expression of the other, consequently resulting in a transformed phenotype. Analyses with microarrays indicate similar patterns of cytokine production by NK1-Tr transfectants and BCCs, but not NK1-FL transfectants. These observations indicate tumor-promoting properties by NK1-Tr, but not NK1-FL. Overall, the oncogenic property of Tac1 in breast cells involves concomitant expression of NK1-Tr and vice versa, consequently leading to the production of cytokines with growth promoting functions.
Keywords: breast cancer, cytokines, bone marrow metastasis, substance P, tachykinin
Despite aggressive treatment, education, and early detection, breast cancer remains a clinical and social dilemma (1). In women, breast cancer accounts for the highest cause of cancer related deaths (1). Although the reasons for deaths may be varied, most patients succumb to bone metastasis (2). Questions regarding the location of breast cancer cells (BCCs) before bone invasion remains a “black box.” It appears that BCCs are located in the bone marrow cavity at low frequency before bone invasion. Early diagnosis of breast cancer and aggressive treatments could nonetheless show cancer resurgence after prolonged remission.
Research studies have showed relevance for neuroendocrine-related molecules in breast cancer development, for example, peptides derived from the Tac1 gene (3-7). These peptides stimulate BCCs through autocrine mechanisms by interacting with neurokinin 1 (NK1) and NK2 receptors (5-11). Tac1 is a single copy gene with seven exons and is alternately spliced into four transcripts, α-, β-, γ-, and δ-PPT-A (12). The PPT-A transcripts produce peptides belonging to the tachykinin family (12). Substance P is the predominant peptide encoded by each of the PPT-A transcripts (12). NK1 and NK2 belong to the family of G protein-coupled, seven-transmembrane receptors (13).
NK1 and Tac1 are constitutively expressed in BCCs (7). This finding contrasts with normal cells in which their expression requires cell stimulation (14). In normal cells, transmembrane NK1 and NK2 appear to be regulated by intracellular crosstalk, negative feedback through the production of specific cytokines, and activation of endopeptidases (14-16). Two forms of NK1 protein are reported in humans, full-length (NK1-FL) and truncated (NK1-Tr) (17). The cytoplasmic end of NK1-Tr lacks 100 residues, a region that functions as the substrate for G protein-receptor kinases (17).
This study tests the hypothesis that BCCs coexpress NK1-Tr and NK1-FL. The hypothesis further states that the presence of NK-1-Tr leads to the expression of Tac1, previously shown to be oncogenic (9). The premise is that coexpressions of peptides from Tac1 and NK-1-Tr in BCCs would facilitate autocrine stimulation by the peptides leading to the production of cytokines with tumor promoting properties (14). The studies were performed with breast cancer cell lines, and the results were verified with primary BCCs from patients with different stages of breast cancer. The method used to select primary BCCs is possible because the normal epithelial cells do not survive.
Materials and Methods
Reagents. Rabbit anti-NK1 was purchased from Sigma. Stromal derived growth factor-1α was purchased from R & D Systems. Horseradish peroxidase-conjugated goat anti-rabbit IgG, FITC-isotype control, and FITC-goat anti-rabbit IgG were purchased from BD Bioscience (San Jose, CA). C-99,994, a NK1 receptor antagonist, was kindly provided by Pfizer (Groton, CT).
Cell Lines and Modified Cells. The following cell lines were purchased from American Type Culture Collection and propagated according to their instructions: Tumorigenic cells produced Substance P at levels ranging between 35 and 68 pg/ml: ZR-75-30, BT474, T47D, MDA-MB-330, DU4475, and BT483; Nontumorigenic cells showed undetectable substance P by ELISA: MCF12A, MCF12F, Hs578Bst, MCF10A, and MCF10-2A. Tac1 knockdown breast cancer cell lines and nontumorigenic cell lines stably expressing βPPT-A have been described (9, 18).
Primary Breast Tissue and Selection Method. Breast tissues were obtained at diagnosis and the hormone profile shown in Table 1. At the time of surgery, patients did not receive any form of anticancer agent. The studies followed guidelines approved by the institutional review board of University of Medicine and Dentistry of New Jersey (Newark). Tissues from four patients (P5-P8) were provided by the Cooperative Human Tissue Network. Expansion of BCCs from surgical tissues was described, and they are hereafter referred as primary BCCs (9). Nontumorigenic cells do not survive during expansion. By RT-PCR, we have not detected NK-1 mRNA in surgical samples from benign tissues (unpublished data). This observation is similar to nontumorigenic cells.
Table 1. Profile of breast cancer patients (P1-P12).
| Patients | Stage | Age, years | ER | PR | HER2 | NK1-TR | DNA Sequencing for NK1-Tr |
|---|---|---|---|---|---|---|---|
| P1-P3 | IIIA | 36-73 | − | − | − | + | P1 |
| P4 | IIIA | 60 | + | + | + | + | P4 |
| P5-P8 | IIA | 38-82 | + | + | − | + | P6, P7 |
| P9 | I | 45 | + | + | − | + | P9 |
| P10 | I | 38 | + | + | + | + | |
| P11 | M0 | 45 | ND | ND | ND | + | |
| P12 | M0 | 40 | ND | ND | ND | + |
Cancer staging followed standard guidelines (43). β-PPT-A and substance P levels were studied with total RNA and cell extracts from surgical tissues (n = 12), as described (3). Mean β-PPT-A molecules per μg total RNA = 25 ± 7 (±SD). Substance P levels = 66 ± 11 pg/ml (±SD). Tissue extracts were prepared by homogenizing ≈0.2 g of tissues in 0.2 ml of PBS containing protease inhibitor cocktail. ER, estrogen receptor; PR, progesterone receptor; HER2, c-erbB-2; ND, not done.
Cloning of NK1-FL and NK1-Tr. Total RNA was extracted from bone marrow stroma, stimulated with 5 ng/ml of stromal derived growth factor-1α and served as the template for amplifying NK1-FL and NK1-Tr cDNA by RT-PCR. The upstream primers were the same and contained BamHI linkers: 5′-ATGGATAACGTCCTCC-3′ (GenBank accession no. M84425). The downstream primers contained HindIII linkers: 5′-AGGTTACACGAGAGGATC-3′ for NK1-FL cDNA (GenBank accession no. M84425) and 5′-CCTGTCATTGAGGCAGCAGTA-3′ for NK1-Tr cDNA (GenBank accession no. M84426). PCR products for NK1-FL (≈1.3 kb) and NK1-Tr (≈0.9 kb) (see Fig. 2B) were cloned into pCR2.1 (Invitrogen) and then sequenced. The DNA inserts were excised with EcoRI and then subcloned in the forward and reverse orientations in pSG5 (Stratagene). Their orientations were verified by restriction mapping and DNA sequencing.
Fig. 2.
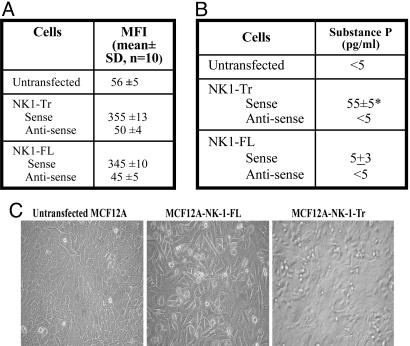
Stable transfection of NK1-Tr and NK1-FL cDNA in MCF12A and MCF10A. (A) The mean fluorescence intensities (MFI, mean ± SD) are shown for transfectants for five cell passages. (B) Substance P production (mean ± SD, n = 10) in the culture media of NK1-FL and NK1-Tr transfectants. (C) Representative confluent cultures for NK1 transfectants. *, P < 0.05 vs. untransfected and NK1-FL transfectants.
Stable Expression of NK1-FL and NK1-Tr in Nontumorigenic Breast Cells. MCF12A and MCF10 were transfected with pSG5-NK1-FL and pSG5-NK1-Tr in both forward and reverse orientations as described (9). Positive cultures were cloned by limiting dilutions, at one cell per 96-well tissue culture plates. At confluence, cells from each well were split into two wells. One plate was screened for positive wells by indirect immunofluorescence using rabbit anti-NK1 and FITC-goat anti-rabbit IgG (see Fig. 3 C and D). Positive clones were expanded and subjected to repeated screening by flow cytometry for NK1 and PCR for NK1 insert by using plasmid DNA (not shown).
Fig. 3.
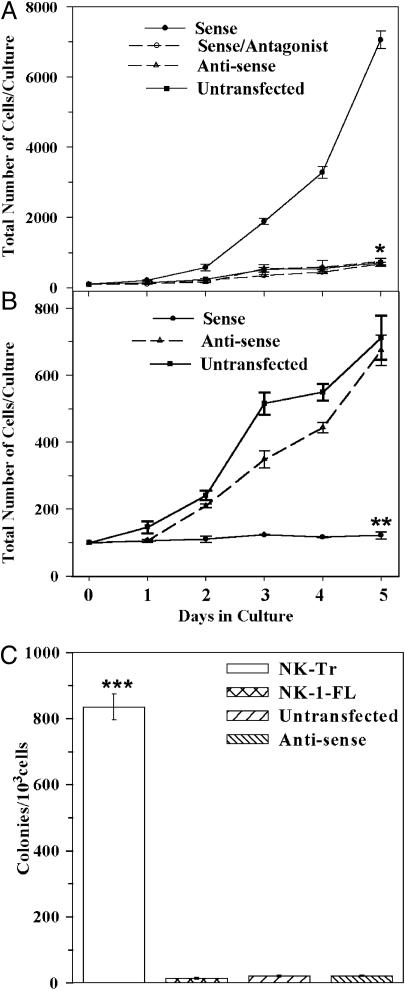
Growths curves and clonogenic assays for nontumorigenic cell lines. MCF10A and MCF12A, untransfected or stably transfected with NK1-Tr (A) or NK1-FL (B) in the sense and antisense orientations. Cells were seeded at 100 per ml in 2 ml of culture media. Transfectants with NK1-Tr cDNA were cultured in the presence or absence of NK1-specific antagonist (10 nM C-99,994). Beginning at day 1, viable cells were counted daily up to day 5. The mean ± SD, n = 10 represent five cell passages per transfectant. (C) Clonogenic assays were performed with cells from five cell passages at 103 cells per ml. At day 5, colonies with >20 cells were counted, and the results were presented as the mean ± SD, n = 10. *, P < 0.05 vs. sense transfectants; **, P < 0.05 vs. sense transfectants; ***, P < 0.05 vs. NK1-FL transfectants and untransfected/antisense.
Northern Analysis. Northern analyses were performed for NK1-FL and NK1-Tr mRNA as described (16). Briefly, 10 μg of total RNA were analyzed with NK1-specific cDNA probe, randomly labeled with 3,000 Ci/mM [α-32P]dATP (Dupont/NEN; 1 Ci = 37 GBq). Bands were normalized by stripping and reprobing with cDNA for 18S rRNA. Hybridizations were detected after 24 h on the Typhoon 9410 Molecular Imager (Amersham Pharmacia). The NK1 cDNA probe spanned the coding sequence of NK1-FL (GenBank accession no. M76675, 1.3 kb). The lower bands were verified as NK1-Tr mRNA, based on rehybridization with a cDNA probe spanning the deleted 300-bp region (17). The 300-bp cDNA probe was prepared with gel-purified fragments obtained from NK1-FL cDNA digested with AluI.
Western Blots. Membrane extracts were prepared by incubating 2 × 106 cells at room temperature for 15 min in 400 μl of 1× lysis buffer (Promega). The membrane fractions were resuspended in 300 μl of 1× PBS and 20 μg was analyzed in Western blots for NK1 as described (16). NK1 was detected with anti-NK1 (1/10,000) and HRP-goat anti-rabbit IgG (1/10,000) primary and secondary antibodies, respectively. HRP was developed with Chemiluminescence Reagent Plus (PerkinElmer Life Sciences).
Microarrays for Cytokines. Media was replaced from confluent BCC cultures, and after 24 h, cell-free media were analyzed with human cytokine protein array II (Ray Biotech, Norcross, GA), as described (18). Briefly, membranes were incubated for 1 h with culture supernatants and then incubated consecutively with biotin-conjugated anti-cytokines and HRP-streptavidin followed by chemiluminescence detection. Background was subtracted in analyses with fresh culture media.
Sequence of NK1 cDNA in BCCs. RT-PCR was performed with total RNA extracted from surgical samples of five random patients and the breast cancer cell lines described above. The PCR fragments were purified, and fragments equivalent to ≈1.2 kb and ≈0.9 kb were cloned into pCR2.1. The DNA inserts were sequenced at the Molecular Core Facility (University of Medicine and Dentistry/New Jersey Medical School) with primers used for RT-PCR. The results were analyzed with gcg wisconsin package version 10 (Accelrys, San Diego).
Clonogenic Assays. Clonogenic assays were performed as described (9). Briefly, cells were resuspended in 1.2% methylcellulose containing the respective culture media. Assays were performed with cells seeded at 103 per ml in 35-mm suspension dishes. Colonies with >25 cells were counted after 1 week of incubation at 37°C.
Statistical Analysis. Data were analyzed by using analysis of variance and Tukey-Kramer multiple comparisons test. A P value <0.05 was considered significant.
Results
NK1, NK2, and PPT-A peptides have been reported to mediate autocrine proliferation of BCCs (3, 9). In normal cells, NK2 acts as negative feedback on NK1 functions (15). Because NK1-Tr lacks residues that could be important for crosstalk with NK2 (19), we premise that NK1-Tr might be the predominant NK1 subtype in BCCs (15).
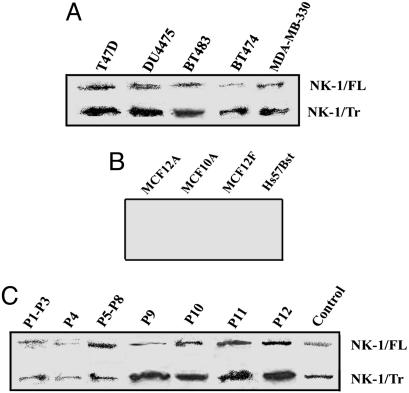
Coexpressions of NK1-FL and NK1-Tr in BCCs. Western blots with membrane extracts from five different breast cancer cell lines showed coexpression of NK1-FL and NK1-Tr (Fig. 1A). Similar studies with four different nontumorigenic breast cells showed no band for NK1 (Fig. 1B). Studies repeated with 12 primary BCCs showed coexpressions of NK1-FL and NK1-Tr (Fig. 1C). In summary, the studies indicate membrane expression of both NK1-FL and NK1-Tr in BCCs (cell lines and patients), whereas neither was detectable in nontumorigenic cell lines.
Fig. 1.
Coexpression of NK1-FL and NK1-Tr cDNA in BCCs. Membrane extracts were examined for NK1 by Western blots in BCCs (A); nontumorigenic breast cell lines (B), and primary BCCs, with DU4475 as control (C).
DNA sequencing of RT-PCR products from five randomly selected primary BCCs (Table 1), and in the malignant cell lines used in this study showed no mutation for NK1-FL and NK1-Tr cDNAs. By Northern analyses, we have verified the expressions of both NK1-FL and NK1-Tr mRNA in three patients (P1, P4, and P11). Discrimination between NK1-FL and NK1-Tr mRNA occurred by two consecutive hybridizations, the firstwith a cDNA common to both transcripts: hybridization revealed two bands equivalent to the predicted sizes for NK-1-FL and NK-1-Tr mRNA. The second hybridization using a cDNA probe equivalent to the missing region of NK1-Tr cDNA resulted in the disappearance of lower bands. In summary, the results demonstrate coexpression of both NK1-FL and NK1-Tr mRNA. There was no evidence of cDNA mutation in breast cancer cell lines and in at least five patients.
Transformation of Nontumorigenic Cells by NK1-Tr. Compared to NK1-FL, NK1-Tr has been reported to have reduced efficiency with respect to its internalization and desensitization (20), but could signal via G-proteins (20, 21). We therefore asked whether over-expression of NK1-Tr in nontumorigenic breast cells cause their transformation. NK1-Tr and NK1-FL expression vectors were stably transfected in MCF12A and MCF10A, and their growth properties studied. By immunofluorescence, we showed similar mean fluorescence intensities, MFI, n = 10 ± SD in five cell passages (Fig. 2A), indicating comparable expressions of NK1-Fl and NK1-Tr. The growth patterns of transfectants are shown for 2-week cultures, which were initiated with 100 cells. Untransfected cells showed contact inhibition (Fig. 2C Left), NK1-FL transfectants were ≈40% confluent (Fig. 2C Center), and NK1-Tr transfectants showed foci formation (Fig. 2C Right).
Increased production of substance P has been reported in malignant cells (7, 9). Using ELISA, we observed a significant (P < 0.05) increase in substance P production for NK1-Tr and minimal increase for NK1-FL transfectants (Fig. 2B). Substance P production was not because of plasmid insertion, because it was undetectable (<5 pg/ml) for transfectants with inserts in the antisense orientation (Fig. 2B). In summary, the results show NK1-Tr inducing foci formation with concomitant increase in substance P levels.
Growth Rates of Nontumorigenic Cells Transfected with NK1-Tr and NK1-FL cDNA. We next compared the proliferation of nontumorigenic cells (MCF10A or MCF12A), untransfected or stably transfected with NK1-Tr or NK1-FL cDNA, in sense and antisense orientations. Growth assays were initiated with 100 cells per ml and daily counts were done for 5 days. The growth of NK1-FL transfectants and untransfected cells were significantly (P < 0.05) lower than NK1-Tr transfectants (Fig. 3 A and B). Transfectants with DNA inserts in the antisense orientations showed similar growth rates as untransfected cells (Fig. 3 A and B, open triangle and filled square). In summary, the results show significant (P < 0.05) increases in the growth of NK1-Tr transfectants, compared to NK1-FL transfectants and untransfected cells.
Autocrine Stimulation of NK1-Tr Receptor by Substance P. NK1-Tr expression in nontumorigenic cells led to the production of substance P and concomitant increase in cell proliferation and foci formation (Figs. 2 and 3). The next set of studies asked whether the increased proliferation of NK1-Tr transfectants could be caused by autocrine stimulation by substance P. We repeated the growth curve studies in the presence or absence of various concentrations of NK1 receptor antagonist (C-99,994). At 10 nM C-99,994, there was optimum and significant (P < 0.05) reduction in cell growth, with no evidence of cell death by trypan blue exclusion (Fig. 3A, open circle). The blunting effects of C-99,994 on cell growth were reversed after the antagonist was washed and the cells were reincubated with fresh media (data not shown).
Foci formation and increased growth rate by NK1-Tr transfectants (Figs. 2C and 3 A and B) were further analyzed for a transforming phenotype in an assay that studied contact-independent growth. Clonogenic assays were performed in methylcellulose matrix (9). Controls were performed with NK1-FL transfectants, transfectants in the antisense orientation, and untransfected MCF12A and MCF10A. In five experiments (n = 10), the total number of colonies were counted after 5 days. The results showed significant (P < 0.05) increase in colonies for NK1-Tr transfectants as compared to NK1-FL transfectants (Fig. 3C). Untransfected cells and transfectants with inserts in the antisense orientations showed few colonies with two to five cells per colony. In summary, the results show contact independence and rapid proliferation of NK1-Tr transfectants, as compared to NK1-FL transfectants.
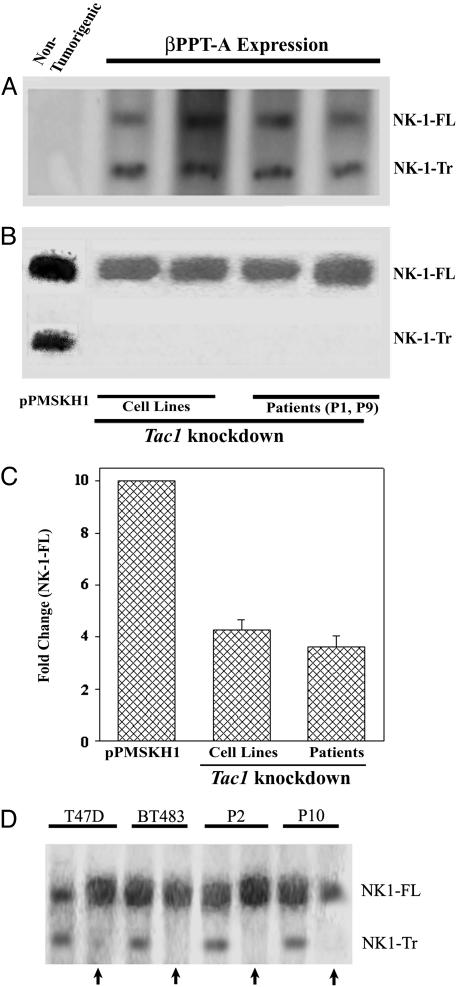
Link Between Tac1 and NK1 Expression. Autocrine stimulation of NK1-Tr by substance P in MCF12A and MCF10A transfectants (Figs. 2B and 3A) led us to ask whether NK1-Tr has oncogenic properties, or whether its effect is secondary to the production of substance P, previously shown to exert oncogenic properties (9). To this end, we asked whether the expression of the Tac1 is responsible for the expression of NK1-Tr receptor in BCCs. To avoid confounds by other molecules in malignant cells, the studies were done with four different nontumorigenic cell lines stably expressing the βPPT-A cDNA (9). Although Northern (Fig. 4A) and Western (Fig. 4D, no arrows) blots showed the products of NK1-FL and NK1-Tr in transfectants, no bands were detected for transfectants with vector alone (data not shown) or untransfected cells (represented in Fig. 4A, left lane).
Fig. 4.
NK1 expression in PPT-A knockdown BCCs and βPPT-A expressing nontumorigenic breast cells (MCF12A, MCF10A, MCF12F, MCF10-2A). Northern analyses for NK1-FL and NK1-Tr mRNA were determined in βPPT-A-expressing cells. (A) Representative untransfected nontumorigenic cell is shown for MCF12A in the left lane. (B) Tac1 knockdown in T47D and BT474 with pPMSKH1-PPT-A. In the left lane, control transfectants with pPMSKH1 alone shown for T47D. (C) The density of NK1-FL is normalized to 10, and fold changes in PPT-A knockdown BCCs are shown for the six breast cancer cell lines described in Materials and Methods. Western blots with anti-NK-1 using membrane extracts from two breast cancer cell lines and two patients are shown. Arrows represent transfectants with pPMSKH1-PPT-A compared to transfectants with pPMSKH1 alone (no arrow).
To further verify a role for the Tac1 in NK1-Tr receptor expression, studies were performed with BCCs (two cell lines and two primary cells). Tac1 expression was knocked down in BCCs with pPMSKH1-PPT-A as described (9). Northern (Fig. 4B) and Western (Fig. 4D, arrows) blots showed dense bands for NK1-FL and light to undetectable bands for NK1-Tr. In summary, we have demonstrated two major points: (i) Nontumorigenic cells stably transfected with βPPT-A coexpressed NK1-FL and NK1-Tr at the levels of mRNA and protein (Fig. 4 A and D). (ii) Knockdown of Tac1 resulted in 5-fold reduction of NK1-FL mRNA and undetectable NK1-Tr mRNA, as compared to untreated BCCs (Fig. 4 B and C). These findings correlate with the corresponding protein (Fig. 4D).
Cytokine Production in NK1-FL and NK1-Tr Transfectants. Because NK1-Tr has shown transforming abilities and its expression requires Tac1 induction (Fig. 4), we next determined whether NK1-Tr receptor can induce cytokines similar to BCCs. Cytokine microarrays compared the production of cytokines in transfectants and the five breast cancer cell lines listed in Materials and Methods. We also analyzed media from untransfected MCF12A and MCF10A. Overall, the results (Table 2) showed similar production of cytokines for NK1-Tr transfectants and BCCs. Comparably low production of cytokines were detected in media from MCF12A, MCF10A, and NK1-FL transfectants.
Table 2. Cytokine microarray analyses in the supernatants of confluent cells.
| Normalized cytokine production
|
||||
|---|---|---|---|---|
| Growth factors | NK1-Tr transfectants | NK1-FL transfectants | BCC | Nontumorigenic breast cells |
| Chemokines | ||||
| RANTES, GRO-α | 9.8 ± 0.1 | 0.12 ± 0.1 | 9.5 ± 0.03 | 0.1 ± 0.01 |
| MCP-1, IL-8 | 8.1 ± 0.01 | 0.2 ± 0.12 | 8.0 ± 0.02 | 0.1 ± 0.001 |
| MIP-1β | 5.2 ± 1 | 0.15 ± 0.01 | 4.5 ± 0.15 | 0.1 ± 0.01 |
| SDF-1α | 8.2 ± 0.2 | 0.2 ± 0.12 | 8.0 ± 0.25 | ND |
| Angiogenin | 10 ± 0.4 | 0.1 ± 0.02 | 9.8 ± 0.1 | 0.1 ± 0.022 |
| Oncostatin M | 10 ± 0.3 | 0.15 ± 0.01 | 9.5 ± 0.13 | 0.1 ± 0.012 |
| Cytokines | ||||
| IL-6 | 10 ± 0.5 | 5.5 ± 0.02 | 10 ± 0.2 | 3.0 ± 0.2 |
| IL-2 | 10 ± 0.3 | 5 ± 0.3 | ND | ND |
| IL-12 | ND | ND | 0.1 ± 0.01 | ND |
| IFN-α, TNF-α, IL-1α | 10 ± 0.4 | 1.5 ± 0.2 | 10 ± 0.5 | ND |
| IL-4, IL-10, IL-13* | 8 ± 1.2 | 1.5 ± 12 | 9 ± 1.5 | ND |
| TGF-β* | 10 ± 1.5 | 2.2 ± 0.14 | 10 ± 0.3 | ND |
| G-CSF, GM-CSF† | 10 ± 1.1 | 1.8 ± 0.04 | 10 ± 0.5 | ND |
Cell-free media were analyzed from breast cancer cell lines (n = 5). Two different cell passages of two nontumorigenic cells stably transfected with NK1-Tr or NK1-FL cDNA expression plasmid (n = 4) or untransfected (n = 4). The densitometric scans of the internal positive controls were normalized to 10, and the unknowns were calculated accordingly and presented as the mean ± SD. ND, none detected; *, antiinflammatory; †, hematopoietic growth factors.
Discussion
This report used both cell lines and primary BCCs (Table 1). Primary cells were selected from surgical breast tissues by using an established coculture method (9). We detected NK1-Tr mRNA, regardless of hormone profile or the stage of breast cancer (Table 1), suggesting that NK1-Tr transcripts might be expressed at an early phase of cancer development. The results show an efficient transforming property of NK1-Tr coding sequence with minimum growth promoting effects of NK1-FL coding region. The absence of mutation in NK1-Tr cDNA from five randomly selected patients, and the transforming property of NK1-Tr cDNA in nontumorigenic cells, suggests mechanisms other than DNA mutation in the oncogenic property of NK1-Tr. A polymorphic site in NK1 (GenBank accession no. BD223571) suggests that polymorphism could be a predisposing factor in NK1-mediated development of breast cancer. Robust analyses of the NK1 gene in clinical samples are required to determine whether DNA mutations, epigenetic changes, and/or polymorphism are involved in breast cancer.
Previous studies show NK1 and NK2 mediating BCC proliferation (3). This finding is in contrast to healthy cells, in which NK2 receptor mediates cell cycle quiescence while regulating the expression of NK1 by intracellular crosstalk (14, 15). Because NK1-FL transfectants did not elicit significant proliferation (Fig. 3B), it was deduced that NK1-Tr could be the dominant NK1 subtype in BCC proliferation. An unanswered question is whether intracellular crosstalk between NK2 and NK1-Tr exists, or whether the missing region of NK1-Tr precludes crosstalk with NK2. Compared to efficient receptor desensitization exhibited by NK1-FL, NK1-Tr shows reduced efficiency. Regardless of these differences, both NK1-Tr and NK1-FL induce pathways incorporating G proteins and activation of protein kinase C (PKC), which has been suggested as a potential cancer target (21, 23). Because NK1 activates PKC, we propose that targeting of NK1 could prevent downstream activation of PKC.
It is unlikely that the omitted residues in NK1-Tr are deleted by posttranslational mechanisms because the respective mRNA was detected (Figs. 1 and 4). Increased transcription rate with early termination is the favored hypothesis because a slower rate might by-pass a false stop codon (NM015727) to generate NK1-FL transcripts. This premise is supported in studies showing only NK1-FL in Tac1 knockdown BCCs (Fig. 4B). Cytokine microarray studies indicated increases in the production of cytokines from NK1-Tr transfectants (Table 2). The levels were similar to those from BCCs (Table 2). In Tac1 knockdown BCCs, cytokine production was significantly decreased (18). This decrease could lead to slower transcription rate of NK1 to generate NK1-FL (Fig. 4B). We propose that NFκB is central to increase in the transcription rate because it can be activated by several of the cytokines shown in Table 2. Furthermore, unpublished studies show NFκB being central to the generation of NK1-Tr mRNA, and the production of substance P in NK1-Tr transfectants (Fig. 2B) allows for a working hypothesis to test whether cytokines produced in NK1-Tr transfectants are secondary to the production of substance P. This premise is based on substance P as an inducer of cytokines with growth-promoting functions (9).
Radiolabeled substance P has been proposed as a potential cancer therapy (5). Its effectiveness would require detailed pharmacological, biochemical, and molecular studies to determine potential differences between NK1-Tr and NK1-FL receptors. It should be noted that the findings reported for NK1-Tr might be similar for other cancers (8, 11, 24, 25). NK1-Tr transfectants and BCCs show similar patterns of cytokine production (Table 2). Several of these cytokines could activate the transcription factor, AP1. Interestingly, substance P receptor antagonist also activates AP1 (23). A benefit for AP1 in the presence of substance P receptor antagonist could be enhanced susceptibility to chemotherapy. This could occur by AP-1 inducing rapid cycling of BCCs. Substance P has been shown to protect against cell death (25). Thus, blocking signaling of NK1-Tr with an antagonist might enhance BCC death. Based on this report, it appears that substance P receptor antagonist could be an adjuvant to current cancer therapies.
The level, transcription vs. posttranscription, at which NK1 could be dysregulated has not been proven in this study. Despite this limit, the present studies serve as an impetus for continued research to treat BCCs at the level of cell membrane with antagonists and intracellularly to examine mediators of cytokines induced by NK1-Tr receptor (Table 2). Because PPT-A peptides could directly and indirectly act as angiogenic factors, targeting NK1-Tr might lead to decrease angiogenesis (26, 27).
The discovery of additional human tachykinins that interact with NK1 (28) adds to the complex role of NK1 in breast cancer development. In summary, this study adds to the potential roles of NK receptors in breast cancer development, and begins to form a network incorporating PPT-A peptides, cytokines, and NK1-Tr in breast cancer research. The observations in this study, if incorporated with previous studies on NK receptors in breast cancer development, show a bidirectional communication between NK1 and Tac1 genes in breast cancer. Constitutively produced PPT-A peptides in BCCs interact with NK receptors to induce the production of cytokines, and perhaps subsequent activation of multiple transcription factors with growth-promoting functions. The intracellular second messengers might converge, through unresolved pathways, to facilitate the expression of NK1-Tr. The transforming property of NK1-Tr receptor (Figs. 2 and 3) and its link to PPT-A gene products are significant findings that warrant future studies, including its role in breast cancer invasion and metastasis.
Acknowledgments
This work was supported by National Cancer Institute Grant CA-89868. This work is partial fulfillment of a Ph.D. thesis (S.H.R.).
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: BCC, breast cancer cell; NK, neurokinin.
References
- 1.Jemal, A., Murray, T., Samuels, A., Ghafoor, A., Ward, E. & Thun, M. J. (2003) CA Cancer J. Clin. 53, 5-26. [DOI] [PubMed] [Google Scholar]
- 2.Mundy, G. R. (2002) Nat. Rev. Cancer 2, 584-593. [DOI] [PubMed] [Google Scholar]
- 3.Singh, D., Joshi, D. D., Hameed, M., Qian, J., Gascón, P., Maloof, P. B., Mosenthal, A. & Rameshwar, P. (2000) Proc. Natl. Acad. Sci. USA 97, 388-393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Grotzer, M. A., Janss, A. J., Fung, K. M., Biegel, J. A., Sutton, L. N., Rorke, L. B., Zhao, H., Cnaan, A., Phillips, P. C., Lee, V. M. Y., et al. (2000) J. Clin. Oncol. 18, 1027-1035. [DOI] [PubMed] [Google Scholar]
- 5.Heppeler, A., Froidevaux, S., Eberle, A. N. & Maecke, H. R. (2000) Curr. Med. Chem. 7, 971-994. [DOI] [PubMed] [Google Scholar]
- 6.Hennig, I. M., Laissue, J. A., Horisberger, U. & Reubi, J. C. (1995) Int. J. Cancer 61, 786-792. [DOI] [PubMed] [Google Scholar]
- 7.Moharita, A., Harrison, J. S. & Rameshwar, P. (2004) Drug Des. Rev. Online 1, 297-302. [Google Scholar]
- 8.Heasley, L. E. (2001) Oncogene 20, 1563-1569. [DOI] [PubMed] [Google Scholar]
- 9.Rao, G., Patel, P. S., Idler, S. P., Maloof, P., Gascon, P., Potian, J. A. & Rameshwar, P. (2004) Cancer Res. 64, 2874-2881. [DOI] [PubMed] [Google Scholar]
- 10.Friess, H., Zhu, Z., Liard, V., Shi, X., Shrikhande, S. V., Wang, L., Lieb, K., Korc, M., Palma, C., Zimmermann, A., et al. (2003) Lab. Invest. 83, 731-742. [DOI] [PubMed] [Google Scholar]
- 11.Palma, C. & Maggi, C. A. (2000) Life Sci. 67, 985-1001. [DOI] [PubMed] [Google Scholar]
- 12.Leeman, S. E. & Ferguson, S. L. (2000) Neuropeptides 34, 249-254. [DOI] [PubMed] [Google Scholar]
- 13.Quartara, L. & Maggi, C. A. (1997) Neuropeptides 31, 537-563. [DOI] [PubMed] [Google Scholar]
- 14.Greco, S. J., Corcoran, K. E., Cho, K. J. & Rameshwar, P. (2004) Front. Biosci. 9, 1782-1793. [DOI] [PubMed] [Google Scholar]
- 15.Bandari, P. S., Qian, J., Oh, H. S., Potian, J. A., Yehia, G., Harrison, J. S. & Rameshwar, P. (2003) J. Neuroimmunol. 138, 65-75. [DOI] [PubMed] [Google Scholar]
- 16.Joshi, D. D., Dang, A., Yadav, P., Qian, J., Bandari, P. S., Chen, K., Donnelly, R., Castro, T., Gascón, P., Haider, A., et al. (2001) Blood 98, 2697-2706. [DOI] [PubMed] [Google Scholar]
- 17.Fong, T. M., Anderson, S. A., Yu, H., Huang, R. R. & Strader, C. D. (1992) Mol. Pharmacol. 41, 24-30. [PubMed] [Google Scholar]
- 18.Oh, H. S., Moharita, A., Potian, J. G., Whitehead, I. P., Livingston, J. C., Castro, T. A., Patel, P. S. & Rameshwar, P. (2004) Cancer Res. 64, 6327-6336. [DOI] [PubMed] [Google Scholar]
- 19.Dery, O., Defea, K. A. & Bunnett, N. W. (2001) Am. J. Physiol 280, C1097-C1106. [DOI] [PubMed] [Google Scholar]
- 20.Li, H., Leeman, S. E., Slack, B. E., Hauser, G., Saltsman, W. S., Krause, J. E., Blusztajn, J. K. & Boyd, N. D. (1997) Proc. Natl. Acad. Sci. USA 94, 9475-9480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Alblas. J., van Etten, I. & Moolenaar, W. H. (1996) EMBO J. 15, 3351-3360. [PMC free article] [PubMed] [Google Scholar]
- 22.Beaujouan, J. C., Torrens, Y., Saffroy, M., Kemel, M. L. & Glowinski, J. (2004) Peptides 25, 339-357. [DOI] [PubMed] [Google Scholar]
- 23.Mackay, H. J. & Twelves, C. J. (2003) Endocrine-Related Cancer 10, 389-396. [DOI] [PubMed] [Google Scholar]
- 24.MacKinnon, A. C., Waters, C., Rahman, I., Harani, N., Rintoul, R., Haslett, C., Sethi, T. (2000) Br. J. Cancer 83, 941-948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dimri, R., Sharabi, Y. & Shoham, J. (2000) J. Immunol. 164, 2479-2486. [DOI] [PubMed] [Google Scholar]
- 26.Pelletier, L., Angonin, R., Regnard, J., Fellmann, D. & Charbord, P. (2002) Br. J. Haematol. 119, 1083-1089. [DOI] [PubMed] [Google Scholar]
- 27.Fan, T. P., Hu, D. E., Guard, S., Gresham, G. A. & Watling, K. J. (1993) Br. J. Pharmacol. 110, 43-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pennefather, J. N., Lecci, A., Candenas, M. L., Patak, E., Pinto, F. M. & Maggi, C. A. (2004) Life Sci. 74, 1445-1463. [DOI] [PubMed] [Google Scholar]
- 29.Singletary, S. E., Allred, C., Ashley, P., Bassett, L. W., Berry, D., Bland, K. L., Borgen, P. I., Clark, G., Edge, S. B., Hayes, D. F., et al. (2002) J. Clin. Oncol. 20, 3628-3636. [DOI] [PubMed] [Google Scholar]