Abstract
Molecules exuded by plant roots are thought to act as signals to influence the ability of microbial strains to colonize the roots and to survive in the rhizosphere. Differential bacterial responses to signals from different plant species may mediate the selection of specific rhizosphere populations. Very little, however, is known about the effects of plant exudates on patterns of bacterial gene expression. Here, we have tested the concept that plant root exudates modulate expression of bacterial genes involved in establishing microbe-plant interactions. We have examined the influence on the Pseudomonas aeruginosa PA01 transcriptome of exudates from two varieties of sugarbeet that select for genetically distinct pseudomonad populations in the rhizosphere. The response to the two exudates showed only a partial overlap; the majority of those genes with altered expression was regulated in response to only one of the two exudates. Genes with altered expression included those with functions previously implicated in microbe-plant interactions, such as aspects of metabolism, chemotaxis and type III secretion, and a subset with putative or unknown function. Use of a panel of mutants with targeted disruptions allowed us to identify previously uncharacterized genes with roles in the competitive ability of P. aeruginosa in the rhizosphere within this subset. No genes with host-specific effects were identified. Homologues of the genes identified occur in the genomes of both beneficial and pathogenic root-associated bacteria, suggesting that this strategy may help to elucidate molecular interactions that are important for biocontrol, plant growth promotion, and plant pathogenesis.
Keywords: Pseudomonas, Rhizosphere colonization
Molecular signaling between microbes and their eukaryotic hosts plays a fundamental role both in pathogenesis and in the establishment of beneficial interactions between the partners. An understanding of these signaling processes and of the functions regulated may have profound implications for the design of new strategies to combat disease or to promote those interactions of benefit to the eukaryotic partner (1). Beneficial plant-microbial interactions in the rhizosphere can result in the promotion of plant health and development. Aggressive colonization and an ability to compete with resident microorganisms predisposes certain soilborne Pseudomonas strains to the establishment of such beneficial interactions (2, 3). Colonization is a multigenic process under the influence of many environmental factors. Molecules exuded by the plant root may act as signals to influence the ability of microbial strains to colonize the root and to survive in the rhizosphere (4-6). It is well established that different plant species can select for specific microbial populations in the rhizosphere (7-11). From recent data, it is now evident that this selection can extend to the plant variety or cultivar level (refs. 12 and 13; Fig. 4, which is published as supporting information on the PNAS web site). Plant selection of particular microbial communities is also thought to depend, at least in part, on the activation of specific patterns of gene expression in the microbe in response to molecular signals secreted from the plant (1).
Despite the general acceptance that plant-derived extracellular signals can influence the behavior of bacteria in the rhizosphere, very little is known about the effects of these signals on the patterns of bacterial gene expression and the role of those genes with altered expression in the plant-microbe interaction process. Here we address this issue through the use of a model system to study plant-microbe interactions that exploits Pseudomonas aeruginosa strain PA01 and two varieties of sugarbeet (Beta vulgaris L.), variety (var.) Celt and var. Roberta (hereafter var. Celt and var. Roberta). The choice of plant hosts was based on previous field experiments in which it was demonstrated that var. Celt and var. Roberta select for different culturable pseudomonad populations in the rhizosphere (ref. 12; Fig. 4). P. aeruginosa is a versatile Gram-negative bacterium known to be an opportunistic pathogen of humans but is also found in association with plants (14-16). Furthermore, we have shown that in a gnotobiotic system, P. aeruginosa PA01 colonizes the roots of var. Celt and var. Roberta to similar levels as the biocontrol bacterium P. fluorescens F113 and can successfully compete with P. fluorescens in colonization under these growth conditions (Table 2, which is published as supporting information on the PNAS web site). The availability of the complete genome sequence for P. aeruginosa PA01 (17) and commercial gene arrays has facilitated investigations of global gene expression in this organism.
Our approach to identifying previously uncharacterized bacterial genes that may play a role in colonization and competition in the rhizosphere has been to examine the global influence of the root exudates from the two varieties of sugarbeet on the transcriptome of P. aeruginosa PA01, followed by functional analysis of (selected) genes with altered expression patterns. (This strategy is depicted in Fig. 5, which is published as supporting information on the PNAS web site.) By analysis of responses to different root exudates, we anticipated that it would be possible to identify genes that might have varietal-specific effects as well as those that might play a role in colonization of both B. vulgaris varieties. The data obtained indicate that the root exudates from var. Celt and var. Roberta had significant, partially overlapping, effects on the pattern of gene transcription. Genes with altered expression included those with functions previously implicated in microbe-plant interactions as well as a number with only putative or unknown function. Functional analysis of a subset of these genes in gnotobiotic tests allowed us to identify previously uncharacterized genes with roles in the competitive ability of P. aeruginosa in the rhizosphere, although no gene giving a host-specific effect has yet been identified. Homologues of the genes identified by using this model system occur in the genomes of both beneficial and pathogenic root-associated bacteria, indicating that this approach may facilitate a broader understanding of the molecular determinants underpinning complex interactions in the rhizosphere.
Materials and Methods
Bacterial Strains and Media. The wild-type bacterial strain used was P. aeruginosa PA01 (provided by B. Iglewski, University of Rochester, Rochester, NY). Tetracycline-resistant transposon insertion mutants generated in this strain (18) were obtained from University of Washington Genome Center (www.genome.washington.edu/uwgc/Pseudomonas/index.cfm). Transposon type, insertion, location, and orientation were confirmed for each of the mutants. The media used were LB broth (1% tryptone/0.5% yeast extract/0.5% NaCl) and CAA broth (2 g/liter Casamino acids/0.36 g/liter K2HPO4/10 mM MgSO4).
Root Exudate Collection. Root exudates were collected separately from sugarbeet (B. vulgaris L.) varieties var. Celt (Syngenta, Landskrona, Sweden) and var. Roberta (KWS, Einbeck, Germany) grown in gnotobiotic (axenic) microcosms comprising 50 ml of sterilized vermiculite in Magenta GA-7 vessels (Sigma). Forty milliliters of 1/10-strength sterile Hoagland's nutrient solution (19) was added to each vessel before sowing of the seeds. Seeds were surface-sterilized for 10 min in sodium hypochlorite (10% wt/wt) and rinsed thoroughly before sowing (25 seeds per vessel) under aseptic conditions. No seeds were sown in the control microcosms. Plant and control microcosms were arranged in a replicate randomized block design and maintained for 16 days at 12°C in a 16-h light/8-h dark regime. The lighting was provided by cool white fluorescent strip lamps mounted 34 cm above the top of the magenta tubs. The wattage was 36 W (240 V), and the photon flux density was 960 μmol·m-2·s-1. After this time, shoots were removed and root exudates and control eluate (liquid from control microcosms) were collected destructively via vacuum filtration. Each vessel was rinsed with 5 ml of sterile distilled water, and the total volume obtained was 10 ml. Plating of both the root material and exudate on standard microbiological medium indicated no microbial contamination was present. Root exudates were filter-sterilized (0.22-μm filter, Millipore) and stored at -20°C in the dark until use. Comparison of HPLC profiles of UV absorbing compounds from freshly isolated, and stored exudates gave no indication of breakdown of these compounds.
Transcriptome Profiling and Microarray Analysis. The influence of the root exudates collected from var. Celt and var. Roberta on the transcriptome of P. aeruginosa PA01 was profiled by using PA01 Affymetrix GeneChips. P. aeruginosa PA01 cultures (initial OD at 600 nm of 0.1) were established in 90 ml of CAA medium supplemented with 10 ml of exudates from var. Celt or var. Roberta or 10 ml of control eluate. Ten milliliters of root exudates were selected for use, because at this level, no discernible effect on bacterial growth was observed. These cultures were grown with shaking at 37°C, and cells were harvested after 6 h when the culture had reached an OD at 600 nm of 0.6. RNA was extracted from the harvested bacterial cells with the RNeasy Midi Column kit (Qiagen Sciences, Germantown, MD), converted to cDNA (RT SuperScript II, Invitrogen), and fragmented, and the fragments were labeled with Biotin-ddUTP (EnzoBioArray terminal labeling kit, Invitrogen). Hybridizations to the GeneChips were carried out at Cambridge University and Affymetrix. The transcriptome profiling experiments was replicated in a miame compliant fashion (20). Microarray data were analyzed by using genedirector software (Version 4.1.5 standard addition, BioDiscovery, El Segundo, CA). Validation of the transcriptome profiling experiments was carried out by semiquantitative RT-PCR on selected candidate genes identified from the transcriptome profiling experiments. P. aeruginosa PA01 was grown to an OD at 600 nm of 0.6 as for the transcriptome profiling experiment. RNA was extracted from the cell pellets (RNeasy Mini Kit, Qiagen Sciences) and cDNA synthesized (First Strand cDNA Synthesis Kit for RT-PCR, Roche) followed by RT-PCR using specifically designed primers. The gene designated PA0005 was used as a control for standardized RNA levels. RT-PCR products were sequenced to confirm amplification of genes of interest.
Functional Genomic Analysis. The growth of the P. aeruginosa mutants was compared with wild type in LB and CAA broth by following the OD at 600 nm. Mutants were also coinoculated with the wild type in LB and CAA broth at a ratio of 1:1, and growth in competition was assessed by plating on LB agar with and without tetracycline (60 μg·ml-1). In both cases, inoculation was with an overnight culture diluted with 1/4 Ringer's solution to OD at 600 nm of 0.001. Only mutants that had growth rates indistinguishable from the wild type in individual cultures and that could compete equally with the wild type in binary cultures were selected for further analysis in plant trials. For plant trials, surface-sterilized seeds were inoculated with each mutant and the wild-type strain both separately and in 1:1 combination. Seeds were soaked in the bacterial suspensions at an inoculum density of 108 colony-forming units per ml and agitated for 5 min before sowing in the gnotobiotic system in Magenta GA-7 vessels. Under these conditions, the level of inoculum on each seed at day 0 of the trial was 106 colony-forming units; in coinoculation experiments, the wild type and mutant were each present on the seed at this level. Four replicates were set up per treatment with 12 seeds sown per Magenta GA-7 vessel. Growth conditions for the gnotobiotic plant trials are as described for the root exudate collection. Nutrient limitation in the gnotobiotic plant system was prevented by the addition of 1/10-strength Hoagland's nutrient solution (2 ml per Magenta vessel) aseptically at day 24 of the trial. One seed or seedling from each replicate was destructively sampled at day 1, 16, 20, 33, and 40 of the trial. Seeds or roots of inoculated plants were harvested and loosely adhering vermiculite was removed before they were placed in 1/4-strength Ringer's solution and vigorously shaken for 5 min. The resulting suspensions were serially diluted in 1/4 Ringer's solution before replicate plating on LB agar with and without tetracycline (60 μg·ml-1). Bacterial numbers were expressed as colony-forming units per seed for day 1, and colony-forming units per gram of dry weight root for later time points. Root and shoot length and plant biomass of plants colonized by PA01 were indistinguishable from those of uninoculated plants of both varieties indicating that PA01 does not have a negative influence on plant health. The insertion of the tetracycline cassette carried by the transposons used to create the mutants (18) did not reduce fitness in P. aeruginosa PA01 because several mutants tested were able to colonize the plant hosts at wild-type levels.
Statistical Analysis. Data from the plant trials was converted to logarithmic values, confirmed as parametric, and analyzed by using a linear model univariate analysis of variance with significant differences calculated at P ≤ 0.05 (least squares deconvolution). Statistical analysis was carried out by using spss software for Macintosh (Version 11.02 for Mac os x, SPSS, Chicago). A WT:mutant colonization ratio of greater than 90:10 was used as a cut-off point to indicate a significantly reduced competition phenotype in the plant assays.
Results and Discussion
Profiling Changes in the P. aeruginosa PA01 Transcriptome in Response to Plant Root Exudates. The first part of our strategy to identify genes involved in plant-microbe interactions was to examine the influence on the transcriptome of P. aeruginosa PA01 of root exudates from sugarbeet varieties Celt and Roberta. Analysis of the transcriptome profiles showed that the expression of the number of genes was significantly altered (P ≤ 0.05) in response to root exudates from the two varieties when compared with the control eluate (Table 1; see also Figs. 6-8, which are published as supporting information on the PNAS web site). A total of 516 genes representing 9.3% of the transcriptome were significantly altered in response to var. Celt exudate with approximately equal numbers being up- and down-regulated. In response to var. Roberta exudate, 451 genes showed altered expression and, again, similar numbers were up- and down-regulated. The response to the two exudates showed only a partial overlap, however. Whereas 134 genes responded to both exudates, the majority of genes with altered expression responded to only one of the two exudates (Table 1). Another six genes showed divergent patterns of transcription in response to the two exudates. A significant proportion of the genes with altered expression (≈70%) encoded either proteins with only putative functions or hypothetical proteins (Table 1) as classified in the P. aeruginosa PA01 genome project database at www.pseudomonas.com (17).
Table 1. Effects of root exudates from sugarbeet var. Celt and Roberta on gene expression in P. aeruginosa PA01.
| No. of P. aeruginosa PA01 genes that are significantly altered in expression in response to root exudates
|
|||||
|---|---|---|---|---|---|
| Level of gene expression relative to control | var. Celt | var. Roberta | var. Celt and var. Roberta | var. Celt but not var. Roberta | var. Roberta but not var. Celt |
| Increased | 259 (162) | 235 (158) | 80 (46) | 179 (55) | 149 (52) |
| Decreased | 257 (206) | 216 (153) | 54 (37) | 197 (45) | 162 (40) |
Gene expression in P. aeruginosa PA01 in response to root exudates was compared with a control eluate (see Materials and Methods) by using Affymetrix GeneChips. Numbers of genes with significantly altered levels of expression (P ≤ 0.05) from the control are shown. Values in parentheses denote the number of genes with putative or unknown function as assigned in the Pseudomonas Genome Project database (www.pseudomonas.com). In total, 134 genes were significantly altered in response to both exudates. Another six genes were up-regulated in response to var. Roberta but down-regulated in response to var. Celt exudates, and these are not included in Table 1. Transcriptome profiling experiments were repeated in a miame-compliant fashion.
Expression Patterns of Genes with Known Function. Many of the genes of known function whose expression was up-regulated in response to the root exudates were involved in aspects of metabolism such as aromatic compound catabolism, energy generation, and amino acid biosynthesis and metabolism (Fig. 1). This finding is perhaps not surprising because monosaccharides, amino acids, and organic acids are thought to be the major constituents of plant root exudates (3, 5, 21). Lugtenberg and Dekkers (21) have shown that utilization of organic acids by P. fluorescens is the nutritional basis governing the ability of this organism to colonize tomato roots. Genes in pseudomonads encoding proteins with functions in nutrient acquisition and in energy generation are up-regulated in the rhizosphere or when bacteria were exposed to the soil environment as revealed by in vivo expression technology-based approaches (22, 23). Differences in expression of these genes in response to the var. Celt and var. Roberta exudates could reflect differences in the levels of metabolites in the exudates. We have established, for example, that the exudates from var. Celt and var. Roberta have different profiles of UV-absorbing compounds as revealed by reverse phase HPLC (Fig. 9, which is published as supporting information on the PNAS web site).
Fig. 1.
A subset of P. aeruginosa PA01 genes with known functions that show significant alteration in expression (at P ≤ 0.05) in response to root exudates from B. vulgaris varieties Celt and Roberta. The first column represents expression levels of genes in the control where +++ indicates high, ++ denotes medium, and + is low. The second and third columns show the relative expression level in response to exudates from var. Celt and var. Roberta. Red represents up-regulation, whereas green denotes down-regulation of gene expression. White indicates genes whose expression levels are not significantly different from the control. Genes represented have a fold change in expression ranging from 1.1 to 7.0.
Interestingly, three genes involved in chemotaxis, cheY encoding a two-component response regulator, cheA encoding a chemotaxis response regulator, and pctA encoding a chemotactic transducer protein, were down-regulated by var. Celt exudate but not significantly altered in expression by var. Roberta exudates (Fig. 1). Again, this finding could reflect differences in levels of key attractants in the two exudates that could conceivably play a role in host selection because chemotaxis plays an important role in successful colonization of the rhizosphere (21, 24-28). Some genes encoding proteins with functions previously implicated in microbial-plant interactions were down-regulated in response to both root exudates (Fig. 1). Genes in this category included algD, involved in alginate biosynthesis, pilK involved in twitching motility, and metE, involved in amino acid biosynthesis and metabolism. The reasons for these effects are unknown. Previously, Rainey (29) has shown that genes involved in type III secretion in P. fluorescens SBW25 are induced in the sugarbeet rhizosphere. In our experiments, genes encoding translocation proteins involved in type III secretion (pscR, pscQ, and pscP) showed a complex pattern of regulation in response to the root exudates. Such complexities in regulation of bacterial gene expression in response to plant root exudates might be expected because exudates comprise a complex mixture of molecular signals and metabolites (15, 21, 30) and potential inhibitory compounds (31, 32).
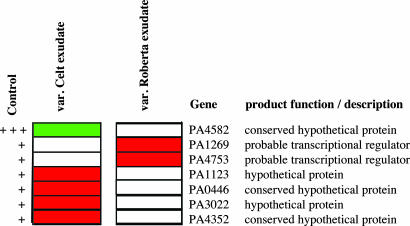
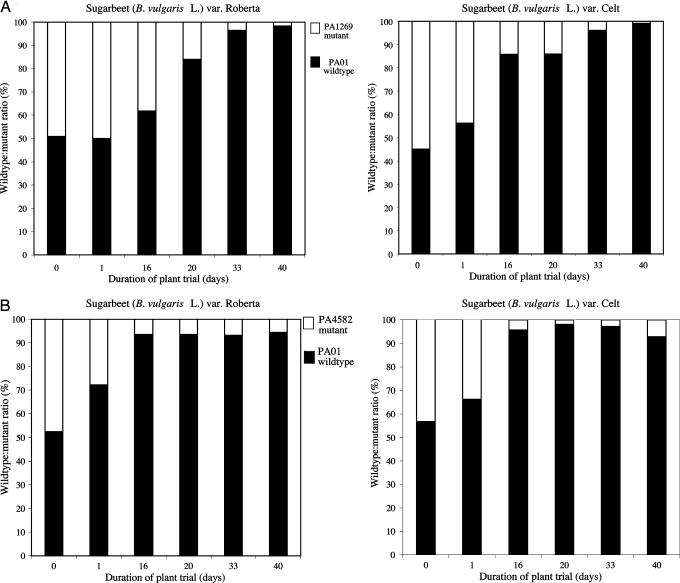
Functional Analysis Identifies Previously Uncharacterized Genes Involved in Bacterial Colonization and Competition in the Rhizosphere. Differential expression was also seen in those genes annotated as having putative or unknown functions (Table 1). A subset of genes that was differentially regulated by the two root exudates was identified. We hypothesized that some of these differentially regulated genes would have roles in colonization of specific B. vulgaris varieties. Seven differentially regulated genes with putative or unknown functions were selected for further functional analysis (Fig. 2). Mutants with defined transposon insertions in each of these genes were obtained from the University of Washington genome center. Five of these mutants (with insertions in PA1269, PA4582, PA4753, PA3022, and PA4352) exhibited growth characteristics indistinguishable from the wild type in LB and CAA broth and were able to compete successfully with the wild type in vitro (data not shown). Colonization and ability to compete in the rhizosphere of var. Celt and var. Roberta were assessed for these mutant strains. In the first round of experiments, these strains were inoculated individually. In the second round of experiments, each mutant strain was examined in competition with the wild type (see Materials and Methods). Each mutant inoculated separately had similar colonization ability and kinetics to the wild type on both varieties. In competition assays, however, three mutants (with insertions in PA1269, PA4582, and PA4753) had reduced ability to compete with the wild type in the rhizosphere of both varieties (Fig. 3; see also Table 3, which is published as supporting information on the PNAS web site). Two distinct phenotypes were observed. Disruption of PA1269 significantly reduced the competitive ability at the later phases of the plant trial (Fig. 3A), whereas the effects of disruption of PA4582 (Fig. 3B) and PA4753 (Fig. 10, which is published as supporting information on the PNAS web site) were seen on younger plants (day 16). PA1269 and PA4753 encode putative transcriptional regulators, whereas PA4582 encodes a conserved hypothetical protein (www.pseudomonas.com). It is predicted that PA4582 is likely to be in an operon with PA4583, and further fine structural analysis is required to rule out any polar effects that may be due to the transposon insertion. All of the other genes whose function was analyzed are not part of an operon. It is noteworthy that although only a relatively small number of mutants were tested, several previously uncharacterized genes with a role in determining the competitiveness of P. aeruginosa PA01 in the rhizosphere were discovered. Using a bioinformatic approach, we identified homologues of these genes in the genomes of other rhizosphere microorganisms, including the beneficial P. fluorescens and the plant pathogen Ralstonia solanacearum. Furthermore, homologues of other P. aeruginosa genes of unknown function that show altered expression were also identified. This finding suggests that our model system may help to elucidate molecular interactions that are important both for biocontrol and plant growth promotion as well as plant pathogenesis.
Fig. 2.
A subset of P. aeruginosa PA01 genes with putative or unknown functions that show significant alteration in expression levels (at P ≤ 0.05) in response to root exudates B. vulgaris varieties Celt and Roberta. The first column represents expression levels of genes in the control where +++ indicates high, ++ denotes medium, and + is low. The second and third columns show the relative expression level in response to exudates from var. Celt and var. Roberta. Red represents up-regulation, whereas green denotes down-regulation of gene expression. White indicates genes whose expression levels are not significantly different from the control. Genes represented have a fold change in expression ranging from 1.1 to 4.4.
Fig. 3.
Mutation of PA1269 (A) encoding a putative transcriptional regulator and PA4582 (B) encoding a conserved hypothetical protein reduces the competitive ability of P. aeruginosa PA01 in the rhizosphere of both sugarbeet varieties Roberta and Celt. Mutant strains were coinoculated with the wild type at a ratio of 1:1 on the sugarbeet seeds, which were then sown in gnotobiotic-based microcosms.
Evaluation and Comparison with Other Techniques to Study Rhizosphere Interactions. The approach we adopted has been successful in allowing identification of several previously uncharacterized genes involved in bacterial colonization in the rhizosphere. Nevertheless, it is important to consider both limitations of this approach and advantages compared with other techniques currently in use to investigate rhizosphere interactions. One limitation of our system is that the transcriptome profiling comparisons are performed from cultures grown in exudate-supplemented CAA medium. This system was chosen to allow extraction of sufficient amounts of RNA for transcriptome analysis. It is possible that some of the effects of the exudates may be overwhelmed or inhibited by components in CAA medium and, consequently, will not appear in our analysis as genes with altered expression. A second limitation is that by using 10 ml of exudate in a 100-ml culture, we may have diluted some components to a level at which they no longer influence bacterial gene expression. A third important observation is that the exudates were recovered from plants growing in a gnotobiotic system. It is known that rhizosphere microbes influence the composition of plant root exudates by affecting root cell leakage, cell metabolism, and plant nutrition status (11). Inoculation and colonization of Arabidopsis by P. fluorescens triggered the up-regulation of plant genes involved in metabolism, signal transduction, stress response, and putative auxin-regulated genes (33). Furthermore, P. aeruginosa makes N-acyl homoserine lactone signaling compounds that are known to elicit the release of exudate compounds from roots (34). Further developments of our approach will include the use of exudates from plants colonized with rhizosphere microflora. A fourth notable point is that although the transcriptome profiling identified genes that were differentially regulated by exudates from different varieties, analysis of mutants with disruptions in some of those genes did not show variety-specific colonization effects. It should be recognized, however, that many genes showed differential expression, and it is likely that host selectivity is a multifactorial response that may be observed only under specific growth conditions, in strains deleted for a number of genes. We consider it very significant that genes only showing alteration in response to a single exudate in our experimental system were compromised for competitive colonization in the rhizospheres of both sugarbeet varieties. Furthermore, an examination of the expression of genes that might be expected from other work to have a role in colonization (see Fig. 2A) shows that the majority of these genes also show alterations in response to only one of the two exudates. In combination, these data illustrate the importance of including more than one variety/species in future experiments designed to identify genes important for rhizosphere function.
In vivo expression technology is now established as a method of analyzing bacterial gene expression in the rhizosphere as an approach for the identification of genes involved in microbe-plant interactions (22, 23, 29, 35). One clear advantage of in vivo expression technology is that investigations are carried out in the appropriate environment in the organism of interest; there is no extrapolation from in vitro model systems. The technique as normally used, however, excludes genes that show significant expression in vitro in minimal medium and identifies only those that are up-regulated in the rhizosphere. In contrast, our transcriptome profiling approach allows for the identification of genes that are both up- and down-regulated irrespective of the level of expression in minimal medium. Because it is plausible that down-regulation of certain genes may be required for successful colonization, the transcriptome profiling approach would appear to offer a broader insight. Thus, the two approaches can be considered complementary methods that contribute to the identification of genes that may be important for microbe-plant interaction.
Transcriptome profiling of bacteria isolated from the roots of plants also offers another strategy to examine the influence of the plant host on bacterial gene expression. However, this approach, as applied to beneficial (in contrast to symbiotic) interactions, is currently limited by technical issues such as the quantity and quality of RNA that can be extracted from bacteria in the rhizosphere. Transcriptome-based studies have explored global gene expression during the symbiotic interaction involving Sinorhizobium meliloti (36, 37). The construction of a dual-genome symbiosis chip housing the genome of S. melitoti and the host plant Medicago truncatula has allowed molecular analysis of the symbiosis to be extended to include gene expression by both partners simultaneously (38). These studies have provided valuable insights into the complex molecular signaling that exist between the prokaryotic and eukaryotic partner.
Concluding Remarks. Transcriptome profiling of bacterial response to exudates has directed our functional genomic analysis of rhizosphere colonization to reveal several previously uncharacterized genes involved in this process. The effects of mutation of these genes are observed only in competition with the wild type and are not seen in individual inoculations. The ability of bacteria to compete in the rhizosphere is nevertheless fundamental to the colonization of beneficial and pathogenic organisms with associated consequences for plant health. Further analysis of these and other mutants will address issues of competition through the use of soil-based microcosm systems and spatial aspects of root colonization through the use of strains carrying autofluorescent protein markers (39). Overall, this work may be expected to broaden our understanding of both beneficial and pathogenic bacterial responses to a plant host and may also help to define whether there are unified principles relevant for the interaction of bacteria with other eukaryotic hosts including humans.
Supplementary Material
Acknowledgments
We thank Claire Adams for useful advice and discussions, Pat Higgins for technical advice, and the European Union Pseudomics project consortia for fruitful discussion and valuable scientific comment. This work was supported, in part, by European Union Grant QLRT-2001-00914 (Pseudomics), together with Higher Education Authority of Ireland Grants PRTI2 and PRTI3 and Science Foundation of Ireland (SFI) Grant 04/BR/B0597. J.M.D. is supported by an SFI Principal Investigator Award.
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviation: var., variety.
References
- 1.Morrissey, J. P., Dow, J. M., Mark, G. L. & O'Gara, F. (2004) EMBO Rep. 5, 922-926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chin-A-Woeng, T. F., Bloemberg, G. V., Mulders, I. H., Dekkers, L. C. & Lugtenberg, B. J. (2000) Mol. Plant-Microbe Interact. 13, 1340-1345. [DOI] [PubMed] [Google Scholar]
- 3.Lugtenberg, B. J., Dekkers, L. & Bloemberg, G. V. (2001) Annu. Rev. Phytopathol. 39, 461-490. [DOI] [PubMed] [Google Scholar]
- 4.Kuiper, I., Kravchenko, L. V., Bloemberg, G. V. & Lugtenberg, B. J. (2002) Mol. Plant-Microbe Interact. 15, 734-741. [DOI] [PubMed] [Google Scholar]
- 5.Simons, M., van der Bij, A. J., Brand, I., de Weger, L. A., Wijffelman, C. A. & Lugtenberg, B. J. (1996) Mol. Plant-Microbe Interact. 9, 600-607. [DOI] [PubMed] [Google Scholar]
- 6.Gough, C., Galera, C., Vasse, J., Webster, G., Cocking, E. C. & Denarie, J. (1997) Mol. Plant-Microbe Interact. 10, 560-570. [DOI] [PubMed] [Google Scholar]
- 7.Press, M. C. & Phoenix, G. K. (2005) New Phytol. 166, 737-751. [DOI] [PubMed] [Google Scholar]
- 8.Kuske, C. R., Ticknor, L. O., Miller, M. E., Dunbar, J. M., Davis, J. A., Barns, S. M. & Belnap, J. (2002) Appl. Environ. Microbiol. 68, 1854-1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Landa, B. B., Mavrodi, O. V., Raaijmakers, J. M., McSpadden Gardener, B. B., Thomashow, L. S. & Weller, D. M. (2002) Appl. Environ. Microbiol. 68, 3226-3237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Smalla, K., Wieland, G., Buchner, A., Zock, A., Parzy, J., Kaiser, S., Roskot, N., Heuer, H. & Berg, G. (2001) Appl. Environ. Microbiol. 67, 4742-4751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yang, C. H. & Crowley, D. E. (2000) Appl. Environ. Microbiol. 66, 345-351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mark, G. L., Morrissey, J., Baysse, C., Sweeney, P. & O'Gara, F. (2004) in IS-MPMI 11th International Congress on Molecular Plant-Microbe Interactions, eds. Tikonovich, I., Lugtenberg, B. J. & Provorov, N. (Am. Phytopathol. Soc., St. Paul), Vol. 4, pp. 585-588. [Google Scholar]
- 13.Mazzola, M., Funnell, D. L. & Raaijmakers, J. M. (2004) Microbial. Ecol. 48, 338-348. [DOI] [PubMed] [Google Scholar]
- 14.Goldberg, J. B. (2000) Trends Microbiol. 8, 55-57. [DOI] [PubMed] [Google Scholar]
- 15.Walker, T. S., Bais, H. P., Deziel, E., Schweizer, H. P., Rahme, L. G., Fall, R. & Vivanco, J. M. (2004) Plant Physiol. 134, 320-331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Anjaiah, V., Cornelis, P. & Koedam, N. (2003) Can. J. Microbiol. 49, 85-91. [DOI] [PubMed] [Google Scholar]
- 17.Stover, C. K., Pham, X. Q., Erwin, A. L., Mizoguchi, S. D., Warrener, P., Hickey, M. J., Brinkman, F. S., Hufnagle, W. O., Kowalik, D. J., Lagrou, M., et al. (2000) Nature 406, 959-964. [DOI] [PubMed] [Google Scholar]
- 18.Jacobs, M. A., Alwood, A., Thaipisuttikul, I., Spencer, D., Haugen, E., Ernst, S., Will, O., Kaul, R., Raymond, C., Levy, R., et al. (2003) Proc. Natl. Acad. Sci. USA 100, 14339-14344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hoaglands, D. R. & Arnon, D. I. (1938) Circ.-Calif. Agric. Exp. Sta. 347.
- 20.Brazma, A., Hingamp, P., Quackenbush, J., Sherlock, G., Spellman, P., Stoeckert, C., Aach, J., Ansorge, W., Ball, C. A., Causton, H. C., et al. (2001) Nat. Genet. 29, 365-371. [DOI] [PubMed] [Google Scholar]
- 21.Lugtenberg, B. J. & Dekkers, L. C. (1999) Environ. Microbiol. 1, 9-13. [DOI] [PubMed] [Google Scholar]
- 22.Silby, M. W. & Levy, S. B. (2004) J. Bacteriol. 186, 7411-7419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ramos-Gonzalez, M. I., Campos, M. J. & Ramos, J. L. (2005) J. Bacteriol. 187, 4033-4041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.de Weert, S., Vermeiren, H., Mulders, I. H., Kuiper, I., Hendrickx, N., Bloemberg, G. V., Vanderleyden, J., De Mot, R. & Lugtenberg, B. J. (2002) Mol. Plant-Microbe Interact. 15, 1173-1180. [DOI] [PubMed] [Google Scholar]
- 25.de Weert, S., Kuiper, I., Lagendijk, E. L., Lamers, G. E. & Lugtenberg, B. J. (2004) Mol. Plant-Microbe Interact. 17, 1185-1191. [DOI] [PubMed] [Google Scholar]
- 26.Kravchenko, L. V. & Makarova, N. M. (1993) Microbiology, 324-327.
- 27.O'Sullivan, D. J. & O'Gara, F. (1992) Microbiol. Rev. 56, 662-676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Walsh, U. F., Morrissey, J. P. & O'Gara, F. (2001) Curr. Opin. Biotechnol. 12, 289-295. [DOI] [PubMed] [Google Scholar]
- 29.Rainey, P. B. (1999) Environ. Microbiol. 1, 243-257. [DOI] [PubMed] [Google Scholar]
- 30.Czarnota, M. A., Rimando, A. M. & Weston, L. A. (2003) J. Chem. Ecol. 29, 2073-2083. [DOI] [PubMed] [Google Scholar]
- 31.Bais, H. P., Prithiviraj, B., Jha, A. K., Ausubel, F. M. & Vivanco, J. M. (2005) Nature 434, 217-221. [DOI] [PubMed] [Google Scholar]
- 32.Park, W., Jeon, C. O., Cadillo, H., DeRito, C. & Madsen, E. L. (2004) Appl. Microbiol. Biotechnol. 64, 429-435. [DOI] [PubMed] [Google Scholar]
- 33.Wang, Y., Ohara, Y., Nakayashiki, H., Tosa, Y. & Mayama, S. (2005) Mol. Plant-Microbe Interact. 18, 385-396. [DOI] [PubMed] [Google Scholar]
- 34.Mathesius, U., Mulders, S., Gao, M., Teplitski, M., Caetano-Anollés, G., Rolfe, B. G. & Bauer, W. D. (2003) Proc. Natl. Acad. Sci. USA 100, 1444-1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rediers, H., Rainey, P. B., Vanderleyden, J. & De Mot, R. (2005) Microbiol. Mol. Biol. Rev. 69, 217-261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ampe, F., Kiss, E., Sabourdy, F. & Batut, J. (2003) Genome Biol. 4, R15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Becker, A., Bergès, H., Krol, E., Bruand, C., Rüberg, S., Capela, D., Lauber, E., Meilhoc, E., Ampe, F., de Bruijn, F. J., et al. (2004) Mol. Plant-Microbe Interact. 17, 292-303. [DOI] [PubMed] [Google Scholar]
- 38.Barnett, M. J., Toman, C. J., Fisher, R. F. & Long, S. R. (2004) Proc. Natl. Acad. Sci. USA 101, 16636-16641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bloemberg, G. V., O'Toole, G.A., Lugtenberg, B. J. & Kolter, R. (1997) Appl. Environ. Microbiol. 63, 4543-4551. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.