Abstract
Alzheimer's disease is the most fatal neurodegenerative disorder wherein the process of amyloid-β (Aβ) amyloidogenesis appears causative. Here, we present the 3D structure of the fibrils comprising Aβ(1–42), which was obtained by using hydrogen-bonding constraints from quenched hydrogen/deuterium-exchange NMR, side-chain packing constraints from pairwise mutagenesis studies, and parallel, in-register β-sheet arrangement from previous solid-state NMR studies. Although residues 1–17 are disordered, residues 18–42 form a β-strand–turn–β-strand motif that contains two intermolecular, parallel, in-register β-sheets that are formed by residues 18–26 (β1) and 31–42 (β2). At least two molecules of Aβ(1–42) are required to achieve the repeating structure of a protofilament. Intermolecular side-chain contacts are formed between the odd-numbered residues of strand β1 of the nth molecule and the even-numbered residues of strand β2 of the (n – 1)th molecule. This interaction pattern leads to partially unpaired β-strands at the fibrillar ends, which explains the sequence selectivity, the cooperativity, and the apparent unidirectionality of Aβ fibril growth. It also provides a structural basis for fibrillization inhibitors.
Keywords: NMR, hydrogen/deuterium exchange
Pathological protein fibril formation can be observed in more than a dozen usually fatal human diseases, including Alzheimer's disease (AD) (1). Therefore, the fibrils' 3D structures are a focus of interest both for a molecular understanding of amyloidogenesis and for the development of innovative therapeutic and diagnostic approaches. Solid-state NMR studies have contributed substantially to the understanding of amyloid fibrils (2, 3). Only one 3D structure of an engineered amyloid fibril has yet been determined (4), owing in part to fibrils' noncrystalline, insoluble, and mesoscopically heterogeneous nature, which makes them difficult to access by established structural techniques such as x-ray crystallography or solution-state NMR.
The major component of AD-associated amyloid plaques is the 39- to 42-residue-long amyloid-β (Aβ) peptide, which is generated from the amyloid precursor protein by the proteolytic activities of β- and γ-secretase (5, 6). The Aβ(1–42) fragment is the dominant Aβ species in the amyloid plaques of AD patients, and, compared with Aβ(1–40), it displays a dramatically increased propensity to form amyloid fibrils in vitro (7–9). Several important structural characteristics of Aβ(1–42) fibrils have been determined (2), establishing that Aβ(1–42) fibrils form a cross-β structure (10) that contains parallel, in-register β-sheets (11). Here, we combined the available structural information on Aβ(1–42) fibrils with detailed amide hydrogen-exchange measurements, pairwise mutagenesis, thioflavin T (ThioT) binding, and high-resolution cryoelectron microscopy to determine a 3D structure of Aβ(1–42) fibrils.
Materials and Methods
H/D Exchange. For the H/D-exchange studies, recombinant 15N-labeled Aβ(1–42) fibrils were used. The fibrils were resuspended in 10 mM Tris DCl (pH* = 7.7) containing 150 mM sodium chloride and D2O as the solvent. At suitable intervals, aliquots of the fibrillar suspension were sedimented at 100,000 × g for 10 min. The pellet was frozen in pure D2O on liquid nitrogen and lyophylized to quench H/D exchange. The fibrils were dissociated in DMSO containing 0.01% trifluoroacetic acid and, within 30 s, diluted 10-fold in DMSO. Immediately thereafter, a series of 80 2D [15N,1H]-correlation spectra were measured for 4 h.
Structure Calculation. For the cyana structure calculation of an Aβ(1–42) pentamer, we used the following upper limit restraints (UPLs): (i) two long-range UPLs per monomer of dmax(F19Cα,G38Cα) = 8.0 Å and dmax(A21Cα,V36Cα) = 8.0 Å from pairwise mutagenesis data, (ii) the intermolecular salt bridge between residues D23 and K28 from pairwise mutagenesis was implemented by the intermolecular UPL of dmax(D23Cγ,K28Nζ) = 2.8 Å, and (iii) H-bond constraints for residues 18–26 and residues 31–42 from H/D exchange were set to be consistent with intermolecular, parallel, in-register β-sheets (11). H bonds were implemented by two UPLs per H bond [dmax(HN,O) = 2.0 Å and dmax(N,C′) = 2.9 Å]. The H bonds also provided backbone torsion angle constraints (–164 ≤ Φ ≤ –74 and 68 ≤ Ψ ≤ 158) for residues 18–26 and residues 31–42.
Further details are provided in Supporting Text, which is published as supporting information on the PNAS web site.
Results and Discussion
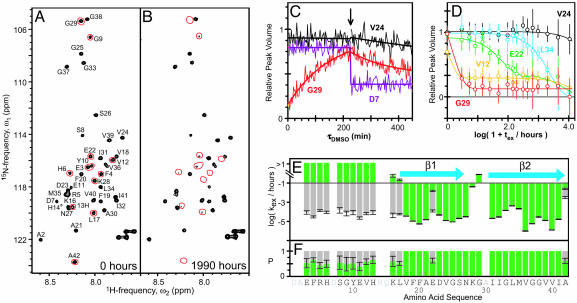
Identification of β-Sheets. For the following structural studies, amyloid fibrils were produced from recombinant 35MoxAβ(1–42) peptide that contains a methionine sulfoxide at position 35 (see Supporting Text). We first identified the sequence-specific positions of regular secondary structure elements, which are stabilized by hydrogen bonds of the peptide backbone. Using the quenched hydrogen-exchange NMR technique (12, 13) for 35MoxAβ(1–42) fibrils (see Supporting Text), the protonation state of most residues could be retained after dissociation into monomers (Fig. 1 C and D), thus allowing for the accurate determination of backbone amide exchange rates. Fig. 1 A and B show the [15N-1H]-correlation NMR spectra of 35MoxAβ(1–42) monomers in DMSO corresponding to fully protonated and partially H/D-exchanged 35MoxAβ(1–42) fibrils, respectively. After exchange of the fibrils in a D2O buffer for 1,990 h, ≈30% of the peaks were absent from the spectrum (Fig. 1B; compare Fig. 1 A). This observation is consistent with previous qHX (quenched hydrogen/deuterium exchange) mass spectrometry studies and the qualitative notion that 35MoxAβ(1–42) fibrils contain a highly protected core structure (14, 15).
Fig. 1.
Sequence-specific determination of regular secondary structure in 35MoxAβ(1–42) fibrils by quenched hydrogen exchange. (A and B) Fast heteronuclear multiple quantum correlation spectra of 15N-labeled monomeric 35MoxAβ(1–42) in d6-DMSO containing 0.01% d1-trifluoroacetic acid corresponding to fully protonated (A) and partially exchanged (B) amyloid fibrils. Sequence-specific chemical shift assignment of peptide backbone amide cross-peaks are indicated. Red lines encircle cross-peaks that show a virtually complete loss of intensity between exchange times tex = 0 and tex = 1,990 h. The shown spectra are a sum of the first 10 spectra recorded at the given tex.(C) Intrinsic exchange curves of 35MoxAβ(1–42) in DMSO. The evolution of the peak volumes versus the time (τDMSO) after dissociation of partially hydrogen-exchanged amyloid fibrils into monomers is shown. The identities are indicated next to each curve. The arrow indicates the time point at which a solution of 50% (mol/mol) D2OinH2O was added to yield a final combined deuteron/proton concentration of 2.0 mol·liter–1. Smooth, solid lines represent the monoexponential fits of the raw data. (D) Plot of peak volumes at τDMSO = 0 min versus logarithm of tex of 35MoxAβ(1–42) fibrils. Data for the residues V12 (yellow), E22 (green), V24 (black), G29 (red), and L34 (cyan) are shown. Smooth, solid lines represent fits of the data to a monoexponential equation (V24, L34, and G29) or to the sum of two monoexponential equations (V12 and E22). Error bars indicate the uncertainty in peak volume due to the noise level in each single measurement of initial peak volumes. (E and F) Plots of the observed exchange rates kex/h–1 (E) and the relative population P (F) against the amino acid sequence of 35MoxAβ(1–42). The blue arrows indicate β-strands.
Fig. 1E shows the plot of the resulting exchange rates against the sequence of Aβ(1–42). In the N-terminal segment of residues 1–17, approximately half of the population of each backbone amide exchanged quickly (≥10 h–1); the other half exchanged slowly (≤10–2 h–1). This observation indicated structural heterogeneity in this segment, which is consistent with previous electron paramagnetic resonance studies (16). Conversely, in the segment of residues 18–42, the exchange was slow and almost exclusively monoexponential (Fig. 1 D and E), which suggests that this C-terminal segment constitutes the core structure of the Aβ(1–42) fibril. Considering residues with exchange rates of ≤10–2 h–1 as strongly hydrogen-bonded, the overall protection pattern (Fig. 1 E and F) suggests the presence of two β-strands, β1 and β2, which consist of residues ≈18–26 and ≈31–42, respectively.
Intersheet Side-Chain Packing. Fiber diffraction studies have established that Aβ fibrils contain a cross-β structure, in which β-sheets run parallel and β-strands run perpendicular to the fibril axis (10). It has previously been demonstrated by solid-state NMR that Aβ(1–40) fibrils and Aβ(1–42) fibrils consist of parallel, in-register β-sheets (11). Given the exact sequence location of the β-sheets as determined here by qHX-NMR (Fig. 1), this finding suggests two β-sheets that run along the fibril axis. Sheet β1 is formed by residues ≈18–26, and sheet β2 is formed by residues ≈31–42, so that corresponding residues of adjacent monomers line up along the fibril. If fully extended, this structure would span ≈90 Å perpendicular to the fibril axis. However, from cryoelectron micrographs, we knew that the protofilaments contained dimensions of only ≈25 × ≈45 Å (Fig. 4E). Therefore, it appeared plausible that the two β-sheets form a parallel β-sandwich that is stabilized by inter-β-sheet side-chain contacts. These geometric restraints reduced the search for the correct side-chain packing essentially to a two-dimensional problem of intersheet side-chain interactions. Considering (i) the nature of β-sheets with two distinct faces, (ii) the shortness of the loop that connects the two β-strands, (iii) the solvent inaccessibility of residue 19 (17), (iv) the experimental evidence that M35 is not part of the intersheet side-chain packing because it is accessible for intermolecular chemical cross-linking to residue 4 (18), and (v) parallel, in-register β-sheets and a salt bridge, which had previously been proposed for Aβ(1–40) fibrils based on solid-state NMR data (19), the following intersheet side-chain interactions appeared likely in Aβ(1–42) fibrils: intersheet contacts formed between residues F19/G38 and A21/V36 and a salt bridge between residues D23 and K28.
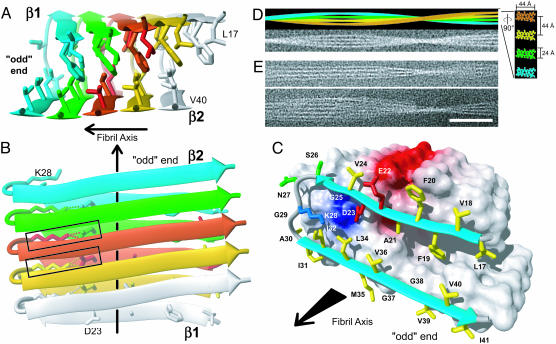
Fig. 4.
The 3D structure of a 35MoxAβ(1–42) fibril. (A and B) Ribbon diagrams of the core structure of residues 17–42 illustrating the intermolecular nature of the inter-β-strand interactions. Individual molecules are colored. For example, the monomer at the odd end is shown in cyan. The β-strands are indicated by arrows, nonregular secondary structure is indicated by spline curves through the Cα atom coordinates of the corresponding residues, and the bonds of side chains that constitute the core of the protofilament are shown. In B, the intermolecular salt bridge between residues D23 and K28 is indicated by dotted lines, and the two salt bridges formed by the central Aβ(1–42) molecule are highlighted by rectangles. (C) van der Waals contact surface polarity and ribbon diagram at the odd end of the 35MoxAβ(1–42) protofilament comprising residues 17–42. The β-sheets are indicated by cyan arrows, and nonregular secondary structure is indicated by gray spline curves. The hydrophobic, polar, negatively charged, and positively charged amino acid side chains are shown in yellow, green, red, and blue, respectively. Positively and negatively charged surface patches are shown in blue and red, respectively, and all others are shown in white. The direction of the fibril axis is indicated by an arrow pointing from even to odd. (D)(Upper) Simulation of a 35MoxAβ(1–42) fibril that consists of four protofilaments colored individually. Lower shows the same fibril in a noisy gray-scale image, which has been blurred corresponding to a resolution of 2 nm. In Right,a ×5-magnified cross section perpendicular to the fibril axis is shown, using the same color code. Dimensions are indicated. To match the experimental twist of the protofilament of the fibril shown in E, a twist angle of 0.45° per molecule was used. (E) Two examples of cryoelectron micrographs of single 35MoxAβ(1–42) fibrils. (Scale bar, 50 nm.)
To test for the presence of these specific interstrand side-chain contacts in Aβ(1–42) fibrils, we used the following pairwise mutagenesis strategy (Fig. 2A): The presumed side-chain interactions in the fibrillar core were disturbed by amino acid substitutions aimed to diminish or alter fibril formation. In a second step, compensating substitutions were introduced to restore the side-chain interaction pattern and, concomitantly, WT-like fibril formation. For these studies, we used the variant 35LAβ(1–42), which can be bacterially expressed and easily purified in substantial amounts (see Supporting Text; compare ref. 20). 35LAβ(1–42) forms fibrils with indistinguishable ultrastructural morphology, and its H/D-exchange pattern is similar to that of 35MoxAβ(1–42) (data not shown).
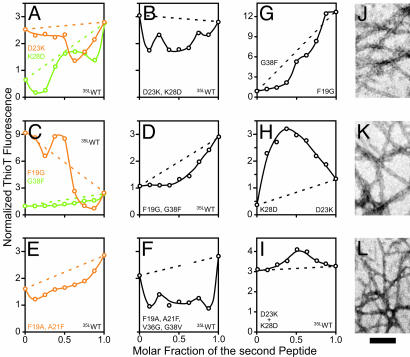
Fig. 2.
Pairwise mutagenesis of 35LAβ(1–42) peptides. (A) Cartoon of intramolecular versus domain swapping-type interaction between monomers in the Aβ(1–42) protofilament that consists of parallel, in-register β-sheets exemplified by the salt bridge formed between the charged residues D23 and K28. Individual Aβ molecules are indicated as colored bars that correspond to a cross section along the protofilament axis through the Cα atom positions of one presumed interacting pair of amino acids. The identity of each variant Aβ peptide is indicated next to the corresponding schemes in rows 1–4. “intra” denotes the intramolecular scenario, and “inter” indicates the domain swapping-type scenario. Red diagonal crosses mark all scenarios that were considered to be incompatible with WT fibril formation. (B–J) Negative staining electron microscopy of 35LAβ(1–42) peptides. The variants are indicated on each image. All electron micrographs were recorded at a nominal magnification of ×72,000. (Scale bar, 100 nm.)
Eight different variants of the 35LAβ(1–42) peptide were generated and allowed to form fibrils for 2 months (see Supporting Text) before they were examined by negative staining EM (Fig. 2 B–J). Whereas nonordered precipitates and spherical aggregates were observed for the two variants K28D and G38F (Fig. 2 B and E), rod-like fibrils were obtained for the variants D23K and F19G (Fig. 2 C and F). Thus, all of these single amino acid variants formed aggregates that were readily distinguishable from WT 35LAβ(1–42) fibrils. In contrast, the complementary variant D23K–K28D formed fibrils that looked like WT (Fig. 2 D and J), suggesting that D23 and K28 form a salt bridge in Aβ(1–42) fibrils. Likewise, the complementary variant F19G–G38F was capable of forming fibrils that looked similar to WT (Fig. 2 G and J), even though the majority of the observed aggregates had the typical morphology of premature fibrils (21, 22) (data not shown). Hence, the residues F19 and G38 appeared to form a hydrophobic interaction pair that connects the two β-strands.
To support the side-chain interaction pattern implied by the F19/G38 permutation, we then generated the double variant F19A–A21F. This peptide formed globular aggregates and some worm-like fibrils (Fig. 2H). The additional introduction of the compensatory substitutions V36G–G38V on strand β2 restored WT 35LAβ(1–42) fibril formation (Fig. 2 I and J). Whereas this pairwise mutagenesis approach simultaneously exchanged two presumed interaction pairs along the β-strands, the F19/G38 cycle inverted a single interaction pair between strands. Thus, two independent sets of Aβ(1–42) variants suggested a close spatial proximity between the pairs of residues F19/G38 and A21/V36. This approach established the register of side-chain contacts in the hydrophobic core of Aβ(1–42) fibrils, which is formed by the odd-numbered residues of the β1-strand and the even-numbered residues of the β2-strand.
Intermolecular Arrangement. With the EM studies (Fig. 2), it is not possible to determine whether the side-chain interactions between the two β-strands are intramolecular or intermolecular. In a conformation with parallel, in-register β-sheet secondary structure, both scenarios could be envisioned (Fig. 2 A). Because the fibril formation of peptide mixtures can distinguish between both scenarios (Fig. 2 A, rows 5–7), we used a cofibrillization assay of binary Aβ peptide mixtures measured by ThioT binding. A relative decrease of the ThioT fluorescence compared with pure Aβ peptides indicates a different fibrillar structure or the presence of fewer fibrils (23).
The equilibrium ThioT-binding capacity of cofibrillized mixtures of WT 35LAβ(1–42) with one of the single-point variants D23K, K28D, F19G, and G38F, or the double variant F19A–A21F, was significantly reduced (Fig. 3 A, C, and E). These data further support the results from the EM studies (Fig. 2 B–J) that these variants are incapable of forming WT-like fibrils and of being incorporated into the final WT structure. In particular, ThioT binding of F19G and D23K was only slightly affected by the presence of a small amount of WT 35LAβ(1–42), whereas WT 35LAβ(1–42) peptide was strongly decreased by these mutants. This finding suggests that the F19G and D23K variants form fibrils of a different molecular structure, which is consistent with the observed morphological differences between the corresponding fibrils (Fig. 2 C, F, and J).
Fig. 3.
Phase behavior of binary 35LAβ(1–42) peptide mixtures. (A–F) Plots of the normalized ThioT fluorescence against the mixing ratio with 35LAβ(1–42), as observed after 2 months of equilibration at room temperature. (G–I) Plots of the normalized ThioT fluorescence against the mixing ratio of two variant Aβ peptides. In A–I, the identities of the peptides present in the mixtures are indicated next to the corresponding y axis on each diagram. Plots corresponding to variants of the β1-strand are shown in orange, plots corresponding to variants of the β2-strand or its preceding loop are shown in green, and plots corresponding to double variants are shown in black. Dotted lines in the corresponding colors indicate the ThioT fluorescence that would be expected if the peptides in the mixture would not interact and thus aggregate independently of each other. The ThioT signal was normalized to the fluorescence signal of the single tyrosine in Aβ(1–42). (J–L) Negatively stained electron micrographs corresponding to the samples at a molar fraction of 0.5 in G–I, recorded at a nominal magnification of ×72,000. (Scale bar, 100 nm.)
To find clues about the intermolecular interaction pattern, the sequence-permutated variants D23K–K28D, F19G–G38F, and F19A–A21F–V36G–G38V were mixed with 35LAβ(1–42). However, even though these peptides formed WT-like fibrils, they did not readily coaggregate with 35LAβ(1–42) (Fig. 3 B, D, and F), suggesting the presence of intermolecular side-chain packing, which forbids the formation of mixed fibrils (Fig. 2 A, row 5). To explore this possibility further, we coaggregated the complementary single variants G38F and F19G, or, alternatively, the variants D23K and K28D, none of which had formed WT-like fibrils (see Fig. 2). At equimolar mixing ratios, a local plateau of ThioT binding is observed for a mixture of G38F and F19G, and a maximum is observed for a mixture of D23K and K28D (Fig. 3 G and H), indicating cooperative fibril formation. Negative staining EM of the equimolar mixtures subsequently confirmed WT-like fibril formation (Fig. 3 J and K; compare Fig. 2J). Thus, this result is consistent with an intermolecular interaction scheme (Fig. 2 A, row 6). We then reasoned that, despite the strong inhibitory effect of D23K or K28D on WT 35LAβ(1–42) fibril formation (Fig. 3A), a mixture of all three peptides should coaggregate, provided that D23K and K28D are present in equimolar amounts (Fig. 2 A, row 7). Indeed, our data confirmed this prediction (Fig. 3 I and L). In summary, these mixing experiments yielded a consistent data set, which suggests that Aβ(1–42) fibrils are stabilized by intermolecular domain swapping-type side-chain interactions.
The direction of the observed intermolecular side-chain interactions is ambiguous, because each molecule in the Aβ fibril has two immediate neighbors. In a β-sheet, all H bonds involving the odd-numbered residues are on one side of an individual β-strand, and those of the even-numbered residues are on the opposite side. Taking the β1-strand of a given Aβ(1–42) molecule as a reference, the “odd” neighbor extends the H-bonding network involving the odd-numbered residues of the β1-strand (Fig. 6A, which is published as supporting information on the PNAS web site), and the “even” neighbor extends the H-bonding network involving its even-numbered residues (Fig. 6B). Aβ16–22m is a synthetic in vitro inhibitor of fibril extension (24) that contains residues 16–22 of Aβ, in which the backbone amide groups of L17, F19, and A21 have been methylated. Binding to the protofilament can therefore only occur on the odd side of the protofilament (Fig. 6). Tight binding of the inhibitor is straight-forward for an odd intermolecular side-chain interaction pattern (Fig. 6A), because extensive hydrophobic contacts of the inhibitor with the protofilament can be formed. Thus, strand β2 most likely forms side-chain contacts with strand β1 of the odd neighbor.
Structure Calculation of Aβ(1–42) Protofilament. The calculation of protein structures from NMR-derived UPLs is a well established technique (25). Here, we apply it toward the determination of a 3D structure of the Aβ(1–42) protofilament (see Table 1 and Supporting Text, which are published as supporting information on the PNAS web site). The UPLs are derived from the presented secondary and tertiary structure information and the previously established structural properties of Aβ(1–42) fibrils (see Supporting Text) [i.e., 4.9-Å intermolecular distance (10, 11) and parallel, in-register β-sheets, which were demonstrated by solid-state NMR (10, 11)], including (i) H-bond restraints identified from H/D-exchange studies, (ii) an intermolecular salt bridge restraint between residues D23 and K28 derived from the mutagenesis studies, (iii) two intersheet distance restraints between F19 and G38, and A21 and V36, as derived from the pairwise mutagenesis studies, and (iv) intermolecular polymer restraints that take the repetitive nature of the protofilament into account. Based on the restraints, the structure calculation was performed for an Aβ(1–42) pentamer. Of 100 randomized starting conformers, the 10 best final structures had minimal constraint violations and small deviations from ideal geometry. The middle molecule of the pentamer had a backbone rms deviation of 1.18 Å relative to the mean coordinates for the peptide segment 18–42 (Fig. 7A, which is published as supporting information on the PNAS web site). It might appear surprising that only three intersheet constraints should suffice to yield a well defined Aβ(1–42) protofilament structure. However, a number of additional test calculations showed that the repetitiveness of the fibril restricts the conformational space of each individual Aβ(1–42) molecule to a narrow range of possible conformations (Table 1 and Supporting Text). Therefore, we concluded that the overall 3D structure of the Aβ(1–42) protofilament is defined unambiguously.
The 3D Aβ(1–42) Protofilament Structure. The 3D structure of the Aβ(1–42) protofilament consists of two stacked, intermolecular, parallel, in-register β-sheets that perpetuate along the fibril axis (Fig. 4 A and B). The residues L17, F19, and A21 of sheet β1 mediate the hydrophobic intermolecular contacts with the even-numbered residues of sheet β2. The loop region of residues 27–30 is connected to sheet β1 by means of the intermolecular salt bridge D23–K28, which also forms contacts to residues I32 and L34 of sheet β2 (Fig. 4C; see also Fig. 8, which is published as supporting information on the PNAS web site). These tertiary interactions are distinct from a previously proposed model of Aβ(1–40) fibrils (19). There, the side-chain interactions are intramolecular, and their register deviates by ≈10 Å. As a consequence, the hydrophobic side-chain packing in the core of the proposed Aβ(1–40) protofilament structure is entirely different (Fig. 9, which is published as supporting information on the PNAS web site; for a detailed comparison see Supporting Text).
In cryoelectron micrographs of the 35MoxAβ(1–42) fibrils, the protofilaments show dimensions of ≈25 × ≈45 Å, with an average spacing of ≈45 Å (Fig. 4E). To test whether our protofilament structure can explain the morphology of Aβ(1–42) fibrils, we assembled 1,200 Aβ molecules into a model fibril (Fig. 4D Upper) by using molmol (26). In this model, the 44-Å-wide nodes correspond to the length of the β-strands, and the 24-Å thickness of the protofilament corresponds to the β-sheet sandwich as shown in Fig. 4. Manipulation of Fig. 4D Upper by adding noise and blurring to a resolution of 2 nm resulted in an image (Fig. 4D Lower) that is strikingly similar to the experimental cryoelectron micrographs of frozen-hydrated 35MoxAβ(1–42) (Fig. 4E). Hence, the dimensions predicted by the protofilament structure are in good agreement with mesoscopic experimental observations. In contrast to previous model predictions for Aβ(1–40) (19), these data suggest the presence of only one Aβ(1–42) molecule per protofilament unit length.
Between the protofilaments, a 15-Å-wide area with low electron density is observed that appears to extend along the entire fibril (Fig. 4E). The molecular details that lead to this regular spacing are currently unclear. However, it has been reported that residue 4 can form specific intermolecular cross-links with methionine 35 in Aβ(1–42) fibrils (18), suggesting that the structurally heterogeneous peptide segment 1–16 (see Fig. 1 E and F) is involved in interprotofilament binding and organization.
Fibril extension most likely occurs at the tip of the protofilaments. The domain swapping-type interactions between Aβ(1–42) molecules imply the presence of distinct surfaces at the opposing ends: In contrast to the even end (Fig. 10, which is published as supporting information on the PNAS web site), a hydrophobic cleft is formed by strand β1 and strand β2 of the template at the odd end of the protofilament (Fig. 4C). An incoming Aβ monomer could initially bind by means of the contiguous hydrophobic stretch of residues 17–21. This complex could be further stabilized by the formation of the intermolecular salt-bridge and backbone H bonds. However, the newly added monomer could still dissociate, because the β2-strand has substantially fewer interactions with the template than β1. Hence, the last added Aβ monomer would be stabilized permanently only by the addition of the next monomer, suggesting a sequence-selective, cooperative mechanism of Aβ fibril extension that follows first-order kinetics. Recently, apparent unidirectional Aβ(1–40) fibril growth with a constant slow rate was directly observed (27). Therefore, key experimental observations on fibril growth are in good agreement with the molecular mechanism suggested here, based on the structure of the Aβ(1–42) protofilament. It appears evident that this mechanism also provides a rationale for the activity of Aβ peptide analog inhibitors comprising the Aβ peptide segment of residues 17–21 (24, 28). They would presumably bind to the hydrophobic stretch of residues 17–21 (Fig. 4C), thereby blocking this site and preventing incoming Aβ molecules from extending the fibril (see above and Fig. 6).
Biological Activity of Aβ. During the presented structural investigation, morphological variants of Aβ(1–42) aggregates were obtained that all share a virtually identical amino acid sequence. Because all deviations from WT morphology can be rationalized based on the presented structure, this set of variants appeared suitable for a structure–function study of Aβ(1–42). The 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide-formazan assay (MTT assay) is a widely used assay of cell viability and cellular response to Aβ. To determine the cytotoxicity of the 35LAβ(1–42) variants, we used this assay in the CNS cell line B12, which is susceptible to Aβ toxicity (29) (see also Supporting Text). Table 2, which is published as supporting information on the PNAS web site, summarizes the results: All variants displayed a detectable neurotoxic response. With the exception of the variant K28D, the most toxic peptides all formed fibrils (Fig. 2 B–J and Table 1) and displayed toxicity similar to synthetic Aβ(25–35), which was used as a reference. A lower but still significant toxicity was observed for the variants F19G and G38F, which form rod-like fibrils and seed-like (21) oligomers (Fig. 2). Finally, the variants F19A–A21F and F19G–G38F, which form mostly premature fibrils, displayed the lowest toxicity. These data suggest that mature Aβ fibrils, as well as some types of oligomers, are neurotoxins.
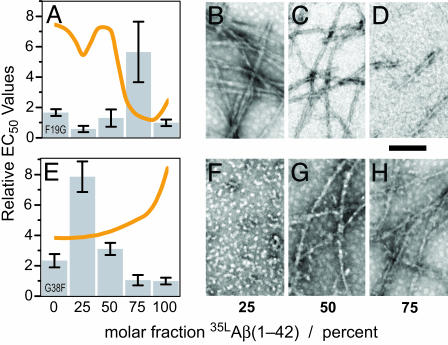
To further investigate the structure–toxicity relationship of Aβ aggregates, we next asked how mixing of structurally incompatible peptides might affect toxicity. Fig. 5A shows the relative EC50 toxicity values for various ratios of F19G and WT 35LAβ(1–42). Two of the ratios displayed near WT toxicity. However, at 75% WT 35LAβ(1–42), the EC50 value was increased 6-fold compared with pure WT peptide, indicating a significantly reduced toxicity. This sample contained very few short fibrils (Fig. 5D), which supports the previous notion that fibrils are toxic. In contrast, the exclusively oligomer-forming variant G38F possesses a significant toxicity, which can be abolished by the presence of a small amount of WT peptide (Figs. 5E and 2D). These data suggest that a specific oligomer tertiary structure is required for G38F toxicity. At higher fractions of WT peptide, fibrils (Fig. 5 G and H) were observed, which were again more toxic.
Fig. 5.
Morphology and neurotoxicity of Aβ(1–42) peptides. (A and E) Bar diagrams of the relative EC50 values of binary mixtures of F19G (A) and G38F (E) with 35LAβ(1–42) as obtained by the 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide assay using B12 cells. The orange lines indicate the relative ThioT fluorescence of the corresponding samples. (B–D and F–H) Negative stained electron micrographs of the individual peptide mixtures in A and E are shown to the right of the corresponding bar diagrams. The molar fraction of WT 35LAβ(1–42) is indicated below. (Scale bar, 100 nm.)
The structure–toxicity relationship indicates that Aβ amyloid fibrils contain an intrinsic toxicity that correlates with their morphology (e.g., more regular and longer fibrils tend to be more toxic). This observation, which is in agreement with previous observations on various aggregation stages of 35MoxAβ(1–42) (21), supports the concept that neuronal degeneration in AD is caused by the deposition of Aβ amyloid (30). However, our data also suggest that oligomers that are similar to small diffusible ligands (ADDLs, or Aβ-derived diffusible ligands) (31) or protofibrils (32) are toxic and may play a central role in AD. Such oligomeric structures, which seem to be involved in a variety of human amyloid diseases, appear to have common structural features that are distinct from amyloid fibrils (33).
The presented structure of Aβ(1–42) protofilament explains the sequence selectivity, the cooperativity, and the unidirectionality of Aβ fibril growth and also provides a structural explanation for the working mechanism of current Aβ fibrillization inhibitors. Furthermore, it shows a correlation between neurotoxicity and various conformational entities of Aβ that might be involved in the amyloidogenesis of AD. These studies may therefore open an avenue toward a comprehensive molecular understanding of amyloidogenesis of AD and structure-assisted development of potent anti-AD drugs and AD diagnostic markers.
Supplementary Material
Acknowledgments
We thank N. Soder (F. Hoffmann–La Roche) for producing 15N-labeled 35MoxAβ(1–42). R.R. is a Pew Scholar. This work was supported in part by the National Institutes of Health.
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: Aβ, amyloid-β; AD, Alzheimer's disease; ThioT, thioflavin T; UPL, upper limit restraint.
Data deposition: The coordinates of the fibrillar core structure of Aβ(1–42) have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 2BEG).
References
- 1.Selkoe, D. J. (2003) Nature 426, 900–904. [DOI] [PubMed] [Google Scholar]
- 2.Tycko, R. (2004) Curr. Opin. Struct. Biol. 14, 96–103. [DOI] [PubMed] [Google Scholar]
- 3.Ritter, C., Maddelein, M. L., Siemer, A. B., Luhrs, T., Ernst, M., Meier, B. H., Saupe, S. J. & Riek, R. (2005) Nature 435, 844–848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nelson, R., Sawaya, M. R., Balbirnie, M., Madsen, A. O., Riekel, C., Grothe, R. & Eisenberg, D. (2005) Nature 435, 773–778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Masters, C. L., Simms, G., Weinman, N. A., Multhaup, G., McDonald, B. L. & Beyreuther, K. (1985) Proc. Natl. Acad. Sci. USA 82, 4245–4249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kang, J., Lemaire, H. G., Unterbeck, A., Salbaum, J. M., Masters, C. L., Grzeschik, K. H., Multhaup, G., Beyreuther, K. & Muller-Hill, B. (1987) Nature 325, 733–736. [DOI] [PubMed] [Google Scholar]
- 7.Jarrett, J. T., Berger, E. P. & Lansbury, P. T., Jr. (1993) Biochemistry 32, 4693–4697. [DOI] [PubMed] [Google Scholar]
- 8.Burdick, D., Soreghan, B., Kwon, M., Kosmoski, J., Knauer, M., Henschen, A., Yates, J., Cotman, C. & Glabe, C. (1992) J. Biol. Chem. 267, 546–554. [PubMed] [Google Scholar]
- 9.Riek, R., Guntert, P., Dobeli, H., Wipf, B. & Wuthrich, K. (2001) Eur. J. Biochem. 268, 5930–5936. [DOI] [PubMed] [Google Scholar]
- 10.Kirschner, D. A., Abraham, C. & Selkoe, D. J. (1986) Proc. Natl. Acad. Sci. USA 83, 503–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Balbach, J. J., Petkova, A. T., Oyler, N. A., Antzutkin, O. N., Gordon, D. J., Meredith, S. C. & Tycko, R. (2002) Biophys. J. 83, 1205–1216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Alexandrescu, A. T. (2001) Pac. Symp. Biocomput., 67–78. [DOI] [PubMed]
- 13.Hoshino, M., Katou, H., Hagihara, Y., Hasegawa, K., Naiki, H. & Goto, Y. (2002) Nat. Struct. Biol. 9, 332–336. [DOI] [PubMed] [Google Scholar]
- 14.Kheterpal, I., Zhou, S., Cook, K. D. & Wetzel, R. (2000) Proc. Natl. Acad. Sci. USA 97, 13597–13601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang, S. S., Tobler, S. A., Good, T. A. & Fernandez, E. J. (2003) Biochemistry 42, 9507–9514. [DOI] [PubMed] [Google Scholar]
- 16.Torok, M., Milton, S., Kayed, R., Wu, P., McIntire, T., Glabe, C. G. & Langen, R. (2002) J. Biol. Chem. 277, 40810–40815. [DOI] [PubMed] [Google Scholar]
- 17.Kheterpal, I., Williams, A., Murphy, C., Bledsoe, B. & Wetzel, R. (2001) Biochemistry 40, 11757–11767. [DOI] [PubMed] [Google Scholar]
- 18.Egnaczyk, G. F., Greis, K. D., Stimson, E. R. & Maggio, J. E. (2001) Biochemistry 40, 11706–11714. [DOI] [PubMed] [Google Scholar]
- 19.Petkova, A. T., Ishii, Y., Balbach, J. J., Antzutkin, O. N., Leapman, R. D., Delaglio, F. & Tycko, R. (2002) Proc. Natl. Acad. Sci. USA 99, 16742–16747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dobeli, H., Draeger, N., Huber, G., Jakob, P., Schmidt, D., Seilheimer, B., Stuber, D., Wipf, B. & Zulauf, M. (1995) Bio/Technology 13, 988–993. [DOI] [PubMed] [Google Scholar]
- 21.Seilheimer, B., Bohrmann, B., Bondolfi, L., Muller, F., Stuber, D. & Dobeli, H. (1997) J. Struct. Biol. 119, 59–71. [DOI] [PubMed] [Google Scholar]
- 22.Bohrmann, B., Adrian, M., Dubochet, J., Kuner, P., Muller, F., Huber, W., Nordstedt, C. & Dobeli, H. (2000) J. Struct. Biol. 130, 232–246. [DOI] [PubMed] [Google Scholar]
- 23.LeVine, H., III (1999) Methods Enzymol. 309, 274–284. [DOI] [PubMed] [Google Scholar]
- 24.Gordon, D. J., Sciarretta, K. L. & Meredith, S. C. (2001) Biochemistry 40, 8237–8245. [DOI] [PubMed] [Google Scholar]
- 25.Wuthrich, K. (1986) NMR of Proteins and Nucleic Acids (Wiley, New York).
- 26.Koradi, R., Billeter, M. & Wuthrich, K. (1996) J. Mol. Graphics 14, 51–55. [DOI] [PubMed] [Google Scholar]
- 27.Ban, T., Hoshino, M., Takahashi, S., Hamada, D., Hasegawa, K., Naiki, H. & Goto, Y. (2004) J. Mol. Biol. 344, 757–767. [DOI] [PubMed] [Google Scholar]
- 28.Bieler, S. & Soto, C. (2004) Curr. Drug Targets 5, 553–558. [DOI] [PubMed] [Google Scholar]
- 29.Behl, C., Davis, J. B., Lesley, R. & Schubert, D. (1994) Cell 77, 817–827. [DOI] [PubMed] [Google Scholar]
- 30.Hardy, J. & Selkoe, D. J. (2002) Science 297, 353–356. [DOI] [PubMed] [Google Scholar]
- 31.Lambert, M. P., Barlow, A. K., Chromy, B. A., Edwards, C., Freed, R., Liosatos, M., Morgan, T. E., Rozovsky, I., Trommer, B., Viola, K. L., et al. (1998) Proc. Natl. Acad. Sci. USA 95, 6448–6453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hartley, D. M., Walsh, D. M., Ye, C. P., Diehl, T., Vasquez, S., Vassilev, P. M., Teplow, D. B. & Selkoe, D. J. (1999) J. Neurosci. 19, 8876–8884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kayed, R., Head, E., Thompson, J. L., McIntire, T. M., Milton, S. C., Cotman, C. W. & Glabe, C. G. (2003) Science 300, 486–489. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.