Abstract
We have identified mutant alleles of two sporophytically acting genes, HAIKU2 (IKU2) and MINISEED3 (MINI3). Homozygotes of these alleles produce a small seed phenotype associated with reduced growth and early cellularization of the endosperm. This phenotype is similar to that described for another seed size gene, IKU1. MINI3 encodes WRKY10, a WRKY class transcription factor. MINI3 promoter::GUS fusions show the gene is expressed in pollen and in the developing endosperm from the two nuclei stage at ≈12 hr postfertilization to endosperm cellularization at ≈96 hr. MINI3 is also expressed in the globular embryo but not in the late heart stage of embryo development. The early endosperm expression of MINI3 is independent of its parent of origin. IKU2 encodes a leucine-rich repeat (LRR) KINASE (At3g19700). IKU2::GUS has a similar expression pattern to that of MINI3. The patterns of expression of the two genes and their similar phenotypes indicate they may operate in the same genetic pathway. Additionally, we found that both MINI3 and IKU2 showed decreased expression in the iku1-1 mutant. IKU2 expression was reduced in a mini3-1 background, whereas MINI3 expression was unaltered in the iku2-3 mutant. These data suggest the successive action of the three genes IKU1, IKU2, and MINI3 in the same pathway of seed development.
Keywords: autoregulation, endosperm development
Seed development involves a complex of processes, including the expansion and growth of the maternal integuments of the ovule and the development of the diploid zygote after the union of the maternal egg cell with one of the two sperm cells delivered to the embryo sac by the pollen tube. It also involves the development of a triploid endosperm after the union of the two nuclei of the homodiploid central cell of the embryo sac with the second sperm cell (1). In eudicots such as Arabidopsis, endosperm development progresses through phases of syncytial growth, cellularization, and cell death. The syncytial phase is characterized by successive divisions of the triploid nuclei without cytokinesis (2). The endosperm cytoplasm is initially compartmentalized into nuclear cytoplasmic domains (3), and subsequently cellularization occurs after the eighth round of syncytial mitoses, initially in the region surrounding the embryo, and proceeding toward the chalazal region (4). Viable seed formation results from the integrated growth and development of the genetically diverse integument, embryo, and endosperm tissues.
Major seed controls are provided by genes that define the development of the maternal integument and the new-generation embryo and endosperm. A number of mutations have been described that impair integument development (5, 6), and genes disrupting embryo pattern formation have also been described (7-9). Endosperm development controls are represented by the FIS loci, MEA, FIS2, and FIE, as well as MSI1 (10-12). These genes code for proteins that are components of a chromatin-associated polycomb complex that prevents endosperm development before double fertilization. Genes controlling cytokinesis in endosperm and/or embryo have also been described (4). Clearly, the endosperm plays a key role in controlling seed size in both Arabidopsis, a dicot, and in maize, a monocot.
In both monocots and dicots, when the relative dosage of maternal and paternal genomes is perturbed, the endosperm and seed size are affected (13, 14). Arabidopsis, a diploid plant pollinated with tetraploid pollen, produces large seeds. This cross generates tetraploid endosperm (2 ♀:2 ♂) with paternal genome excess rather than the normal triploid endosperm produced by diploid parents. A different endosperm and seed result is generated when a tetraploid plant is pollinated with diploid pollen generating maternal genomic excess (4 ♀:1 ♂), and the pentaploid endosperm results in smaller seeds than normal. Apart from the ploidy level and parental genome representation, endosperm development is also subject to differential expression of many genes that depends on their parent of origin.
The gametophytic expression of the fis class genes presents one example of imprinting with expression limited to alleles present in the maternal genome (11, 15, 16). The phenotypes of embryo arrest and endosperm overproliferation of fis mutants can be overcome by pollination with hypomethylated pollen (16, 17), resulting in viable seed of normal size. In contrast, when a hypomethylated male genome is introduced into ovules carrying normal FIS alleles, seed is reduced in size relative to wild type (16, 17). These results with hypomethylated paternal genomes may reflect the disturbance of methylation-dependent genomic imprinting of critical endosperm specific genes.
At the genetic level, Garcia et al. (18) described two mutants in Arabidopsis, haiku1 (iku1) and haiku2 (iku2), which show a recessive sporophytic mode of action causing reduction in endosperm growth accompanied by precocious cellularization and reduced seed size. Combinations of the iku mutants with the transparent testa glabra 2 (ttg2) mutant (19), which has defective seed integuments, cause even greater reduction of seed size, indicating the integument and endosperm growth are both regulators of seed size (20).
To identify genes involved in endosperm proliferation and cellularization, we screened for mutants with reduced seed size. We identified two small seed sporophytic mutants, one allelic to the iku2 locus and the second defining a new locus, MINI3. We report the cloning of these two genes and describe their expression patterns. Our work suggests that IKU1, IKU2, and MINI3 operate in the same pathway controlling seed size. Using reporter gene constructs, we show that MINI3 is expressed in developing endosperm when derived from either parent, indicating this regulator of seed development is not imprinted.
Materials and Methods
Plant Materials and Growth Conditions. Arabidopsis ecotypes used in this study were Landsberg erecta (Ler) and Columbia. Plants were grown in pots with compost soil under continuous artificial light at 20°C in growth chambers. The artificial light was achieved by using an incandescent light source producing a fluence of 150 microeinstein (1 einstein = 1 mol of photons).
The Salk-insertion lines, Salk_032380, Salk_077422, Salk_073260, and Salk_013627, which have a T-DNA insertion in genes At3g19670, At3g19680, At3g19700, and At3g19710, and Salk_050364, which has a T-DNA insertion in gene At1g55610 (AtWRKY10), were obtained from the Arabidopsis Biological Resource Center.
iku2-1 and iku2-2 (18) were crossed to iku2-3.
Mutagenesis and Mutant Identification. The FIS2/fis2-2 heterozygote in the Ler background was the starting material for mutagenesis (11). One gram of seeds was mutagenized with ethylmethane-sulfonate (EMS), as described (21), and ≈300-500 seeds were sown in each pot. Approximately 50 surviving plants that grew from the mutagenized seed (M1 plants) in each pot were bulk-harvested and the seed stored separately. The seed of M1 plants (M2 seed) was planted, and small M3 seeds selected under a dissecting microscope.
Microscopy. Mature seeds of Ler, mini3, and iku2-3 were photographed and captured as digital images under a dissection microscope. Developing seeds were cleared (2). Specimens were examined with a Leica (Deerfield, IL) microscope by using differential interference contrast optics or bright field. Nuclei were counted by using images displayed on a video monitor attached to the microscope. Nuclei from between 10 and 15 ovules were counted. The embryos of 15-day-old seeds were dissected out and photographed. For scanning electron microscopy, dissected ovaries were attached with colloidal graphite to a copper stub, frozen under vacuum, and examined according to Craig and Beaton (22).
DNA Preparation, PCR, and Plant Transformation. Plant DNA preparation and PCR reactions were carried out as described by Bell and Ecker (23).
Plants were transformed by floral dipping (24). The seeds from these dipped plants (T1 seeds) were screened with kanamycin at 50 mg/liter. To verify that plants contained the transgene, the primers 5′-GAGGCTATTCGGCTATGA-3′ and 5′-ACTTCGCCCAATAGCAG-3′ from the NPTII gene were used for amplification.
Genetic Mapping and Cloning of IKU2 and MINI3. See Supporting Text, which is published as supporting information on the PNAS web site, for details (Fig. 7, which is published as supporting information on the PNAS web site).
RT-PCR. We used RT-PCR to demonstrate the expression pattern of MINI3 and IKU2. The primers for MINI3 RT-PCR are 5′-GCTCTCACTGTCCTCAATGGAG-3′ and 5′-TTCCGGTGAAGACAATGCAGCG-3′. The primers for IKU2 are 5′-CGTGTGAGACAAGCGTTAGC-3′ and 5′-GAGGAGACTTGTCCGTGCAT-3′. The expected PCR products from cDNAs are shorter than from genomic DNA, because both sets of primers are spanning introns. For the control gene FORMALDEHYDE DEHYDROGENASE (FDH), primers 5′-TGGGAAACCCATTTATCACTTCA-3′ and 5′-CAGCAAGTCCAACAGTGCCAAG-3′ were used (25). To determine the tissue specificity of gene expression in Ler, 50-100 mg of tissue was harvested from leaves, stem, buds, and open flowers, including 3-day-old siliques. Total RNA was extracted by using an RNeasy plant mini kit (Qiagen, Valencia, CA). Total RNA (0.5 μg) was used for RT-PCR for 30, 35, and 40 cycles.
To check the gene expression in iku1, iku2, mini3-1, and Ler, 2- to 3-day postfertilization siliques were harvested and extracted for total RNA. Four micrograms of total RNA was treated with 2 units of RQ1 RNase-Free DNase for 60 min at 37°C in 20 μl of total volume (Promega), inactivated by the addition of 2 μl of RQ1 DNase Stop Solution (Promega), and heated at 65°C for 10 min. RT-PCR was performed by using the Promega Access RT-PCR kit. The 20-μl PCR mix contained 4 μl of 5× avian myeloblastosis virus (AMV)/Thermus flavus (Tfl) buffer, 0.8 μl of dNTPs (5 mM), 1 μl of each primer (20 μM), 1 unit of AMV reverse transcriptase, and 1 unit of Tfl DNA polymerase (Promega). The thermal cycler program includes an initial incubation of 45 min at 48°C and 2 min at 94°C, followed by 19-29 cycles. Each cycle consisted of 30 s at 94°C, 60 s at 55°C, and 40 s at 68°C, followed by 5 min at 68°C and 5 min at 25°C. PCR products were visualized on a 1% agarose gel with ethidium bromide. The number of cycles was varied for each gene to ensure the amplification was within the linear range. To test MINI3 and IKU2 expression in different mutants, we isolated at least three independent RNA samples from iku1, iku2, mini3-1, and Ler and carried out RT-PCR for each gene at least six times.
AtWRKY10::GUS and At3g19700::GUS Constructs. A HindIII fragment spanning from -2,357 bp to +143 bp of the AtWRKY10 gene was inserted in-frame with a GUS reporter gene in the pBI101-2 binary plasmid (Clontech). A fragment spanning from -808 bp to +8 bp of the At3g19700 gene was amplified with PCR by using primers 5′-TTTACGTACGTGTTGGTGGTGAAGCTT-3′ and 5′-CGCGGATCCGGAGCATTGTTCTCTACGTCGG-3′. This PCR product was cut with HindIII and BamHI and inserted in-frame in front of a GUS reporter gene in the pBI101-3 binary plasmid.
Ler or C24 was transformed with the constructs. After GUS staining (26), ovules or flowers of the transgenic plants were cleared with lactophenol and observed with differential interference contrast microscopy.
DNA Sequencing and Protein Sequence Analysis. To compare the genomic sequences of mini3-1 and Ler in the AtWRKY10 coding region, five sets of PCR primers were designed covering the whole coding region of AtWRKY10. To compare the genomic sequences of iku2-3 and Ler in At3g19700, six sets of PCR primers were designed covering the whole coding region of At3g19700. The PCR products were sequenced with an Applied Biosystems Model 370A DNA Sequencer using fluorescent dye-labeled dideoxy terminators. Protein sequence comparison was performed by using blast searches and multiple sequence alignments were performed with the clustal w 1.8 program (27).
Results
Isolation of Two Small Seed Mutants. We mutagenized seeds heterozygous for the fis2 allele, so that we could screen for small seed mutants in either a homozygous FIS2/FIS2 or a heterozygous FIS2/fis2-1 background. Three M2 mutants bred true for small seed size in the M3 generation; one was unstable in subsequent generations. One of the two stable mutants proved, in complementation tests, to be an allele of the IKU2 locus (iku2-3) (18). The other mutant, mini3-1, defined a previously unidentified locus controlling seed size.
Both iku2-3 and mini3-1 homozygotes produced seeds significantly smaller than parental control seeds in both weight and size (Table 1, Fig. 1 a-c). Pollination with Ler pollen produced seed of Ler size, showing that both mutations were recessive and sporophytic in action. Plants homozygous at both loci produce seeds not significantly different in size and weight to the seeds of plants homozygous for either one of the mutant alleles, indicating that MINI3 and IKU2 may operate in the same pathway (Table 1).
Table 1. Seed size of Ler, homozygous mini3-1, iku2-3 mutants and mini3-1/iku2-3 double mutants.
| Genotype | Weight, mg, 1,000 seeds | Dimensions (length × width), mm |
|---|---|---|
| mini3-1 | 11.8 ± 0.64 | 0.343 ± 0.029 × 0.225 ± 0.022 |
| iku2-3 | 10.08 ± 0.49 | 0.311 ± 0.023 × 0.211 ± 0.019 |
| mini3-1/iku2-3 | 10.58 ± 0.31 | 0.301 ± 0.025 × 0.221 × 0.022 |
| Ler | 20.25 ± 056 | 0.480 ± 0.037 × 0.295 ± 0.020 |
Fig. 1.
Phenotypes of mini3-1 and iku2-3. (a) Ler dry seed, (b) mini3-1 dry seed, (c) iku2-3 dry seed, (d) mature Ler embryo, (e) mature mini3-1 embryo, and (f) mature iku2-3 embryo. (Scale bars, 0.05 mm.)
The embryos in seeds of iku2-3 or mini3-1 homozygotes were smaller than the embryos in control Ler plants (Fig. 1 d-f). After germination, mutant seedlings were smaller than the control Ler seedlings, but during development, the mutant plants grew to a size equivalent to the controls. The number of rosette leaves in the mutants at the time of flowering was similar to that in Ler plants (Table 2), and both mutants were comparable to the Ler control in silique size and morphology and in the number of seeds per silique (Table 2).
Table 2. Characteristics of mini3-1 and iku2-3.
| Genotype | Number of rosette leaves at flowering | Length of siliques, mm | Seeds per silique | Germination, % |
|---|---|---|---|---|
| mini3-1 | 8.06 ± 1.30 | 10.3 ± 0.38 | 54.1 ± 3.25 | 99.36 |
| iku2-3 | 8.15 ± 0.90 | 9.20 ± 0.35 | 53.55 ± 4.30 | 99.40 |
| Ler | 8.2 ± 1.38 | 9.92 ± 0.47 | 54.1 ± 3.25 | 99.50 |
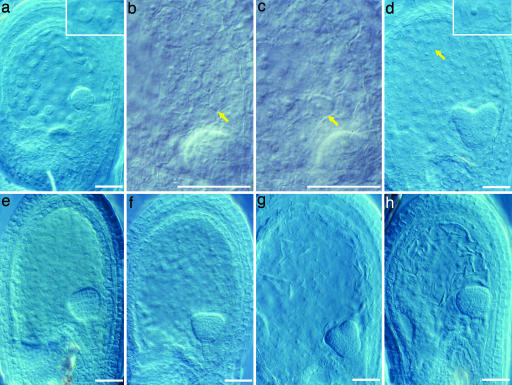
Cytological examination of the developing seeds of Ler and of the two small seed mutants at 48 hr postpollination showed the embryos of all three genotypes were at the octant stage of development. The mutants had a similar number of nuclei in the syncytial endosperm and were not significantly different from Ler (48.1 ± 3.8 for mini3-1, 42.3 ± 7.8 for iku2-3, and 52.8 ± 10.2 for Ler). The developing seeds were of comparable size in all three genotypes at 48 hr. By 72 hr postpollination, the developing seeds of the mutants were significantly smaller than those of the Ler ecotype. At 72 hr, the endosperm of the Ler ovules was still syncytial and contained 175.3 ± 26.4 nuclei (Fig. 2a). The endosperm of ≈20% of the mini3-1 ovules had cellularized at this time. The average total number of cellularized and uncellularized endosperm nuclei, 136.9 ± 34.0, was less than in wild type (Fig. 2b). The endosperm of 25% of the iku2-3 ovules had also cellularized. The iku2 ovules contained 144.5 ± 32.5 endosperm nuclei of both types (Fig. 2c). The result for Ler ovules differs from data reported by Boisnard-Lorig et al. (2), who scored ≈200 nuclei in Ler at 30-60 hr. The discrepancy almost certainly results from differences in growth conditions affecting the rate of endosperm development.
Fig. 2.
Cytological phenotypes of mini3-1 and iku2-3 developing seeds. (a) Seventy-two-hour-old Ler seed with globular stage embryo and uncellularized endosperm. (Inset) Free nuclei. (b) Seventy-two-hour-old mini3-1 seed with globular stage embryo and cellularized peripheral endosperm. Arrow shows cell walls of endosperm. Embryo is not in focus. (c) Seventy-two-hour-old iku2-3 seed with globular-stage embryo and cellularized peripheral endosperm. Arrow shows cell walls of endosperm. Embryo is not in focus (d) Ninety-six-hour-old Ler seed with heart stage embryo and cellularized endosperm. (Inset) Cell walls of endosperm. (e) Ninety-six-hour-old mini3-1 seed with triangular embryo and cellularized endosperm. (f) Ninety-six-hour-old iku2-3 seed with triangular embryo and cellularized endosperm. (g) Ninety-six-hour-old fis2 seed with early-heart stage embryo and uncellularized endosperm. (h) Ninety-six-hour-old fis2/mini3-1 seed with late globular stage embryo and uncellularized endosperm. (Scale bars, 0.05 mm.)
The control Ler ovules did not develop cellularized endosperm until 96 hr postpollination, when they contained 194.5 ± 9.5 nuclei (Fig. 2d). At this time, the mutant seeds had not increased in size beyond their 72-hr postpollination size, whereas Ler seeds were considerably larger than their 72-hr counterparts (Fig. 2 a-c). Embryo development was slower in the two mutants relative to the Ler control. Ler reached the midheart stage of embryo formation at 96 hr, whereas both mutant embryos were at the triangular or early-heart stage (Fig. 2 d-f).
At 120 hr postpollination, Ler embryos were at the late-heart or torpedo stage, and the endosperm had fully cellularized with 409.3 ± 35.5 cells, whereas the iku2-3 and mini3-1 ovules had cellularized with only 168.9 ± 39.3 and 250.9 ± 52.5 endosperm cells, respectively. The embryos of the two types of mutant seeds were at the late-heart stage of development and were considerably smaller than the Ler embryos.
The smaller embryos of the mutants had cotyledon cell sizes (iku2-3 126.3 ± 7.3 μm2 and mini3-1 139.1 ± 8.4 μm2), not significantly different from the cotyledon cells of Ler embryos (132.3 ± 11.0 μm2). We assume that the mature embryos of the mutants contained fewer cells than the Ler embryos (Fig. 1 d-f), but otherwise they had normal embryo morphology. The cytological details of the small embryo need to be investigated further. The size of cells of the outer integument was correlated with embryo size, Ler having cells with an average area of 825.2 ± 113.2 μm2; iku2-3, 490.1 ± 53.1 μm2; and mini3-1, 541.4 ± 41.6 μm2.
We examined the interaction of the sporophytically acting iku2-3 and mini3-1 loci with the gametophytically acting fis2 locus. In plants homozygous for mini3-1 and heterozygous FIS2/fis2-1, a ratio of one viable to one shriveled seed (198:209) was observed. The viable seeds contained cellularized endosperm, and the shriveled seeds contained noncellularized endosperm and aborted. The developing fis2-1 ovules in the mini3-1 homozygotes were smaller than fis2-1 ovules at the comparable stage of development in homozygous MINI3 plants (Fig. 2 g and h). These results show that mini3-1 is epistatic to fis2-1 in regard to the seed size phenotype, and that the fis2-1 allele is epistatic to the mini3-1 mutant with respect to endosperm development. Because of lack of cellularization and clumping of nuclei the number of endosperm nuclei in fis2, mini3-1 double mutants could not be counted.
We obtained similar results in crosses involving iku2-3 and FIS2/fis2-1. Garcia et al. (18) reported similar outcomes in crosses between MEA/mea plants and plants homozygous for iku1 or iku2.
Cloning of the IKU2 and MINI3 Genes. We cloned the IKU2 and MINI3 genes by using chromosome-walking methodology (see Fig. 7, which is published as supporting information on the PNAS web site, for details). IKU2 was located in a 27-kb region of chromosome 3 containing five annotated genes. Analysis of the seed phenotypes of Salk insertion lines in four of these five genes showed that Salk-073260, which had an insertion 200 bp downstream of the ATG in the first exon of At3g19700, showed a small seed phenotype. At3g19700 encodes a leucine-rich repeat transmembrane protein kinase.
We sequenced three alleles of At3g19700 (iku2-1, iku2-2, and iku2-3; see Fig. 8, which is published as supporting information on the PNAS web site, for details). The coding region of the gene in iku2-1 has a single nucleotide substitution (G to A), resulting in the introduction of a stop codon at amino acid residue 49. In iku2-2, there is a 35-base deletion from base 408 to 442, causing amino acid deletions and a reading frame shift. In the iku2-3 allele, there is a single-nucleotide substitution of G to A in the conserved kinase domain, which is predicted to lead to a lysine replacing an arginine residue conserved in other leucine-rich repeat kinases.
MINI3 mapped to a 43-kb region in chromosome 1 (see Fig. 9, which is published as supporting information on the PNAS web site, for details). Segments spanning this region were introduced into homozygous mini3-1/mini3-1 plants. Only those plants that carried the gene At1g55600 resulted in normal size seeds (see Supporting Text for details). This gene is predicted to encode a WRKY transcription factor, AtWRKY10, which contains a predicted WRKY domain at its N-terminal end, along with a zinc-finger-like motif located between amino acid residues 301 and 370 (see Supporting Text for details) (28). The mini3-1 allele has a single G to A substitution in the zinc-finger motif of the WRKY domain converting a glutamic acid to a lysine residue. mini3-2, in the Salk line_050364, has an insertion following amino acid residue 14 in the third exon of the gene. This line also produced small seeds.
AtWRKY10 contains five exons and four introns (see Fig. 10, which is published as supporting information on the PNAS web site, for detail). We identified three putative W-box elements in the promoter within 300 base pairs of the translation start, one identical to the consensus motif TTGACC and the others containing four of the core bases (ATGACG and CTGACA).
MINI3::GUS plants containing a translational fusion showed that expression of MINI3 occurred in the fertilized ovule, and the male gametophyte. RT-PCR analysis was consistent with the MINI3::GUS pattern. mRNA was detected in the ovules of young siliques but not in the leaves or in the stem of the inflorescence (Fig. 3a). Buds containing pollen grains and prepollinated ovules also contained MINI3 mRNA. In unfertilized ovules, the nuclei of the egg and central cell did not show any reporter gene expression, but GUS activity was found in 12- to 96-hr-old postpollination ovules (Fig. 4 b-e). GUS expression was nuclear localized and at the 72- to 96-hr stage expression was found in the globular triangular stage embryo as well as in the endosperm nuclei. There was high expression in the embryo-surrounding region of the endosperm (Fig. 4d). No expression was found in the full-heart stage embryo at 110 hr postpollination.
Fig. 3.
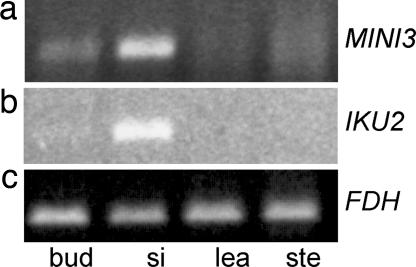
Expression of MINI3 and IKU2. cDNA from flower buds prepollination (bud), young siliques (0- to 3-day fertilized siliques) (si), cauline leaves (lea), and bolt stem without cauline leaves (ste) was used for PCR amplification. (a) RT-PCR of MINI3. Bands are seen in bud and si. (b) RT-PCR of IKU2. Bands are seen in si. (c) RT-PCR of FDH. All tissues give bands.
Fig. 4.
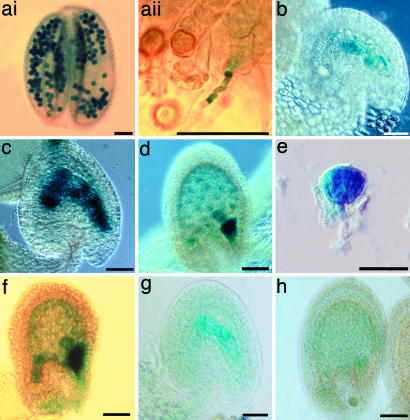
MINI3::GUS and IKU2::GUS activity. (ai) Mature pollen showing GUS activity in homozygous MINI3::GUS transgenic plants. (aii) MINI3::GUS pollen germination on Ler stigma, showing a blue pollen tube. (b) MINI3::GUS developing seed showing GUS activity in two endosperm nuclei 12 hr postfertilization. (c) MINI3::GUS developing seed showing GUS activity in eight endosperm nuclei 24 hr postfertilization. (d) MINI3::GUS developing seed showing GUS activity in endosperm nuclei, globular embryo, and embryo surrounding region (ESR) in the seed 72 hr postfertilization. (e) MINI3::GUS developing seed showing GUS activity in a triangular embryo 96 hr postfertilization. (f)Ler developing seed showing paternal MINI3::GUS activity in endosperm nuclei, embryo, and ESR 72 hr post-fertilization. (g and h) IKU2::GUS activity in developing seeds 12 and 48 hr postfertilization. (Scale bars, 0.05 mm.)
In addition to the expression in the developing ovule, GUS activity was present in pollen grains and some pollen tubes (Fig. 4 ai and aii). In a hemizygous MINI3::GUS plant, ≈50% of the pollen showed GUS activity. We did not find GUS expression in any other part of the plant. In the fertilized ovules, endosperm nuclei showed GUS staining from the two nuclei stage onwards.
Equivalent patterns of expression occurred in the developing ovules whether the MINI3::GUS reporter gene was delivered as a paternal (Fig. 4f) or maternal allele (Fig. 4 b--e); there was no evidence of differential parent-of-origin activity of this gene. This expression pattern is consistent with the fact that mini3 is a sporophytic recessive mutation.
The IKU2::GUS reporter gene also showed activity in the endosperm nuclei of developing seeds (Fig. 4 g and h). The intensity of GUS staining was much lower than in MINI3::GUS plants (Fig. 4 c and d). RT-PCR analysis detected mRNA in the ovules of young siliques but not in the leaves or in the stem of the inflorescence (Fig. 3b); this confirmed the GUS activity patterns. No GUS expression has been found in other parts of the plant, including the embryo and the male gametophyte. In some experiments, we detected weak PCR bands from bud RNA.
Expression Levels of MINI3 and IKU2 in Mutant Genotypes Indicate That These Seed Development Genes Operate in a Single Pathway. Expression levels of MINI3 and IKU2 were scored in homozygous mutants; both showed decreased expression in an iku1-1 background (Fig. 5 a and b). We found that IKU2 expression is reduced in a mini3-1 background (Fig. 5a), whereas MINI3 expression levels remain at wild-type level in the iku2-3 mutant (Fig. 5b). These data indicate that the three genes may be acting in the same regulatory pathway controlling seed development (Fig. 5e).
Fig. 5.
Expression of MINI3 and IKU2 in mutant siliques. (a) RT-PCR of IKU2. IKU2 is repressed in mutant siliques of iku1-1, iku2-3, and mini3-1. (b) RT-PCR of MINI3. MINI3 is repressed only in iku1-1 mutant siliques. (c) RT-PCR of FDH. (d) Total RNA loading. (e) IKU1, MINI3, and IKU2 operate in a single pathway.
MINI3 expression was characterized further by using the MINI3::GUS construct. When MINI3 plants carrying the MINI3::GUS transgene were pollinated with either MINI3 or mini3-1 pollen, the level of GUS staining at 48 and 72 hr pollination was lower in the endosperm fertilized with MINI3 pollen than with the mini3-1 pollen. The endosperm derived from MINI3 pollen fertilization of MINI3/MINI3 carried three doses of the wild-type gene (Fig. 6b); greater GUS activity was found in endosperm derived from plants fertilized with mini3-1 pollen (Fig. 6a), which had only two doses of the wild-type allele together with one of the mutant allele. However, as detected by RT-PCR, the level of transcript of mini3 is lower than that of MINI3 presumably due to the lower stability of the mutant transcript (see Discussion).
Fig. 6.
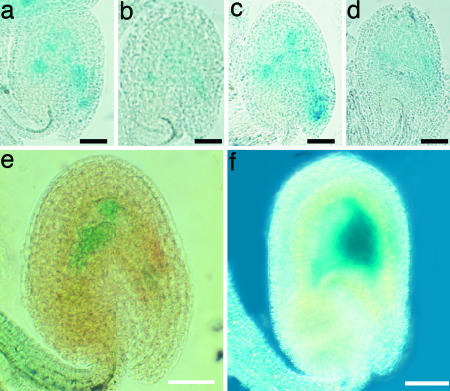
Autorepression of MINI3 and MINI3::GUS activity in fie-2 autonomous seeds. (a and b)Ler ovule showing higher GUS activity when pollinated with mini3-1 (a) than with MINI3 (b). (c and d) mini-3 ovule showing higher GUS activity when pollinated with mini3-1 pollen (c) than with MINI3 pollen (d). (e) A fie-2 autonomous seed showing MINI3::GUS activity in two endosperm nuclei 72 hr postemasculation. (f) fie-2 autonomous seed showing MINI3::GUS activity in endosperm nuclei 96-hr postemasculation. (Scale bars, 0.05 mm.) b and d represent separate experiments with different staining periods and exposures, and thus the intensity of GUS in these figures is not directly comparable.
In a reciprocal experiment, MINI3::GUS was introduced from the pollen parent into either MINI3 or mini3-1 homozygous plants. Higher activity of the transgene was again found in the endosperm, which carried two doses of the mini3-1 allele and one dose of MINI3, compared with endosperm with three MINI3 alleles. When a homozygous mini3-1 plant carrying the GUS transgene was pollinated with either mini3-1 pollen or MINI3 pollen, the triploid endosperm, which contained three mini3-1 alleles, showed stronger GUS activity (Fig. 6c) than the endosperm which carried two mini3-1 alleles and one MINI3 allele (Fig. 6d).
These results show an inverse correlation between the activity of the MINI3::GUS reporter gene, and the dosage of the MINI3 allele and are consistent with the reports of WRKY class genes repressing their own expression (29, 30).
MINI3 expression was also characterized in the autonomous endosperm of fis mutant plants. We generated fie-2 heterozygous plants carrying the MINI3::GUS transgene. MINI3::GUS activity was detected during fie-2 autonomous endosperm development (Fig. 6 e and f).
Discussion
The screen for small seed mutants identified a new allele of iku2 (18) and an allele of another gene involved in seed developmental processes, MINI3. These two genes have been cloned based on map location. We suggest they are important regulators of seed size, and both IKU2 and MINI3 are likely to correspond to two seed size quantitative trait loci (QTLs) defined in the cross of Ler and Cape Verde ecotypes (31). IKU2 and MINI3 as well as IKU1 (15) mutant loci have similar small seed phenotypes and similar patterns of altered seed development producing embryos smaller than wild-type and displaying precocious cellularization of the endosperm. The mutants also displayed maternal integument tissues with smaller cell dimensions than those of wild-type. Because embryo cell size is not altered in the mutants but embryo size is smaller, the small seed must contain fewer cells. Garcia et al. (18) also found that embryo cell size is not altered in iku1 and iku2 mutants. When iku2-3 and mini3-1 were pollinated with wild-type pollen, normal seed size was restored, and the seed coat appeared to be of wild-type phenotype even when the maternal plant was homozygous for the mutant genes, indicating there is no maternal effect on seed size in iku class mutants. In contrast, transparent testa glabra 2 (ttg2) (19) mutants with defective seed integument had maternal effects on seed size and endosperm development (20).
IKU1, IKU2, and MINI3 have sporophytic modes of action in seed development, suggesting these genes are not likely to be imprinted. Genes showing imprinting during seed development would show a deviation from a 3:1 wild type:mutant ratio. Our genetic data show the expected Mendelian segregations and consistent with these data the MINI3::GUS reporter gene does not show any parent of origin specific expression in the developing seed. The autonomous initiation of endosperm development and ovule growth in the fis mutants indicate that FIS proteins act as repressors of ovule and seed development, normally released by the double fertilization event. The expression of MINI3::GUS in a fie mutant background was activated in autonomous endosperm development as well as in endosperm development resulting from double fertilization.
The molecular identity of IKU1 is not known. MINI3 encodes a transcription factor of the WRKY class, corresponding to the WRKY10 gene (28). WRKY class transcription factors are restricted to the plant kingdom, and ≈80 different genes have been defined in Arabidopsis. These transcription factors are known to regulate loci involved in a range of cellular activities by binding to W-box motifs in the target genes (32, 33). In addition, some WRKY genes have been shown to have an autoregulatory function (29, 30). For example, the WRKY18 gene contains a cluster of WRKY-binding sites in its promoter, and its protein product acts as a negative regulator of its own activity. WRKY10 belongs to this subclass of WRKY transcription factors. Consistent with autorepression, we found expression of the MINI3::GUS reporter construct to be higher in endosperm of homozygous mini3-1 plants carrying the construct than in either heterozygous or homozygous MINI3 plants carrying the construct. However, RT-PCR of WRKY10 mRNA on mini3-1 silique RNA did not show higher expression of MINI3 transcripts than in MINI3 siliques. The mini3-1 mutation may cause the mRNA to become unstable.
IKU2 encodes a leucine-rich repeat receptor kinase, a large family of genes with roles in signal transduction pathways in plant development and metabolism. Receptor kinases contain an extracellular domain, a transmembrane domain, and a kinase domain. More than 600 different kinases of this type are known in Arabidopsis. Just over 200 carry leucine-rich repeats in the extracellular domain (34), but only 10 have been annotated with known functions (35). IKU2 is most closely related to EXTRA SPOROGENOUS CELLS (EXS), encoding another leucine-rich repeat kinase that also confers a small seed phenotype and pollen sterility (36).
There are examples of a WRKY protein regulating a receptor-like protein kinase (37). A cluster of W-box elements in the Arabidopsis RLK4 promoter region was shown to be recognized by both purified WRKY18 protein and salicylic acid-induced W-box-binding activities (37). Our results suggest that MINI3 might positively regulate IKU2 through binding to the putative W-box motif identified in the IKU2 promoter.
Genetic and molecular analyses of these mutants allow us to construct a developmental pathway of seed size in Arabidopsis (Fig. 5e). The seed size of the double mutant iku2-1/iku2-1, mini3-1/mini3-1 was similar to the seed size of homozygous mutant alleles of each single locus, suggesting that the genes may lie in the same pathway. This conclusion was supported by gene expression studies. IKU2 and MINI3 are expressed in similar tissues and at similar times in development. When the expression of the IKU2 and MINI3 genes was analyzed in the mutant backgrounds iku1-1, iku2-3, and mini3-1 the expression patterns indicated that all three genes operate in a single pathway, with IKU1 regulating both MINI3 and IKU2 and MINI3 regulating IKU2.
Supplementary Material
Acknowledgments
We thank Bjorg Sherman and Yi Suo for technical help and Aneta Ivanova for help with experiments and helpful discussions.
Conflict of interest statement: No conflicts declared.
References
- 1.Faure, J. E., Rotman, N., Fortuné, P. & Dumas, C. (2002) Plant J. 30, 481-488. [DOI] [PubMed] [Google Scholar]
- 2.Boisnard-Lorig, C., Colon-Carmona, A., Bauch, M., Hodge, S., Doerner, P., Bancharel, E., Dumas, C., Haseloff, J. & Berger, F. (2001) Plant Cell 13, 495-509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Olsen, O. A. (2001) Annu. Rev. Plant Physiol. Plant Mol. Biol. 52, 233-267. [DOI] [PubMed] [Google Scholar]
- 4.Sorensen, M. B., Mayer, U., Lukowitz, W., Robert, H., Chambrier, P., Jurgens, G., Somerville, C., Lepiniec, L. & Berger, F. (2002) Development (Cambridge, U.K.) 129, 5567-5576. [DOI] [PubMed] [Google Scholar]
- 5.Golden, T. A., Schauer, S. E., Lang, J. D., Pien, S., Mushegian, A. R., Grossniklaus, U., Meinke, D. W. & Ray, A. (2002) Plant Physiol. 130, 808-822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sieber, P., Gheyselinck, J., Gross-Hardt, R., Laux, T., Grossniklaus, U. & Schneitz, K. (2004) Dev. Biol. 273, 321-334. [DOI] [PubMed] [Google Scholar]
- 7.Busch, M., Mayer, U. & Jurgens, G. (1996) Mol. Gen. Genet. 250, 681-691. [DOI] [PubMed] [Google Scholar]
- 8.Takada, S., Hibara, K., Ishida, T. & Tasaka, M. (2001) Development (Cambridge, U.K.) 128, 1127-1135. [DOI] [PubMed] [Google Scholar]
- 9.Torres-Ruiz, R. A., Lohner, A. & Jurgens, G. (1996) Plant J. 10, 1005-1016. [DOI] [PubMed] [Google Scholar]
- 10.Ohad, N., Margossian, L., Hsu, Y. C., Williams, C., Repetti, P. & Fischer, R. L. (1996) Proc. Natl. Acad. Sci. USA 93, 5319-5324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chaudhury, A. M., Ming, L., Miller, C., Craig, S., Dennis, E. S. & Peacock, W. J. (1997) Proc. Natl. Acad. Sci. USA 94, 4223-4228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kohler, C., Hennig, L., Bouveret, R., Gheyselinck, J., Grossniklaus, U. & Gruissem, W. (2003) EMBO J. 22, 4804-4814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lin, B. Y. (1984) Genetics 107, 103-115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Scott, R. J., Spielman, M., Bailey, J. & Dickinson, H. G. (1998) Development (Cambridge, U.K.) 125, 3329-3341. [DOI] [PubMed] [Google Scholar]
- 15.Kinoshita, T., Yadegari, R., Harada, J. J., Goldberg, R. B. & Fischer, R. L. (1999) Plant Cell 11, 1945-1952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Luo, M., Bilodeau, P., Dennis, E. S., Peacock, W. J. & Chaudhury, A. (2000) Proc. Natl. Acad. Sci. USA 97, 10637-10642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Adams, S., Vinkenoog, R., Spielman, M., Dickinson, H. G. & Scott, R. J. (2000) Development (Cambridge, U.K.) 127, 2493-2502. [DOI] [PubMed] [Google Scholar]
- 18.Garcia, D., Saingery, V., Chambrier, P., Mayer, U., Jurgens, G. & Berger, F. (2003) Plant Physiol. 131, 1661-1670.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Debeaujon, I., Nesi, N., Perez, P., Devic, M., Grandjean, O., Caboche, M. & Lepiniec, L. (2003) Plant Cell 15, 2514-2531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Garcia, D., Gerald, J. N. F. & Berger, F. (2005) Plant Cell 17, 52-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chaudhury, A. M., Letham, S., Craig, S. & Dennis, E. S. (1993) Plant J. 4, 907-916. [Google Scholar]
- 22.Craig, S. & Beaton, C. D. (1984) J. Microsc. 182, 102-105. [Google Scholar]
- 23.Bell, C. J. & Ecker, J. R. (1994) Genomics 19, 137-144. [DOI] [PubMed] [Google Scholar]
- 24.Clough, S. J. & Bent, A. F. (1998) Plant J. 16, 735-743. [DOI] [PubMed] [Google Scholar]
- 25.Finnegan, E. J., Sheldon, C. C., Jardinaud, F., Peacock, W. J. & Dennis, E. S. (2004) Curr. Biol. 14, 911-916. [DOI] [PubMed] [Google Scholar]
- 26.DeBlock, M. & DeBrouwer, D. (1992) Plant J. 2, 261-266. [Google Scholar]
- 27.Higgins, D. G. & Sharp, P. M. (1989) Comput. Appl. Biosci. 5, 151-153. [DOI] [PubMed] [Google Scholar]
- 28.Eulgem, T., Rushton, P. J., Robatzek, S. & Somssich, I. E. (2000) Trends Plant Sci. 5, 199-206. [DOI] [PubMed] [Google Scholar]
- 29.Robatzek, S. & Somssich, I. E. (2002) Genes Dev. 16, 1139-1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen, C. & Chen, Z. (2002) Plant Physiol. 129, 706-716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Alonso-Blanco, C., Blankestijn-de, V. H., Hanhart, C. J. & Koornneef, M. (1999) Proc. Natl. Acad. Sci. USA 96, 4710-4717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Eulgem, T., Rushton, P. J., Schmelzer, E., Hahlbrock, K. & Somssich, I. E. (1999) EMBO J. 18, 4689-4699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang, Z. L., Xie, Z., Zhou, X., Casaretto, J., Ho, T. H. D. & Shen, Q. J. (2003) Plant Physiol. 134, 1500-1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shiu, S. H. & Bleecker, A. B. (2001) Sci. STKE 113, RE22. [DOI] [PubMed] [Google Scholar]
- 35.Dievart, A. & Clark, S. E. (2004) Development (Cambridge, U.K.) 131, 251-261. [DOI] [PubMed] [Google Scholar]
- 36.Canales, C., Bhatt, A. M., Scott, R. & Dickinson, H. (2002) Curr. Biol. 12, 1718-1727. [DOI] [PubMed] [Google Scholar]
- 37.Du, L. & Chen, Z. (2000) Plant J. 24, 837-847. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.