Abstract
Renal cell carcinomas (RCC) commonly retain wild-type but functionally inactive p53, which is repressed by an unknown dominant mechanism. To help reveal this mechanism, we screened a diverse chemical library for small molecules capable of restoring p53-dependent transactivation in RCC cells carrying a p53-responsive reporter. Among the compounds isolated were derivatives of 9-aminoacridine (9AA), including the antimalaria drug quinacrine, which strongly induced p53 function in RCC and other types of cancer cells. Induction of p53 by these compounds does not involve genotoxic stress and is mediated by suppression of NF-κB activity. In contrast to agents that target IκB kinase 2, 9AA and quinacrine can effectively suppress both basal and inducible activities of NF-κB, representing inhibitors of a previously undescribed type that convert NF-κB from a transactivator into a transrepressor, leading to accumulation of inactive nuclear complexes with unphosphorylated Ser-536 in the p65/RelA subunit. p53 function in RCC can be restored by ectopic expression of a superrepressor of IκB as effectively as by 9AA-derived compounds. These findings suggest that the complete or partial repression of p53 observed in many tumors can be the result of constitutive activation of NF-κB. The results demonstrate, in principle, the possibility to kill cancer cells selectively through simultaneous inhibition of NF-κB and activation of p53 by a single small molecule and suggest anticancer applications for the well known antimalaria drug quinacrine.
Keywords: anticancer treatment, apoptosis, chemical library, quinacrine
The protein p53 controls genetic stability and reduces the risk of cancer through induction of growth arrest or apoptosis in response to DNA damage or deregulation of proto-oncogenes (1). The efficacy of p53 as a tumor-preventing factor is reflected by the high frequency of p53 loss, in at least 50% of human tumors, due to inactivating mutations (2). Understanding the mechanisms of functional inactivation of wild-type p53 in human tumors, for example, by overexpression of natural antagonists of p53, Mdm2, or the viral protein E6, helps to define prospective targets for treating cancer by restoring p53 function (3).
We have recently shown that renal cell carcinomas (RCC), the most frequent and least curable type of kidney cancer, maintain wild-type but functionally inactive p53 (4). The mechanism of p53 repression in RCC is dominant, and therefore “druggable,” and different from that of all reported cases of p53 repression in tumors, suggesting the existence of an as-yet-unknown molecular target for restoring p53 function in cancer. As an approach to finding such factor(s), we have isolated a set of compounds that can restore p53 function in RCC and strongly activate p53 in many other types of cancer cells. Among the most effective compounds from this set were derivatives of 9-aminoacridine (9AA), including an old-known antimalaria drug quinacrine (QC). Analysis of the molecular mechanisms of action of 9AA and QC showed that p53 activation by these compounds occurred through the inhibition of constitutively active NF-κB in tumor cells.
Constitutively active NF-κB signaling, an attribute of chronic inflammation and the property of many tumors, provides selective advantages to tumor cells, probably by inhibiting apoptosis and promoting proliferation by stimulating expression of antiapoptotic factors and cytokines (5). Here we show that inhibition of tumor suppressor p53 is another important benefit to tumors that have constitutively active NF-κB, opening the possibility of simultaneous inhibition of NF-κB and activation of p53 by a single small molecule.
Materials and Methods
The DiverSet library of 34,000 chemical compounds and focused libraries of structural analogues of selected hits were provided by Chembridge. Cell lines with p53- or NF-κB-responsive luciferase or β-gal reporters were described in refs. 4 and 6. For screening chemicals, 2 × 104 RCC45-based p53 reporter cells were plated per well of 96-well plates in 200 μl of phenol-red-free RPMI medium 1640 with 10% FCS. After overnight incubation, 0.2 μl of compounds in DMSO were added per well to final concentrations of 5-15 μM. Equal amounts of DMSO and doxorubicin (0.2-2 μM) were used as controls. After 24 h, lysis buffer with o-nitrophenyl-β-d-galactopyranoside was added and β-gal activity was estimated colorimetrically by a multiwell plate reader. Compounds that induced the reporter stronger than doxorubicin (the only drug tested that was capable of weakly activating the p53-dependent reporter in RCC) were considered to be primary hits. The description of other experimental tools and protocols is provided in Supporting Materials and Methods, which is published as supporting information on the PNAS web site.
Results and Discussion
9AA and QC Activate p53 Through an Unusual Mechanism. To test whether p53 reactivation in RCC is achievable in principle, we expressed increasingly high levels of wild-type p53 in RCC-derived cell lines with lentiviral transduction. At a certain level of expression, p53 became simultaneously cytotoxic and capable of inducing the p53 reporter (Fig. 1A), presumably by depleting a factor that inhibits p53, thus justifying the isolation of small molecules capable of reactivating p53 in RCC.
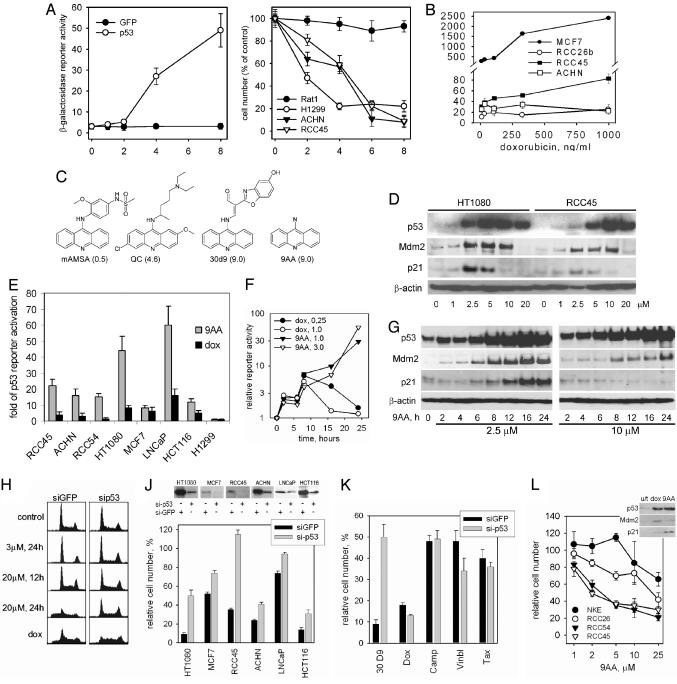
Fig. 1.
Chemicals that activate p53 in RCC. (A) Restoration of p53-mediated transactivation in RCC cells is accompanied by their death. p53-responsive reporter activity in RCC45ConALacZ cells [Left, o-nitrophenyl-β-d-galactopyranoside (ONPG) staining at 48 h] and cell survival after lentivirus-mediated transduction of p53 and GFP. (B) Choice of readout cells and setting of selection criteria. MCF7 (a non-RCC control) and RCC cells, all containing the ConALacZ reporter, were treated with doxorubicin (dox) for 24 h (ONPG reaction normalized to protein concentration). (C) Identification of 9-AA through chemical screening of compounds that activate p53 in RCC cells. 9AA derivatives (13 compounds), plus the original hit s30d9, 9AA, QC, and amsacrine were tested in a dose-dependent assay for activation of the p53 reporter in RCC45ConALacZ cells. Bars represent the relative activity of each compound, calculated as the maximal fold increase of p53 activation induced by the compound relative to the effect of 1 μM dox (average of three experiments). (D) Dose dependence of the effect of 9AA on p53 and p53-transcriptional targets, determined by the Western method in cells treated for 16 h with the indicated amounts of 9AA. (E) 9AA activates p53 more strongly than dox in the majority of tumor cells tested. The p53-responsive reporter activity in cells treated with 9AA (1-10 μM) or dox (0.2-2 μM) for 24 h. Data are presented as relative reporter induction by the most effective dose of each chemical. (F and G) Kinetics of activation of a p53-responsive reporter (F) and expression of p53-dependent targets (G) by 9AA. (H) Effect of 9AA on the cell cycle in HT1080-sip53 or HT1080-siGFP cells (dox, 2 μM, 24 h). (J) Relative survival of the indicated isogenic pairs of tumor cells differing in p53 expression (Upper shows a Western analysis of p53 levels) due to transduction of lentiviral constructs expressing shRNA against p53. (J Lower) The relative numbers of cells that survived 48 h of 9AA treatment (2 μM) are shown compared with an untreated control. (K) p53 dependence of the cytotoxicity of different drugs. The experiment described in J was done by using 9AA (1-10 μM) dox (0.1-1 μM), camptothecin (camp, 0.16-1.6 μM), vinblastine (vinbl, 0.1-1 μM), or taxol (tax, 0.06-0.6 μM). Bars are plotted for the doses of the drugs that demonstrated the highest difference in sensitivity between p53-plus and p53-minus cells. (L) 9AA is more toxic for RCC than for normal kidney epithelium (NKE) cells. Cell numbers were estimated 72 h after treatment with the indicated concentrations of 9AA. The box shows Western analysis of the indicated proteins in lysates of NKE cells treated with dox (1 μM) or 9AA (5 μM) for 16 h.
Twenty-eight compounds were isolated from a diverse library of chemicals that effectively activated p53 reporter in RCC45 cells (see Materials and Methods). The most active compound, 30d9, was seven times stronger than doxorubicin (Fig. 1B), causing a 22-fold induction of the reporter. A structure-activity relationship study done with 59 structural analogues of 30d9 defined 9AA as the structure primarily responsible for this effect. Treatment with 9AA induced stabilization and accumulation of p53 protein (Fig. 6, which is published as supporting information on the PNAS web site) accompanied by a dose-dependent increase in levels of p21 and Mdm2 proteins that are encoded by p53-responsive genes (Fig. 1D). No increase in p53 mRNA expression was found (data not shown).
There were two known drugs among the derivatives of 9AA tested: the anticancer agent amsacrine (m-AMSA), an inhibitor of topoisomerase II, and the anti-malaria drug QC. Between these two, only QC was an effective inducer of p53 transactivation in RCC (Fig. 1C). All of the biochemical and cellular effects described below were identical for 9AA and QC.
9AA activates p53-mediated transcription more strongly than DNA-damaging agents, not only in RCC, but also in the majority of other types of tumor cells that have wild-type p53 (Fig. 1E). The strongest induction of p53 by 9AA was observed in human fibrosarcoma HT1080 cells, which were used in further assays along with RCC.
Activation of p53 response by 9AA had unusually slow kinetics. Although the peak of doxorubicin-induced p53 reporter activity in HT1080 cells was at 8-12 h, the effect of 9AA on reporter activity and on the induction of endogenous p53 targets p21 and Mdm2 reached its maximum at 24 h (Fig. 1 F and G).
Depending on the dose, 9AA caused either p53-dependent growth arrest or apoptosis (3 μM or 20 μM, respectively, Fig. 1H). Low doses of 9AA did not cause apoptosis even after longer incubation times (up to 48 h), coinciding with the strong elevation of p21, whereas high doses of the compound induced apoptosis with no prior growth arrest and without p21 induction (Figs. 1 D, G, and H). These effects were p53-dependent as evidenced by comparison with p53-deficient cells (Fig. 1H). Consistently, 9AA was less toxic to p53-deficient cells, as judged by analysis of a panel of isogenic pairs of cell lines differing in their p53 status due to small hairpin RNA (shRNA)-mediated p53 gene knockdown. Differential toxicity varied among the cell lines tested, with the maximal range observed in HT1080 cells (Fig. 1J; see also Fig. 7, which is published as supporting information on the PNAS web site), possibly reflecting different degrees of p53 knockdown and/or the existence of a p53-independent mechanism of 9AA-mediated cell killing (see below). Importantly, the p53 dependence of cytotoxicity differentiated 9AA from camptothecin, doxorubicin, taxol, and vinblastine, which were equally or even more toxic than 9AA to p53-deficient cells (Fig. 1K), suggesting that 9AA kills cells through a mechanism different from that used by conventional chemotherapeutic drugs.
RCC-derived cell lines were more sensitive to 9AA than normal kidney epithelial cells (Fig. 1L), consistent with the generally higher susceptibility of tumor cells to the cytotoxic effects of activated p53.
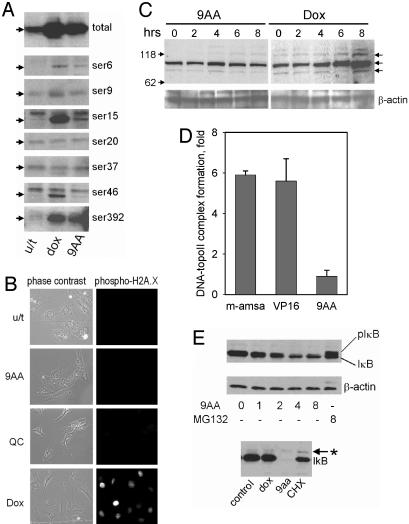
In contrast to DNA-damaging drugs, 9AA and QC did not induce the phosphorylation of p53 (Fig. 2A); Ser-392 that was increasingly phosphorylated after 9AA treatment is the substrate for protein kinase CK2 and not by DNA damage-inducible kinases. Neither 9AA nor QC induced phosphorylation of histone H2A.X (Fig. 2B), a hallmark of DNA damage, nor did either activate the DNA breakage-dependent kinases ATM or ATR (Fig. 2C). Although the 9AA derivative amsacrine causes DNA damage by poisoning topoisomerase II (7), we found no such activity of 9AA in a direct in vitro assay (Fig. 2D). These observations indicated that 9AA activates p53 through a mechanism different from those activated by DNA damage, although the DNA intercalating genotoxic activity was postulated as the basis of toxicity of 9AA and its derivatives (7, 8).
Fig. 2.
9AA and QC activate p53 in an unusual way. (A) Phosphorylation status of p53 in RCC45 cells treated with 9AA (5 μM) or doxorubicin (dox, 1 μM) for 16 h. Western analysis of total protein lysates was done by using antibodies against p53 (DO1) and against specific sites of phosphorylation in p53. (B) Immunofluorescent detection of phosphorylated histone H2A.X in HT1080 cells treated with equally toxic concentrations of 9AA, QC (10 μM), or dox (1 μM). (C) Western blot analysis of lysates of ACHN cells treated with 9AA (10 μM) or dox (1 μM) with antibodies against phosphorylated substrates of ATM/ATR (Cell Signaling Technology, Beverly, MA). (D) Effect of 9AA, amsacrine, and etoposide on topoisomerase II activity in vitro.(E) The amount and phosphorylation status of IκB-α after treatment with 9AA. Western analyses of lysates of PC-3 cells (Upper) treated with 9-AA (10 μM, 1-8 h) or with the proteasome inhibitor MG-132 (10 μM, 8 h) or of MCF7 cells treated with dox (1 μM), 9AA (10 μM), or cycloheximide (10 μg/ml) by using antibodies specific for both phosphorylated and unphosphorylated forms of IκBα.
p53 stabilization might be achieved by disrupting the binding of p53 to Mdm2 (9), a major mediator of p53 degradation. However, shRNA-mediated knockdown of Mdm2 did not lead to p53 reactivation in RCC (4), allowing us to exclude Mdm2 targeting as the mechanism of the p53-activating effect of 9AA.
The accumulation of unphosphorylated p53 is the property of the inhibitors of proteasomes. However, 9AA does not have this activity as shown by using a direct in vitro assay (data not shown) and by monitoring the effect of 9AA on the level of IκBα, another target of proteasomal degradation (10): 9AA treatment had the opposite effect to the proteasomal inhibitor MG132, leading to a decrease or even complete disappearance of IκBα (Fig. 2E).
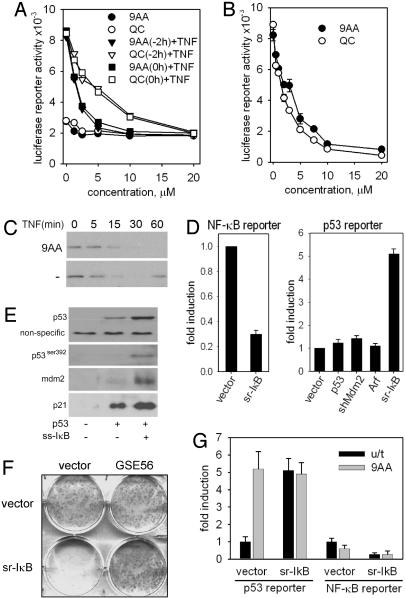
9AA and QC Induce p53 by Inhibiting NF-κB. IκBα inhibits NF-κBby anchoring it in the cytoplasm; it is encoded by an NF-κB-inducible gene, acting as part of a negative feedback regulatory loop (11). A 9AA-induced decrease in IκBα could be explained either by increased degradation of the protein or by inhibition of NF-κB-dependent transcription. To distinguish between these possibilities, we monitored NF-κB-response in the cells treated with different concentrations of compounds added alone or combined with the NF-κB-inducing cytokine TNF. Both 9AA and QC showed a strong dose-dependent suppression of basal and TNF-induced NF-κB reporter activities (Fig. 3A) and blocked TNF-stimulated induction of IκBα (Fig. 3B). This effect was quite specific, because 9AA and QC did not change the activity of several other transcription factors, including the androgen receptor, N-Myc, CLOCK/Bmal, and Smad (data not shown). The results of global gene expression profiling by microarray hybridization confirmed the dual effect of 9AA as an inducer of p53 and an inhibitor of NF-κB transcription (see Materials and Methods; see also Tables 1 and 2, which is published as supporting information on the PNAS web site).
Fig. 3.
9AA activates p53 through inhibition of the NF-κB pathway. (A) H1299-NF-κB-Luc cells were treated with different concentrations of 9AA and QC 2 h before or simultaneously with TNF (10 ng/ml). Reporter activity was measured after 6 h of 9AA treatment. (B) Basal activity of an NF-κB-dependent reporter was measured in lysates of H1299-NF-kBLuc cells treated with different concentrations of 9AA or QC for 24 h. (C) 9AA (10 μM) inhibits reconstitution of the level of IκB protein after TNF treatment (10 ng/ml). (D) sr-IκB inhibits NF-κB transcriptional activity (Left) and activates p53 (Right) in RCC cells. Luciferase activity in ACHN cells cotransfected with p21-ConALuc and the indicated plasmids. Normalization in both assays was done by cotransfection of the pCMV-LacZ plasmid. (E) Effects of NF-κB inhibition on the activity of the p53 pathway. H1299 cells were transfected with p53 alone or p53 with sr-IκB. Levels of p53 (total, with DO1 antibody, and p53 phosphorylated at Ser-392), IκB, p21, and Mdm2 were analyzed by Western blot 36 h after transfection. (F) The toxicity of NF-κB for tumor cells is p53-dependent. A photograph of plates with colonies of HT1080 cells, control (vector) or transduced with GSE56, after 7 days in puromycin after transduction with lentiviral vectors expressing sr-IκB or insert-free vector (both conferring puromycin resistance). (G) Luciferase activity in ACHN cells cotransfected with the NF-κB-responsive reporter pNF-κBLuc and sr-IκB. Twelve hours after transfection, the cells were split and treated with 9AA (10 μM). NF-κB-dependent reporter activity was measured 6 h after treatment, and p53-dependent reporter activity was measured 24 h after treatment. Normalization was done by cotransfection of the pCMV-LacZ plasmid (for cells transfected by different plasmids) or by determining protein concentrations (for 9AA-treated and untreated cells).
Are the effects of 9AA on p53 activation and NF-κB repression interrelated and which is the primary event? We could readily exclude the possibility that p53 activation drives NF-κB repression because all of the effects of 9AA on the NF-κB pathway were seen in p53-deficient (H1299, PC3) and in p53 wild-type (HT1080, RCC) cells. To explore the alternative model (NF-κB repression by 9AA drives p53 activation), we analyzed p53 activity in the cells with NF-κB inhibited by genetic approach. To suppress NF-κB activity, we used IκB superrepressor (sr-IκB), a stable IκB mutant lacking both phosphorylation sites (10). Transduction of this mutant into RCC ACHN cells resulted in a three-fold inhibition of NF-κB reporter activity (Fig. 3D), consistent with the constitutive activity of NF-κB in RCC cell lines (12). Importantly, the activity of a p53-responsive reporter was increased up to five times upon transduction of sr-IκB (Fig. 3D). A similar effect was observed in another RCC line, RCC54, as well as in non-RCC HT1080 cells (data not shown). Remarkably, sr-IκB activated p53 in RCC, whereas such direct regulators of p53 activity, as Arf, shRNA to Mdm2, or p53 itself failed to do it (Fig. 3D). Moreover, ectopic expression of sr-IκB led to the accumulation of p53, a consequent increase in p21 and Mdm2, and an increase in p53 phosphorylated at Ser-392, all seen after 9AA treatment (Figs. 2A and 3E).
Similar to treatment with 9AA, ectopic expression of sr-IκB was toxic to HT1080 and RCC45 cells, reflecting their addiction to constitutively active NF-κB, and, again similar to 9AA, this toxicity was p53-dependent and was greatly reduced by expression of either anti-p53 shRNA or by of the dominant-negative p53 mutant protein GSE56 (Fig. 3F). These results provide a plausible explanation for otherwise puzzling reports indicating resistance to apoptosis of cells selected for sr-IκB expression (13, 14). This resistance probably resulted from inactivation of p53 function.
Because stable expression of sr-IκB interfered with RCC cell viability, the effect of 9AA on cells with repressed NF-κB was tested in transient transfection experiments, in which the introduction of sr-IκB was combined with either NF-κB- or p53-responsive reporters. 9AA could not further activate a p53-dependent reporter in the presence of sr-IκB (Fig. 3G), indicating that its p53-activating effect is indeed mediated through inhibition of NF-κB and that NF-κB activity is responsible for the repression of p53 activity in RCC and other types of tumor cells.
The only known mechanism of mutual negative regulation of p53 and NF-κB is their competition for CBP/p300 transcriptional coactivators (15). We overexpressed CBP/p300 ectopically and traced their effects on p53 activity in HT1080 and ACHN cells, alone and in combination with sr-IκB (Fig. 8, which is published as supporting information on the PNAS web site), concluding that the NF-κB-mediated inhibition of p53 activity in these cells cannot be explained by depletion of transcriptional coactivators and must be due to another mechanism. The dominant nature of p53 repression (4) and the unusually slow kinetics of p53 derepression upon NF-κB inhibition in RCC suggest that the NF-κB-mediated p53 repression is due to an inhibitory factor encoded by an NF-κB-responsive gene, which is depleted when NF-κB is not active. It is likely that identification of their direct molecular target(s) of 9AA and QC will help to uncover new important regulatory components of NF-κB- and p53-dependent signaling that provide interactions between these two major stress-responsive pathways.
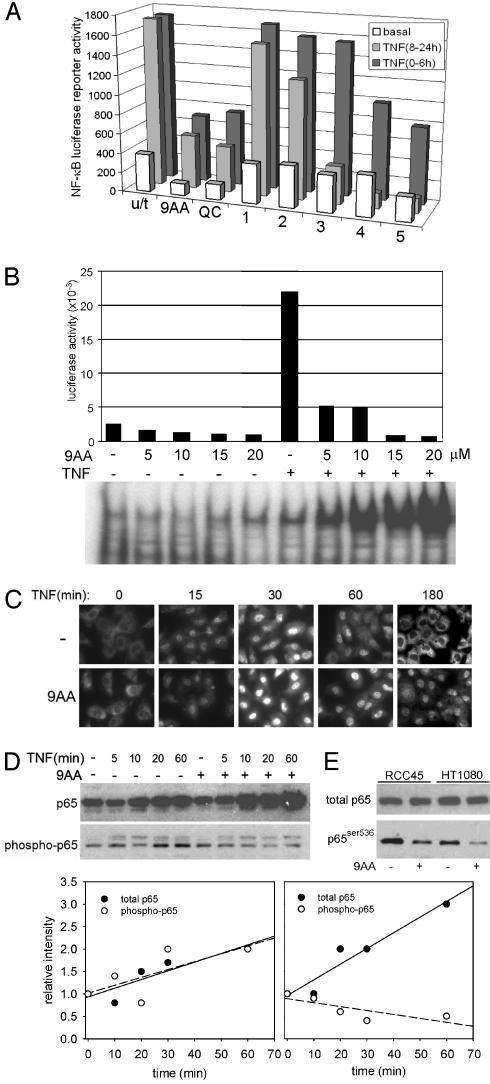
9AA and QC Represent a Previously Uncharacterized Type of NF-κB Inhibitor. Because NF-κB appeared to be the primary target of 9AA, we focused on the mechanism of 9AA-mediated NF-κB suppression. 9AA and QC were capable of inhibiting both basal and TNF-induced activities of NF-κB in H1299 (Fig. 4 A and B), RCC45, RCC54 (Table 2), and other cells. Inhibition of the basal activity of NF-κB is a unique property of 9AA compounds, as known NF-κB inhibitors, sulfasalazine (16), capsaicin (17), and Bay11-7082 (18), tested side-by-side with 9AA, did not reduce the basal activity of an NF-κB (Fig. 4A), which explains why they were not capable of activating p53 (data not shown). Moreover, 9AA and QC can inhibit NF-κB when added before, simultaneously with, or after TNF stimulation (Figs. 3A and 4A), whereas known NF-κB inhibitors required several hours of pretreatment to be effective.
Fig. 4.
Effects of 9AA on the NF-κB pathway. (A) Comparison of the effects of 9AA and QC with those of known inhibitors of NF-κB. H1299-NF-κBLuc cells were treated with 9AA (10 μM), QC (10 μM), and inhibitors of NF-κB: sulfasalzine, capsaicin, and Bay11-7082 (1-3). Inhibitors of phosphatidylinositol 3-kinase (wortmannin and Ly294002; 4 and 5) were also assayed, because their effects on NF-κB have been demonstrated. Several effective concentrations were used, based on published data, and the highest are presented. TNF (10 ng/ml) was added simultaneously (0-6 h) with inhibitors or 8 h after inhibitors (8-24 h). Luciferase activity was measured after 6 or 24 h and normalized to protein concentrations. (B) 9AA causes nuclear accumulation of inactive NF-κB. H1299-NF-κBLuc cells were treated with different concentrations of 9AA and TNF (10 ng/ml). After 6 h, cytoplasmic and nuclear extracts were isolated and used for luciferase or gel-shift assays, respectively. 9AA retards the exit of p65 complexes from the nuclei. (C) Immunofluorescent staining of p65 subunit of NF-κB in HT1080 cells treated with 9AA (10 μM) and TNF (10 ng/ml). (D) 9AA decreases the phosphorylation of p65 in response to TNF. (Upper Left) Western analysis of total cell lysates of HT1080 cells, treated with TNF (10 ng/ml) in the presence or absence of 9AA (10 μM) for the indicated periods of time. The same membrane was probed with antibodies against total p65 and against phospho-536 of p65. (Lower) Quantitation of the experiment presented in Upper Left by using Bio-Rad quantityone software. (E) Western analysis with antibodies against phosphor-Ser-536 of p65 in complexes immunoprecipitated from HT1080 or RCC45 cells by anti-p65 antibodies.
Another distinction between 9AA and other NF-κB inhibitors was its paradoxical effect on the DNA binding by NF-κB. Simultaneously with the inhibition of TNF-stimulated NF-κB-dependent transcription, 9AA significantly increased the binding of NF-κB to DNA that correlated with the nuclear accumulation of p65-containing NF-κB complexes (Fig. 4B). As opposed to NF-κB inhibitors that lock transcription complexes in the cytoplasm by inhibiting the phosphorylation and degradation of IκBα (11, 19, 20), 9AA and QC not only allowed the TNF-induced nuclear translocation and accumulation of NF-κB, but greatly prolonged the time of its presence in nuclei (Fig. 4C). Thus, 9AA and QC act by converting NF-κB to a transcriptionally inactive state that becomes trapped in the nucleus. This mechanism seems to be more relevant to antiinflammatory properties of QC, which was used previously to treat rheumatoid disease, than the inhibition of phospholipase A2 (PLA2), traditionally considered to be the QC target (21). It is noteworthy that other PLA2 inhibitors we tested (thioetheramide-PC, arachidonyl trifluoromethyl ketone, and others) could neither induce p53 nor inhibit NF-κB (data not shown).
Several factors can affect the activity of NF-κB, including composition of the complex (such as the stoichiometry of p65 and p50 subunits and the presence of transcriptional coactivators or corepressors), posttranslational modifications of components of the complexes (i.e., phosphorylation or acetylation of p65), or modification of histones (i.e., deacetylation or phosphorylation) in chromatin near sites of initiation of NF-κB-dependent transcription (5). Although treatment with 9AA did not significantly change the composition of NF-κB complexes, as judged by gel-shift assays with antibodies to different components (Fig. 9, which is published as supporting information on the PNAS web site), it did reduce the proportion of phosphorylated Ser-536 in the p65 subunit of NF-κB, both basally and after TNF-dependent induction (Fig. 4 D and E). This modification of p65 by IκB kinase 1 or other kinases is essential for the NF-κB activity in several cell systems, and lack of this phosphorylation can convert NF-κB into a transrepressor that acts by recruiting histone deacetylases (HDAC) (22, 23). Remarkably, this modification of p65 was reported to be responsible for inhibition of p53 in human T-lymphotrophic virus-infected T lymphocytes, mediated by the tax protein (ref. 24 and references therein). These observations suggest that inhibition of Ser-536 phosphorylation could be the primary mechanism of 9AA-mediated inhibition of NF-κB. In accordance with this model, the inhibition of HDAC activity by trichostatin A (TSA) resulted in a strong activation of NF-κB-dependent transcription that could no longer be blocked by 9AA (Fig. 10, which is published as supporting information on the PNAS web site). Ser-536 is phosphorylated by numerous kinases, including IκB kinase 1, NIK, TBK1, PKC, and PKA (25), each of which could be the target for 9AA. For example, IκB kinase 1 is responsible for basal NF-κB activity in several cell types (26) and is involved in the control of NF-κB activity at the promoter level (27). Moreover, a deficit in IκB kinase 1 activity increases the half-life of p65 in nuclei (28). All these traits are affected by 9AA treatment, making this kinase a primary suspect for being the 9AA target.
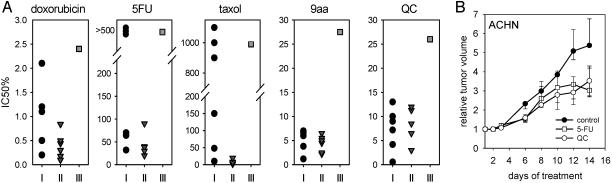
Perspectives of Antitumor Applications of QC. Because RCC is one of the most drug-resistant tumors, we assessed whether 9AA-based compounds would be more potent in killing RCC cells than conventional chemotherapeutic agents by comparing doxorubicin, taxol, and 5-fluorouracil with 9AA and QC in a set of RCC and non-RCC-derived tumor cells (six of each type) and in normal kidney epithelium. The average IC50 in RCC was higher than in non-RCC cells for all chemotherapeutic agents used and near the IC50 of normal cells. However, the IC50 of 9AA and QC for RCC cells were in the same range as for non-RCC cells (Fig. 5A). Moreover, QC inhibited the growth of tumor xenografts formed by s.c. injection of ACHN cells into nude mice. QC showed the same antitumor effect as 5-fluorouracil but without the significant weight loss (up to 20%) that accompanied treatment with this drug (Fig. 5B). We also noted that the p53-inducing activity of QC is not limited to tumor cells in culture but can be also detected in short-term organ cultures of surgically removed human RCCs (see Fig. 11, which is published as supporting information on the PNAS web site).
Fig. 5.
9AA and QC as prospective anti-RCC agents. (A) Comparison of IC50 doses of 9AA, QC, and several anticancer agents in different RCC and non-RCC cells. The IC50 for each cell line and each drug was determined as described in Supporting Materials. Each point represents the IC50 of particular types of cells, which are grouped as follows: (i) circles, RCC cell lines (ACHN, RCC9, RCC13, RCC29, RCC45, and RCC54); (ii) triangles, non-RCC cell lines (MCF7, HT1080, H1299, U20S, LNCaP, and HCT116); (iii) squares, normal kidney cells (NKE). (B) Antitumor activity of QC. ACHN cells (107) were inoculated under the skins of nude mice. When tumors reached 5 mm in diameter, daily i.p. administration of QC (50 mg/kg) or 5-fluorouracil (35 mg/kg) was started. Tumor sizes are presented as the fold increase in tumor volume.
In conclusion, 9AA can be viewed as the prototype of a previously uncharacterized class of bitargeted anticancer drugs that attack simultaneously and in the desirable direction of two important stress-responsive pathways. The ability to simultaneously inhibit NF-κB and activate p53 makes 9AA-based compounds potentially useful against tumors that, like RCC, maintain wild-type p53 in a state that is completely or partially repressed by constitutively active NF-κB (12, 29). Presence of QC among these compounds, the drug with favorable pharmacological and toxicological properties and a long history of human use as an antimalaria and antiarthritis agent (30), opens the opportunity of rapid clinical evaluation of this approach.
Supplementary Material
Acknowledgments
We thank Preet Chaudhary and Elena Feinstein for discussions and advice; Peter Chumakov, Inder Verma, and Nickolay Neznanov for recombinant constructs; and Marieta Cepec for help in preparing the manuscript. This work was supported by National Institutes of Health Grants CA60730 and CA75179 (to A.V.G.).
Conflict of interest statement: No conflicts declared.
Abbreviations: NKE, normal kidney epithelium; 9-AA, 9-aminoacridine; QC, quinacrine; RCC, renal cell carcinomas; shRNA, small hairpin RNA; sr-IκB, IκB superrepressor.
References
- 1.Prives, C. & Hall, P. A. (1999) J. Pathol. 187, 112-126. [DOI] [PubMed] [Google Scholar]
- 2.Olivier, M., Hussain, S. P., Caron de Fromentel, C., Hainaut, P. & Harris, C. C. (2004) IARC Sci. Publ. 157, 247-270. [PubMed] [Google Scholar]
- 3.Gudkov, A. V. (2005) in The p53 Tumor Suppressor Pathway and Cancer, ed. Zambetti, G. (Springer, New York), Vol. 2.
- 4.Gurova, K. V., Hill, J. E., Razorenova, O. V., Chumakov, P. M. & Gudkov, A. V. (2004) Cancer Res. 64, 1951-1958. [DOI] [PubMed] [Google Scholar]
- 5.Orlowski, R. Z. & Baldwin, A. S., Jr. (2002) Trends Mol. Med. 8, 385-389. [DOI] [PubMed] [Google Scholar]
- 6.Neznanov, N., Neznanova, L., Kondratov, R. V., Burdelya, L., Kandel, E. S., O'Rourke, D. M., Ullrich, A. & Gudkov, A. V. (2003) J. Biol. Chem. 278, 3809-3815. [DOI] [PubMed] [Google Scholar]
- 7.Zwelling, L. A., Hinds, M., Chan, D., Mayes, J., Sie, K. L., Parker, E., Silberman, L., Radcliffe, A., Beran, M. & Blick, M. (1989) J. Biol. Chem. 264, 16411-16420. [PubMed] [Google Scholar]
- 8.Sohn, T. A., Bansal, R., Su, G. H., Murphy, K. M. & Kern, S. E. (2002) Carcinogenesis 23, 949-957. [DOI] [PubMed] [Google Scholar]
- 9.Vassilev, L. T., Vu, B. T., Graves, B., Carvajal, D., Podlaski, F., Filipovic, Z., Kong, N., Kammlott, U., Lukacs, C., Klein, C., et al. (2004) Science 303, 844-848. [DOI] [PubMed] [Google Scholar]
- 10.Baldi, L., Brown, K., Franzoso, G. & Siebenlist, U. (1996) J. Biol. Chem. 271, 376-379. [DOI] [PubMed] [Google Scholar]
- 11.Panwalkar, A., Verstovsek, S. & Giles, F. (2004) Cancer 100, 1578-1589. [DOI] [PubMed] [Google Scholar]
- 12.Oya, M., Takayanagi, A., Horiguchi, A., Mizuno, R., Ohtsubo, M., Marumo, K., Shimizu, N. & Murai, M. (2003) Carcinogenesis 24, 377-384. [DOI] [PubMed] [Google Scholar]
- 13.Ryan, K. M., O'Prey, J. & Vousden, K. H. (2004) Cancer Res. 64, 4415-4418. [DOI] [PubMed] [Google Scholar]
- 14.Fujioka, S., Sclabas, G. M., Schmidt, C., Niu, J., Frederick, W. A., Dong, Q. G., Abbruzzese, J. L., Evans, D. B., Baker, C. & Chiao, P. J. (2003) Oncogene 22, 1365-1370. [DOI] [PubMed] [Google Scholar]
- 15.Webster, G. A. & Perkins, N. D. (1999) Mol. Cell Biol. 19, 3485-3495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Muerkoster, S., Arlt, A., Witt, M., Gehrz, A., Haye, S., March, C., Grohmann, F., Wegehenkel, K., Kalthoff, H., Folsch, U. R. & Schafer, H. (2003) Int. J. Cancer 104, 469-476. [DOI] [PubMed] [Google Scholar]
- 17.Kim, C. S., Kawada, T., Kim, B. S., Han, I. S., Choe, S. Y., Kurata, T. & Yu, R. (2003) Cell. Signaling 15, 299-306. [DOI] [PubMed] [Google Scholar]
- 18.Cahir-McFarland, E. D., Carter, K., Rosenwald, A., Giltnane, J. M., Henrickson, S. E., Staudt, L. M. & Kieff, E. (2004) J. Virol. 78, 4108-4119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lenz, H. J. (2003) Cancer Treat. Rev. 29, Suppl, 1, 41-48. [DOI] [PubMed] [Google Scholar]
- 20.Greten, F. R. & Karin, M. (2004) Cancer Lett. 206, 193-199. [DOI] [PubMed] [Google Scholar]
- 21.Al Moutaery, A. R. & Tariq, M. (1997) Digestion 58, 129-137. [DOI] [PubMed] [Google Scholar]
- 22.Zhong, H., May, M. J., Jimi, E. & Ghosh, S. (2002) Mol. Cell 9, 625-636. [DOI] [PubMed] [Google Scholar]
- 23.Zhang, W. & Kone, B. C. (2002) Am. J. Physiol. 283, F904-F911. [DOI] [PubMed] [Google Scholar]
- 24.Pise-Masison, C. A. & Brady, J. N. (2005) Front. Biosci. 10, 919-930. [DOI] [PubMed] [Google Scholar]
- 25.Viatour, P., Merville, M. P., Bours, V. & Chariot, A. (2005) Trends Biochem. Sci. 30, 43-52. [DOI] [PubMed] [Google Scholar]
- 26.Li, X., Massa, P. E., Hanidu, A., Peet, G. W., Aro, P., Savitt, A., Mische, S., Li, J. & Marcu, K. B. (2002) J. Biol. Chem. 277, 45129-45140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yamamoto, Y., Verma, U. N., Prajapati, S., Kwak, Y. T. & Gaynor, R. B. (2003) Nature 423, 655-659. [DOI] [PubMed] [Google Scholar]
- 28.Lawrence, T., Bebien, M., Liu, G. Y., Nizet, V. & Karin, M. (2005) Nature 434, 1138-1143. [DOI] [PubMed] [Google Scholar]
- 29.Oya, M., Ohtsubo, M., Takayanagi, A., Tachibana, M., Shimizu, N. & Murai, M. (2001) Oncogene 20, 3888-3896. [DOI] [PubMed] [Google Scholar]
- 30.Wallace, D. J. (1989) Semin. Arthritis Rheum. 18, 282-296. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.