Abstract
Short RNA regulatory molecules, microRNAs, and short interfering RNAs participate in a range of developmental gene networks by base-pairing with their target sequences. Consistent with these findings, genes required for the biogenesis and function of short interfering RNAs and microRNAs, dicer (dcr-1 in Caenorhabditis elegans) and argonaute homologs, are essential for development in diverse organisms, including C. elegans. We demonstrate that genes required for the function of short RNAs synergize with the retinoblastoma tumor suppressor homolog lin-35 in negative regulation of the nuclear divisions in the intestine of C. elegans. The level of cyclin E (cye-1) expression is critical for nuclear divisions in the intestine and is elevated in double mutants in lin-35 and RNA interference pathway genes. We propose that RNA interference-related pathways cooperate with retinoblastoma in transcriptional repression of endogenous genes, an example being cyclin E.
Keywords: cyclin E
RNA interference (RNAi) was originally discovered in Caenorhabditis elegans as a mechanism of posttranscriptional gene silencing induced by exogenous dsRNA (1). It is now known that RNAi-related mechanisms play roles in inhibition of target mRNA translation (2), mRNA degradation (3), or repression of transcription (4). Processing of dsRNA in the first step of RNAi and processing of all C. elegans microRNA (miRNA) precursors requires dcr-1 (5–7). Dcr-1 is accompanied by different Argonaute proteins, Rde-1 and Alg-1/2, in the RNAi and miRNA pathways, respectively (6, 8). Surprisingly, both RNAi and miRNA pathway genes affect RNAi-induced transcriptional gene silencing (RNAi-TGS) in the soma of C. elegans (9).
A majority of human cancers have a mutation inactivating the retinoblastoma (Rb) pathway (10, 11). Three Rb-related genes are expressed in vertebrate cells, but only one member of this family exists in C. elegans (12). In both types of organisms Rb-related activities are thought to control cell proliferation and development primarily by suppressing transcription at promoters near their binding sites. The specificity of Rb proteins is mediated by their association with the E2F family of DNA-binding proteins, which heterodimerize with the DP protein (13). Proteins associated with the Rb protein can inhibit transcription by modifications of chromatin. Specifically, Rb complexes are associated with histone deacetylases and chromatin remodeling proteins (13, 14). Further, in differentiating cells, some Rb-containing complexes are associated with histone methyltransferase Suv39h, which catalyzes H3 lysine 9 (H3-K9) methylation, promoting heterochromatin formation (15, 16).
RNAi-mediated gene silencing can also function through repressive chromatin modifications in some organisms (4, 17). In fission yeast, short interfering RNAs pair in a complex of proteins with nascent transcripts to direct modifications of chromatin (18). Similar processes are thought to be active in plants (17). In C. elegans, RNAi-induced transcriptional transgene silencing has been observed in both the soma (9) and germ line (19). In both cases silencing depended on the hpl-2 gene that encodes a homolog of the heterochromatin protein 1 (20) that binds methylated H3-K9 (21). The extent of transcriptional control through the activities of short RNAs in vertebrate cells is uncertain. Mutations in genes important for RNAi have been reported to activate transcription (22, 23). However, in most cases this effect is probably caused by misregulation of the miRNA pathway where chromatin modifications and transcriptional activation are indirectly affected by posttranscriptional control of other genes. This type of activity is consistent with recent studies indicating that genes encoding miRNAs can function in both oncogenic (24, 25) and tumor-suppressive roles (26).
In this study, we demonstrate that nuclear divisions in the intestine of C. elegans are controlled synergistically by the Rb and RNAi pathways. It appears that this regulation is mediated through control of transcription of the cyclin E gene, probably by chromatin modifications. We believe that this type of inter-play between the RNAi and Rb regulation in gene expression might be general.
Materials and Methods
C. elegans Strains. Worms were maintained on nematode growth medium plates seeded with OP50 bacteria. The following strains were used: LGI, unc-13(e1091) lin-35(n745), cye-1(eh10)/dpy14(e188); LGII, lin-4(e912); LGIII, rde-4(ne299), lin-12(n137); LGV, rde-1(ne300), JR672; LGX, alg-1(gk214), elt-2::gfp/LacZ; not-integrated transgenic strains KM32, KM34, and KM25.
RNAi by Feeding. Feeding C. elegans HT115(DE3) bacteria containing L4440 plasmid with portions of genes of interest cloned between T7 promoters was done as described (9). Primers used for gene fragment PCR amplifications were as follows: alg-1, forward, atgtccggcgggccgcaatatttgcc, reverse, tcaaatagaaactaaattaaatgcc; lin-35, forward, ggttcggaatcggcgtgg, reverse, gattttgaggaactataggtc; and dcr-1, forward, ggtaagagctgatttacaatg, reverse, gaatctttaatcggtctacga.
Adult worms of elt-2::gfp or cye-1::gfp (KM32) reporter strains were placed on bacteria expressing dsRNA, and their L4 or adult progeny were used for counting intestinal nuclei or evaluating intensity of GFP expression.
DAPI Staining. Adult worms were dissected and fixed with paraformaldehyde on the poly-l-lysine-covered slides, DAPI was applied with mounting media (Vector Laboratories).
Microscopy. Worms expressing GFP were put on 2% agarose pads, and DAPI-stained worms were fixed on poly-l-lysine slides and viewed with Zeiss Axoplan 2 microscope at ×10 or ×40 lens magnification by using the appropriate filter. openlab 3.1.7 software (Improvision, Lexington, KY) was used for capturing images, and the same exposure time was used for capturing images to be compared in the same experiment.
Chromatin Immunoprecitipation (ChIP). For preparation of protein lysates ≈300–500 μl of packed L3-L4 staged animals was used for each immunoprecipitation. Formaldehyde fixing, lysate preparation, and sonication was done according to ref. 27. ChIP was performed with anti-dimethyl-histone H3(Lys-79) antiserum (07-366, Upstate Biotechnology, Lake Placid, NY) according to the manufacturer's protocol.
RT-PCR and Real-Time PCR. Tri Reagent (Molecular Research Center, Cincinnati) was used for total RNA preparation. RNA was DNase-treated with DNA-free reagent (Ambion, Austin, TX). C. elegans RNA (0.5–1 μg) was used in 20 μl of RT reaction with random hexamer primers (Ambion) or gene-specific primers. RT reaction (2 μl 1:10 diluted) was added into 25 μl of real-time PCR. The TaqMan system (Applied Biosystems) was used for cDNA amplification, and SYBR green dye (Qiagen, Valencia, CA) was used for the amplification of DNA in ChIP assays. Reactions were run in triplicate on an Applied Biosystems Prism 7000 Real-Time PCR machine. Relative fold changes were calculated by using the 2–ΔΔCt method. TaqMan probe and primer sets were designed by using primer express software (Applied Biosystems), and probes for mature mRNAs were designed to span exon–exon junction. The sequences used were as follow: for cye-1 mRNA, forward primer, cgggacggctgctctatttata, reverse, gatgagcaaaatcgatgcactta, probe, ctaaatacgaggaaatttatccac; and for ama-1 mRNA, forward primer, gatgatccgatgaatgatggaaag, reverse, cggtatgatggttgatagcgacc, probe, aggtcgcaggtggatg. Primers used for the detection of the antisense RNA at cye-1 locus were as follows: primer 1, tgtctgacatctccggag; primer 2, tagattcaaatcgcgcgctc; primer 3, agtcgttatcttcctatttatttg; and primer 4, acacgttcagccgtcctt. Primers 3 and 4 were used for cye-1 ChIP PCR amplification, and control act-3 primers were gcagaaggaaatcaccgctcttg and gcgatgatcttgatcttcatgg.
Results and Discussion
Down-regulation of alg-1/2 by RNAi results in a range of developmental abnormalities (6). The most severe phenotype in worms lacking both alg-1 and alg-2, or, both maternal and zygotic complements of dcr-1, is embryonic lethality (6). However, careful examination of the phenotypes in dcr-1 and alg-1/2 hypomorphic mutants generated by RNAi revealed possible roles of these genes in regulation of cell division.
Postembryonic divisions in several intestinal cells of C. elegans include nuclear divisions that are not followed by cytokinesis. In addition, DNA endoreplication occurs at the end of each larval molt, leading to 32C (C or chromatin value is a multiple of the normal haploid genome) ploidy of gut cells in the adult animals (28). Expression of the elt-2::gfp reporter (29) in the intestinal nuclei provides a convenient system for studying regulation of nuclear divisions (30–32). C. elegans larvae are born with 20 intestinal nuclei, and the number reaches 30–34 in the adult animals, resulting from 10–14 postembryonic nuclear divisions during the L1 molt (28). Mutants of cyclin D, cyd-1, do not increase the number of intestinal nuclei, whereas double mutants for lin-35 (Rb homolog) and cki-1 (p21 and p27 homolog) show excessive divisions, resulting in the numbers of the nuclei reaching 70–80 per animal (30).
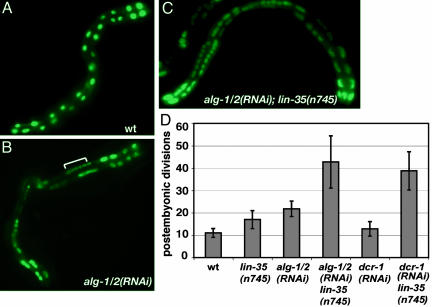
We examined the number of intestinal nuclei in alg-1/2(RNAi) animals where RNAi was introduced by feeding (33). alg-1/2(RNAi) resulted in an increase in the number of postembryonic nuclear divisions from 10 in WT to 20 in the mutant (Fig. 1 B and D). This increase was greatly potentiated when worms mutant for lin-35(n745) were subject to alg-1/2 RNAi with the numbers of intestinal nuclei reaching 70, resulting from 50 nuclear divisions (Fig. 1 C and D). We next tested whether dcr-1(RNAi) would have a similar effect. Because dcr-1 is required for RNAi, only weak down-regulation of its activity can be achieved by dcr-1(RNAi). However, global maturation of miRNAs is inhibited in dcr-1(RNAi) worms (6). When worms were subjected to dcr-1(RNAi) by feeding we did not observe any obvious abnormalities, including increase in the intestinal nuclear divisions (Fig. 1D). However, lin-35(n745) worms treated with dcr-1(RNAi) showed a large increase in the number of intestinal nuclei, similar to that of lin-35(n745);alg-1/2(RNAi) double mutants (Fig. 1D). We also observed extra nuclei in dcr-1;lin-35 and alg-1/2;lin-35 worms when these were scored with other intestinal reporters or nontransgenic worms stained with DAPI (Fig. 2A and data not shown). Thus, the observed increase is not specific for the transgenic strain used.
Fig. 1.
dcr-1 and alg-1/2 negatively regulate nuclear divisions in the intestine. (A) The elt-2::gfp reporter strain shows 30 intestinal nuclei. (B) The number of intestinal nuclei increases in alg-1/2(RNAi) animals because of extra divisions of some nuclei (bracket). (C) The number of intestinal nuclei increases dramatically in lin-35(n745);alg-1/2(RNAi) animals. For A–C ×10 lens magnification was used. (D) Quantification of postembryonic nuclear divisions in the intestine (number of nuclei in adult worms after subtraction of 20 nuclei present in L1) in different genetic backgrounds. For all figures, intestinal nuclei were counted in 15–30 progeny of several worms subjected to RNAi by feeding, and data for each genotype are presented as a mean number ± SD. RNAi feeding experiments were repeated at least three times.
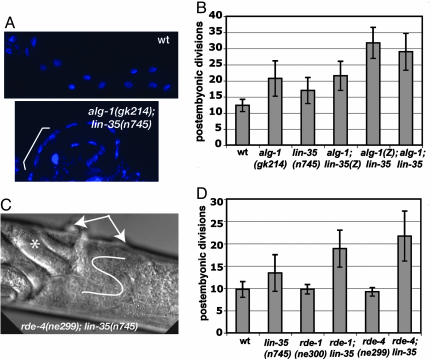
Fig. 2.
miRNA and RNAi pathway genes synergize with lin-35(Rb). (A) Dissected intestines of WT (Upper) and alg-1(gk214);lin-35(n745) double mutant (Lower) adult worms were stained with DAPI. The bracket in Lower highlights the increased number of nuclei. A ×10 lens magnification was used, and exposure times were identical. (B and D) Quantification of postembryonic intestinal nuclear divisions in different genetic backgrounds with elt-2::gfp transgenic strains; alg-1(Z) and lin-35(Z) indicate that only the zygotic complement of the gene product was missing, whereas the maternal contribution was still present. (C) A differential interference contrast microscopy image of the midsection of the representative rde-4(ne299);lin-35(n745) mutant worm at ×40 lens magnification. Ventral is top; double vulva protrusions are shown by arrows, and abnormal gonad migration with two U-turns is indicated by the white line. Note the accumulation of the late-stage embryos (*) caused by egg-laying defects.
An alg-1(gk214) deletion mutant has recently become available from the Gene Knockout Consortium (www.celeganskoconsortium.omrf.org). We examined intestinal divisions in this mutant by using the elt-2::gfp transgene and found an increase similar to alg-1/2(RNAi) (Fig. 2B). In the double alg-1(gk214) and lin-35(n745) mutant worms, the number of the intestinal nuclei was further increased (Fig. 2B). We found that Lin-35 is maternally required for the negative regulation of cell divisions in the intestines of the progeny, whereas Alg-1 is required zygotically (Fig. 2B).
Because both Dcr-1 and Alg-1/2 are essential for miRNA processing, these results indicate that miRNAs might play a role in the regulation of intestinal nuclear divisions. Further, loss-of-function mutations in the lin-4 miRNA gene have been shown to affect nuclear divisions in the intestine by up-regulation of Lin-14 (34). Thus, loss of lin-4 miRNA could account for some of the alg-1/2 phenotype. However, the number of intestinal divisions in alg-1/2(RNAi) worms considerably exceeds that observed in lin-4(e912) null mutant (Fig. 6, which is published as supporting information on the PNAS web site). Further, because alg-1/2(RNAi) does not result in complete inactivation of the alg-1 and alg-2 genes and consequently lin-4 production is not completely silenced, it is unlikely that the observed phenotype of alg-1/2 is caused solely by the loss of lin-4 miRNA. It is possible that the loss of other miRNAs whose synthesis depends on Alg-1/2 could contribute in an indirect fashion to the increase in intestinal nuclear divisions. Some of those miRNAs might synergyze with lin-4 in regulation of Lin-14.
It is also possible that other RNAi-related processes contribute to the increase in nuclear divisions in alg-1/2(RNAi) worms. For example, endogenous ALG-1/2 RNA-induced silencing complexes might contain not only miRNAs, but also endogenous short interfering RNAs (35), and such complexes might be directly involved in the initiation of silencing at the level of transcription. This activity could account for the role of alg-1/2 in regulation of nuclear divisions in the intestine and in the RNAi-TGS process of transgene silencing described recently (9).
The proposed effect of alg-1/2 on nuclear divisions through a RNAi-TGS pathway suggests that classical RNAi pathway genes might have a role in regulation of cell division. Null mutants in the RNAi pathway genes, rde-1(ne300) and rde-4(ne299), do not have any obvious developmental phenotypes (36), and they do not affect nuclear divisions in the intestine as single mutants (Fig. 2D). However, we observed a synergistic effect leading to an increased number of nuclei when either rde-1(ne300) or rde-4(ne299) was combined with lin-35(n745) (Fig. 2D). One possible explanation for these genetic interactions is that both the Rb and RNAi pathways are regulating common targets at a transcriptional level.
Genetic interaction between the RNAi pathway and Rb in C. elegans is not limited to the control of intestinal nuclear divisions. alg-1(gk214);lin-35(n745) double mutant worms are very sick, and the strain is barely viable, with brood size of 1–10 per animal. Most of the eggs die inside the mothers. Those worms surviving to adulthood are smaller in size, have egg-laying defects, and have defects in the germ line. rde-4(ne299);lin-35(n745) and rde-1(ne300);lin-35(n745) double mutant strains resemble alg-1(gk214);lin-35(n745), but are more viable. We also observe gonad migration defects (Fig. 2C) and bi-vulva animals at a low frequency (6%) in the rde-4(ne299);lin-35(n745) strain.
We wanted to examine which step in the cell cycle is regulated by lin-35 and RNAi pathway genes. Lin-35, the C. elegans Rb protein homolog, is an important negative regulator of the G1 → S transition and one of the major targets of the Cyd-1/Cdk-4 complex (30). In mammals and flies, cyclin E is one of the major targets transcriptionally activated by E2F and negatively regulated by the Rb protein (37, 38). Cyclin E is essential for development in flies and mammals (39, 40). It plays a central role in the cell cycle regulation of endoreplicating tissues. In these tissues, oscillation of cyclin E is required for cycles of DNA replication: increases in cyclin E levels cause entry into S phase, and decreases are required for the reloading of prereplication complexes (41). Therefore, either cyclin E loss-of-function mutations or overexpression disrupt endocycles in Drosophila (41). Mice with knockout mutations in both cyclin E genes display defects in endoreplicating cells in the placenta and in megakaryocytes (40).
Because endoreplication occurs in the intestinal cells of C. elegans, we tested for a possible role of cye-1 (cyclin E) in regulation of postembryonic nuclear divisions in the intestine. The maternal contribution of cyclin E is essential for C. elegans development (42, 43). cye-1(RNAi) has been reported to cause embryonic arrest and early larval arrest (42, 43), whereas cye-1 homozygous mutants (progeny of heterozygous mothers) grow up to become sterile adults (42). Fay and Han (42) observed that cye-1 mutants display defects in some postembryonic divisions and also are defective in DNA endoreplication in the intestinal cells. Because postembryonic nuclear divisions in the intestine have not been analyzed in cye-1 mutants, we examined this process in cye-1(eh10) homozygous and heterozygous animals in a transgenic elt-2::gfp background. Interestingly, postembryonic intestinal nuclear divisions are very rare in cye-1(eh10) mutants, with most adult worms containing only the 20 nuclei present at hatching (Fig. 3A). This defect is apparently not rescued by maternal contributions of cyclin E, and this finding contrasts with the modest dependence on zygotic cyclin E expression of other lineages examined by Fay and Han (42). A gain-of-function mutation in Cdc25.1, a phosphatase that activates cyclinE/Cdk2 kinase, has been shown to cause ectopic nuclear divisions exclusively in the intestinal cell lineage (32). Thus, it is likely that cyclin E levels are rate-limiting for nuclei divisions in the intestine. Consistent with this possibility, we were not able to detect large increases in the number of cell divisions in response to alg-1(RNAi);lin-35(RNAi) or dcr-1(RNAi);lin-35(RNAi) in other lineages under conditions where we readily detected dramatic increases in the number of intestinal nuclei (Supporting Results, which are published as supporting information on the PNAS web site).
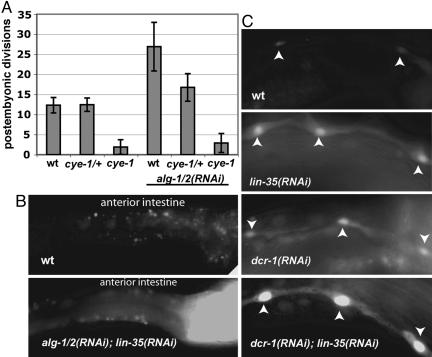
Fig. 3.
The level of cyclin E is critical for nuclear divisions in the intestine and is affected by lin-35 and RNAi pathways. (A) Quantification of postembryonic intestinal divisions in WT, cye-1(eh10) heterozygous, and homozygous animals in the absence (Left) and presence (Right) of alg-1/2(RNAi). (B) Expression of cye-1::gfp (KM32) reporter in the anterior intestine of adult WT type (Upper) or alg-1/2(RNAi);lin-35(RNAi)(Lower) animals. (C) Expression of cye-1::gfp reporter in the hypodermal seam cells (arrowheads) of adult WT worms (Top) or worms treated with RNAi against the indicated genes. Exposure times in all images in B and C were identical.
We tested whether the effect of alg-1/2 in promoting nuclear divisions in the intestine depended on cyclin E. Indeed, the cye-1(eh10) mutation completely suppressed the alg-1/2(RNAi) phenotype in the intestine (Fig. 3A), and worms heterozygous for the cye-1 mutation did not respond to alg-1/2(RNAi) to the same extent with increases in nuclei number as WT worms. These results are consistent with cyclin E being limiting for nuclear divisions in the intestine and with the possible regulation (direct or indirect) of cyclin E by short RNAs.
We hypothesized that cye-1 transcription might be negatively regulated by Lin-35 and RNAi pathways. As a prediction of this model, expression of a cye-1::gfp reporter (43) was examined and expression of cyd-1::gfp and cdk-4::gfp reporters was tested in control experiments. These latter two reporters were not affected under the conditions described below (data not shown). cye-1::gfp expression was significantly elevated in the lin-35(RNAi);alg-1/2(RNAi) (15 of 15 animals scored) or lin-35(RNAi);dcr-1(RNAi) (20 of 20) adult animals (Fig. 3 B and C) compared with transgenic worms not treated with RNAi (16 of 16 animals had very low cye-1::gfp expression as shown in Fig. 3 B and C Upper). Most notably, the increased expression of cyclin E reporter was observed in the intestine (Fig. 3B, compare Upper and Lower) and in the hypodermal seam cells (Fig. 3C, compare WT and mutants). We conclude that dcr-1 and lin-35 synergize in repressing cye-1 transcription as cye-1::gfp derepression was enhanced in double dcr-1(RNAi);lin-35(RNAi) animals compared with dcr-1(RNAi) and lin-35(RNAi) single mutants (Fig. 3C). Enhanced expression of cye-1::gfp reporter in the seam cells in response to dcr-1(RNAi);lin-35(RNAi) indicates that regulation of cye-1 expression by RNAi and Rb is not limited to the intestinal cells. Seam cells with high levels of cyclin E do not undergo ectopic cell divisions consistent with the suggestion that intestinal cells are more sensitized to cyclin E levels.
Interestingly, in Drosophila, both cyclin E mutations or overexpression of cyclin E cause defects in the endoreplication of extremely polyploid nuclei (≈2,048 C) in salivary glands (41). These defects are consistent with finding that oscillations of cyclin E levels are required for endocycles (41). Whereas intestinal nuclei in cye-1(eh10) mutants have dramatically reduced amounts of DNA, as has been reported (42), we have not observed significant changes in DNA content in intestinal nuclei of alg-1(gk214);lin-35(n745) mutant worms where increases in the number of nuclei in the gut are most significant (Fig. 2A). It is possible that C. elegans intestinal cells respond to the increase in cyclin E levels by continuing mitotic division before the endocycles.
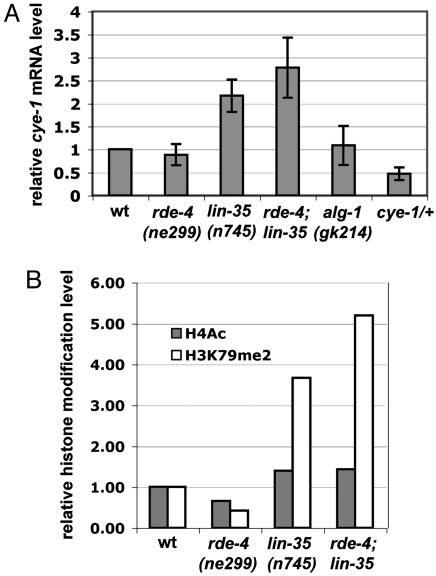
To further confirm the effect of lin-35 and RNAi pathway genes on cye-1 expression, we performed real-time RT-PCR experiments to evaluate the expression level of endogenous cye-1 in various genetic backgrounds at the time of occurrence of nuclear divisions in the intestine (larval stage L1). cye-1 mRNA expression was elevated in lin-35(n745) mutants and further enhanced in the double rde-4(ne299);lin-35(n745) mutants (Fig. 4A), which is consistent with our genetic data. We were not able to test the alg-1(gk214);lin-35(n745) double mutant strain by RT-PCR analysis because of its limited viability.
Fig. 4.
Molecular analysis of the regulation of cye-1 expression suggests a direct role for the RNAi pathway. (A) Real-time RT-PCR analysis of the expression levels of cye-1 mRNA in different mutant backgrounds. Levels of cye-1 mRNA were normalized to ama-1 mRNA levels. Results of three independent experiments are shown as mean relative numbers ± SD. (B) Quantification of the results of ChIP experiments by real-time PCR with primers 3 and 4 and the indicated antibodies. For each DNA sample PCR with actin-specific primers was used for normalization of the signal. Relative ratios are presented where PCR signal from control DNA samples from WT worms is taken as 1. Sequences for primers used are in Materials and Methods.
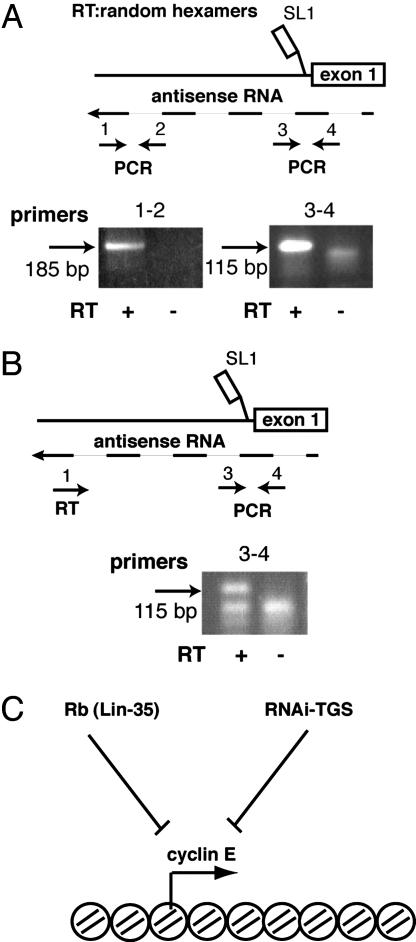
Because Rb is a known negative regulator of cyclin E transcription in other systems it is likely to directly affect cye-1 expression in C. elegans as well. We have recently shown that both alg-1 and RNAi pathway genes are involved in the RNAi-TGS of the transgenic arrays (9), and it is possible that cye-1 is directly regulated by the RNAi-TGS pathway. Human cyclin E2 locus has been included in the list of genes with proven bidirectional transcription that may generate dsRNA (44). We used RT-PCR to detect any transcripts arising from the known 5′ regulatory region (43) in the cye-1 promoter 600 bp upstream of the SL1 transsplice acceptor site. We detected RNA from the upstream and downstream promoter regions when the RT reaction was primed with random hexamers (Fig. 5A). This transcript has a polarity antisense to cye-1 mRNA as it could be detected only with antisense, and not sense, strand-specific RT primers (Fig. 5B and data not shown). Thus, there could be dsRNA formation between this antisense RNA and cye-1 pre-mRNA at the start site of cye-1 transcription.
Fig. 5.
Antisense transcription at the cye-1 locus. (A and B) Schematic representations of the 600-bp region upstream of the SL1 acceptor site in cye-1 gene and locations of the primers used for RT and PCRs are shown at the top. Results of the RT-PCRs detecting antisense transcription at the cye-1 locus are shown at the bottom. The identity of the bands was confirmed by sequencing. Sequences for primers used are in Materials and Methods. (C) Model proposing negative regulation of cyclin E gene expression by parallel Rb and RNAi-TGS pathways.
Next, we used ChIP to test for possible changes in histone modifications associated with active transcription at the cye-1 start site in different genetic backgrounds. We found that the level of modification by histone H3-K79 dimethylation at the cye-1 start site was enhanced in the lin-35 mutant background and was further enhanced in the lin-35;rde-4 double mutants (Fig. 4B). This chromatin modification is associated with actively transcribed genes (45). Under the same conditions, we were not able to detect an increase in histone H4 acetylation, another modification commonly associated with active transcription. Interestingly, up-regulation of the leukemogenic genes in some types of leukemia was recently correlated with hypermethylation of H3-K79 at the promoter regions of these genes (46).
Our results indicate that an increase in the cyclin E levels in Rb and RNAi double mutant background causes ectopic nuclear divisions in the intestine and suggest that both Rb and RNAi pathways might be directly involved in the negative regulation of cyclin E gene transcription by affecting chromatin modifications (Fig. 5C), although additional indirect inputs into cyclin E regulation and existence of other target genes affecting nuclear divisions cannot be excluded. Specifically, miRNAs might be involved in negative regulation of transcription factors activating cye-1 expression and thus might be partially responsible for observed alg-1/2(RNAi) phenotypes.
This work suggests that the RNAi pathway is tumor-suppressive in C. elegans. Importantly, there are indications that this role for the RNAi-related processes might be conserved in mammals (47, 48). Recent work describing enhanced RNAi in Rb pathway mutants in C. elegans (49) is consistent with our findings. It suggests that the Rb pathway is responsible for the repression of a set of germ line-expressed genes in the somatic tissues of C. elegans. This repression could be at the level of chromatin as shown here for cyclin E and could involve the RNAi-TGS pathway.
Supplementary Material
Acknowledgments
We thank J. Sinskey for excellent technical assistance; R. Chan for advice on ChIP; L. Aleman for advice on real-time PCR; C. Peterson and A. Seila for critical reading of the manuscript; members of the Sharp laboratory for discussions; and C. Mello (University of Massachusetts, Worcester), S. van den Heuvel (Massachusetts General Hospital, Charlestown), M. Krause (National Institute of Diabetes and Digestive and Kidney Diseases, Bethesda), B. Horvitz (Massachusetts Institute of Technology), and A. Pasquinelli (University of California at San Diego, La Jolla) for providing strains and reagents. The alg-1(gk214) deletion mutant strain was provided by the C. elegans Reverse Genetics Core Facility at the University of British Columbia, Vancouver, which is part of the International C. elegans Gene Knockout Consortium. Some strains used in this study were obtained from the Caenorhabditis Genetics Center (University of Minnesota, Minneapolis), which is funded by the National Institutes of Health National Center for Research Resources. This work was supported by U.S. Public Health Service MERIT Award R37-GM34277 from the National Institutes of Health (to P.A.S.), National Cancer Institute Grant PO1-CA42063 (to P.A.S.), Damon Runyon Cancer Research Foundation Fellowship DRG 1724-02 (to A.G.), and Cancer Center Support (Core) Grant P30-CA 14051 from the National Cancer Institute.
Conflict of interest statement: No conflicts declared.
Abbreviations: Rb, retinoblastoma; RNAi, RNA interference; miRNA, microRNA; RNAi-TGS, RNAi-induced transcriptional gene silencing; ChIP, chromatin immunoprecipitation.
References
- 1.Fire, A., Xu, S., Montgomery, M. K., Kostas, S. A., Driver, S. E. & Mello, C. C. (1998) Nature 391, 806–811. [DOI] [PubMed] [Google Scholar]
- 2.Olsen, P. H. & Ambros, V. (1999) Dev. Biol. 216, 671–680. [DOI] [PubMed] [Google Scholar]
- 3.Elbashir, S. M., Lendeckel, W. & Tuschl, T. (2001) Genes Dev. 15, 188–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Volpe, T. A., Kidner, C., Hall, I. M., Teng, G., Grewal, S. I. & Martienssen, R. A. (2002) Science 297, 1833–1837. [DOI] [PubMed] [Google Scholar]
- 5.Bernstein, E., Caudy, A. A., Hammond, S. M. & Hannon, G. J. (2001) Nature 409, 363–366. [DOI] [PubMed] [Google Scholar]
- 6.Grishok, A., Pasquinelli, A. E., Conte, D., Li, N., Parrish, S., Ha, I., Baillie, D. L., Fire, A., Ruvkun, G. & Mello, C. C. (2001) Cell 106, 23–34. [DOI] [PubMed] [Google Scholar]
- 7.Ketting, R. F., Fischer, S. E., Bernstein, E., Sijen, T., Hannon, G. J. & Plasterk, R. H. (2001) Genes Dev. 15, 2654–2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tabara, H., Yigit, E., Siomi, H. & Mello, C. C. (2002) Cell 109, 861–871. [DOI] [PubMed] [Google Scholar]
- 9.Grishok, A., Sinskey, J. L. & Sharp, P. A. (2005) Genes Dev. 19, 683–696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sherr, C. J. & McCormick, F. (2002) Cancer Cell 2, 103–112. [DOI] [PubMed] [Google Scholar]
- 11.Yamasaki, L. (2003) Cancer Treat Res. 115, 209–239. [DOI] [PubMed] [Google Scholar]
- 12.Lu, X. & Horvitz, H. R. (1998) Cell 95, 981–991. [DOI] [PubMed] [Google Scholar]
- 13.Frolov, M. V. & Dyson, N. J. (2004) J. Cell Sci. 117, 2173–2181. [DOI] [PubMed] [Google Scholar]
- 14.Zhang, H. S. & Dean, D. C. (2001) Oncogene 20, 3134–3138. [DOI] [PubMed] [Google Scholar]
- 15.Nielsen, S. J., Schneider, R., Bauer, U. M., Bannister, A. J., Morrison, A., O'Carroll, D., Firestein, R., Cleary, M., Jenuwein, T., Herrera, R. E. & Kouzarides, T. (2001) Nature 412, 561–565. [DOI] [PubMed] [Google Scholar]
- 16.Ait-Si-Ali, S., Guasconi, V., Fritsch, L., Yahi, H., Sekhri, R., Naguibneva, I., Robin, P., Cabon, F., Polesskaya, A. & Harel-Bellan, A. (2004) EMBO J. 23, 605–615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zilberman, D., Cao, X. & Jacobsen, S. E. (2003) Science 299, 716–719. [DOI] [PubMed] [Google Scholar]
- 18.Motamedi, M. R., Verdel, A., Colmenares, S. U., Gerber, S. A., Gygi, S. P. & Moazed, D. (2004) Cell 119, 789–802. [DOI] [PubMed] [Google Scholar]
- 19.Robert, V. J., Sijen, T., van Wolfswinkel, J. & Plasterk, R. H. (2005) Genes Dev. 19, 782–787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Couteau, F., Guerry, F., Muller, F. & Palladino, F. (2002) EMBO Rep. 3, 235–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lachner, M., O'Carroll, D., Rea, S., Mechtler, K. & Jenuwein, T. (2001) Nature 410, 116–120. [DOI] [PubMed] [Google Scholar]
- 22.Muljo, S. A., Ansel, K. M., Kanellopoulou, C., Livingston, D. M., Rao, A. & Rajewsky, K. (2005) J. Exp. Med. 202, 261–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Harfe, B. D., McManus, M. T., Mansfield, J. H., Hornstein, E. & Tabin, C. J. (2005) Proc. Natl. Acad. Sci. USA 102, 10898–10903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.He, L., Thomson, J. M., Hemann, M. T., Hernando-Monge, E., Mu, D., Goodson, S., Powers, S., Cordon-Cardo, C., Lowe, S. W., Hannon, G. J. & Hammond, S. M. (2005) Nature 435, 828–833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.O'Donnell, K. A., Wentzel, E. A., Zeller, K. I., Dang, C. V. & Mendell, J. T. (2005) Nature 435, 839–843. [DOI] [PubMed] [Google Scholar]
- 26.Johnson, S. M., Grosshans, H., Shingara, J., Byrom, M., Jarvis, R., Cheng, A., Labourier, E., Reinert, K. L., Brown, D. & Slack, F. J. (2005) Cell 120, 635–647. [DOI] [PubMed] [Google Scholar]
- 27.Chu, D. S., Dawes, H. E., Lieb, J. D., Chan, R. C., Kuo, A. F. & Meyer, B. J. (2002) Genes Dev. 16, 796–805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lambie, E. J. (2002) BioEssays 24, 38–53. [DOI] [PubMed] [Google Scholar]
- 29.Fukushige, T., Hawkins, M. G. & McGhee, J. D. (1998) Dev. Biol. 198, 286–302. [PubMed] [Google Scholar]
- 30.Boxem, M. & van den Heuvel, S. (2001) Development (Cambridge, U.K.) 128, 4349–4359. [DOI] [PubMed] [Google Scholar]
- 31.Boxem, M. & van den Heuvel, S. (2002) Curr. Biol. 12, 906–911. [DOI] [PubMed] [Google Scholar]
- 32.Kostic, I. & Roy, R. (2002) Development (Cambridge, U.K.) 129, 2155–2165. [DOI] [PubMed] [Google Scholar]
- 33.Timmons, L. & Fire, A. (1998) Nature 395, 854. [DOI] [PubMed] [Google Scholar]
- 34.Ambros, V. & Horvitz, H. R. (1984) Science 226, 409–416. [DOI] [PubMed] [Google Scholar]
- 35.Ambros, V., Lee, R. C., Lavanway, A., Williams, P. T. & Jewell, D. (2003) Curr. Biol. 13, 807–818. [DOI] [PubMed] [Google Scholar]
- 36.Tabara, H., Sarkissian, M., Kelly, W. G., Fleenor, J., Grishok, A., Timmons, L., Fire, A. & Mello, C. C. (1999) Cell 99, 123–132. [DOI] [PubMed] [Google Scholar]
- 37.Geng, Y., Eaton, E. N., Picon, M., Roberts, J. M., Lundberg, A. S., Gifford, A., Sardet, C. & Weinberg, R. A. (1996) Oncogene 12, 1173–1180. [PubMed] [Google Scholar]
- 38.Duronio, R. J. & O'Farrell, P. H. (1995) Genes Dev. 9, 1456–1468. [DOI] [PubMed] [Google Scholar]
- 39.Knoblich, J. A., Sauer, K., Jones, L., Richardson, H., Saint, R. & Lehner, C. F. (1994) Cell 77, 107–120. [DOI] [PubMed] [Google Scholar]
- 40.Sherr, C. J. & Roberts, J. M. (2004) Genes Dev. 18, 2699–2711. [DOI] [PubMed] [Google Scholar]
- 41.Edgar, B. A. & Orr-Weaver, T. L. (2001) Cell 105, 297–306. [DOI] [PubMed] [Google Scholar]
- 42.Fay, D. S. & Han, M. (2000) Development (Cambridge, U.K.) 127, 4049–4060. [DOI] [PubMed] [Google Scholar]
- 43.Brodigan, T. M., Liu, J., Park, M., Kipreos, E. T. & Krause, M. (2003) Dev. Biol. 254, 102–115. [DOI] [PubMed] [Google Scholar]
- 44.Yelin, R., Dahary, D., Sorek, R., Levanon, E. Y., Goldstein, O., Shoshan, A., Diber, A., Biton, S., Tamir, Y., Khosravi, R., et al. (2003) Nat. Biotechnol. 21, 379–386. [DOI] [PubMed] [Google Scholar]
- 45.Sims, R. J., 3rd, Nishioka, K. & Reinberg, D. (2003) Trends Genet. 19, 629–639. [DOI] [PubMed] [Google Scholar]
- 46.Okada, Y., Feng, Q., Lin, Y., Jiang, Q., Li, Y., Coffield, V. M., Su, L., Xu, G. & Zhang, Y. (2005) Cell 121, 167–178. [DOI] [PubMed] [Google Scholar]
- 47.Karube, Y., Tanaka, H., Osada, H., Tomida, S., Tatematsu, Y., Yanagisawa, K., Yatabe, Y., Takamizawa, J., Miyoshi, S., Mitsudomi, T. & Takahashi, T. (2005) Cancer Sci. 96, 111–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lu, J., Getz, G., Miska, E. A., Alvarez-Saavedra, E., Lamb, J., Peck, D., Sweet-Cordero, A., Ebert, B. L., Mak, R. H., Ferrando, A. A., et al. (2005) Nature 435, 834–838. [DOI] [PubMed] [Google Scholar]
- 49.Wang, D., Kennedy, S., Conte, D., Kim, J. K., Gabel, H. W., Kamath, R. S., Mello, C. C. & Ruvkun, G. (2005) Nature 436, 593–597. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.