Abstract
We found that the PCR could be dramatically enhanced by Au nanoparticles. With the addition of 0.7 nM of 13 nm Au nanoparticles into the PCR reagent, the PCR efficiency was increased. Especially when maintaining the same or higher amplification yields, the reaction time could be shortened, and the heating/cooling rates could be increased. The excellent heat transfer property of the nanoparticles should be the major factor in improving the PCR efficiency. Different PCR systems, DNA polymerases, DNA sizes and complex samples were compared in this study. Our results demonstrated that Au nanoparticles increase the sensitivity of PCR detection 5- to 10-fold in a slower PCR system (i.e. conventional PCR) and at least 104-fold in a quicker PCR system (i.e. real-time PCR). After the PCR time was shortened by half, the 100 copies/µl DNA were detectable in real-time PCR with gold colloid added, however, at least 106 copies/µl of DNA were needed to reach a detectable signal level using the PCR reagent without gold colloid. This innovation could improve the PCR efficiency using non-expensive polymerases, and general PCR reagent. It is a new viewpoint in PCR, that nanoparticles can be used to enhance PCR efficiency and shorten reaction times.
INTRODUCTION
PCR is one of the most popular tools in molecular diagnosis. It can amplify DNA samples to a detectable signal level within a short period of time. In theory, a DNA product can be amplified and doubled in each cycle. However, primer–dimer and GC-rich regions of the template and the PCR system's heating/cooling ratio may interfere with the efficiency of the PCR (1). To increase the efficiency and yield of the PCR, two key components of PCR should be considered; reagent and equipment. The reagents include the Taq DNA polymerase, primers and the template (2–5). The Taq DNA polymerase derived from the thermalphilic bacterium Thermus aquaticus has shown the best activity at 72°C (6). Primer design, buffer content and the occurrence of primer–dimer have also been reported to affect PCR efficiency (7,8).
Recently, PCR machines showing rapid heating-cooling responses have been developed (9,10). The thermoelectric heating of the PCR used had to be restricted by the Peltier-effect because of the large thermal flux relative to the slow heating and cooling rates (11). To overcome this limitation, machines were developed (12–14) with increased surface/volume ratio and decreased content volume, such as the capillary and the microchip PCR. The surface-to-volume ratio (S/V ratio) of the PCR chamber has been increased from 1.5 to 8.0 to 20.4 as the PCR machines were developed from conventional PCR equipment to capillary PCR machines to microchip-based PCR equipment. This change, however, increased the efficiency of the PCR only 5- to 10-fold (15), and the improvements to these machines pushed the thermal efficiency to its limit. The nanoparticle is a novel material and has many physical properties which are different from bulk materials. It has been reported that the smaller the nanoparticle the higher it's thermal efficiency (16,17), and other studies have reported that thermal conductivity can be increased by >20% when suspending nanoparticles in water (1–5% v/v) (18). In liquid, nanoparticles can transport heat flux and cause thermal equilibrium with the environment within 10–200 ps. The reasons for this increase in thermal conductivity are believed to include the high S/V ratio, molecular-level layering, the effect of nanoparticle clustering and electron–phonon coupling (16–19).
In this study, both the yield and the efficiency of the PCR in reaction with Au nanoparticles are much higher than in the reaction without Au nanoparticles under the conditions of shortened reaction time or increased heating/cooling rates. In this study, we compared different PCR systems, DNA polymerases, DNA sizes and complex samples. We studied different concentrations of nanoparticles, different heating/cooling rates using different PCR machines and different holding times in denaturation, annealing and extension from an engineering viewpoint. In general, this study demonstrated that Au nanoparticles could enhance the PCR efficiency, and further shortened the PCR cycle time without decreasing the efficiency.
METHODS
Preparation of Au nanoparticles (13 nm)
Au nanoparticles were prepared in an aqueous solution following the chemical reduction method (20). In the synthesizing process, an oil bath was used to reach the boiling temperature in the presence of Hydrogen tetrachloraurate (HAuCl4, 5 mM) and Trisodium citrate dihydrate (1 mM). The Au3+ was reduced to its atom form by a reductant.
The synthesis of gold colloid was conducted under acidic condition, (pH ∼5.5–6.0). To optimize the polymerase reaction condition, the Au colloid solution (1000 µl) was condensed by high-speed centrifugation at 11 000 g for 7 min. Then 800 µl supernatant was replaced with deionized water for decreasing the amount of citrate. The optimal condition for Taq polymerase activity condition is pH ∼7.4–8.3. A water substitute of the DNA template was added to the reaction as the negative control. The final pH was 8.1–8.3.
Preparation of the PCR reagent and the DNA template
Qualified reagents were used to amplify the DNA template by real-time PCR. In 20 µl of solution, there were sufficient reagents for 45 amplification reactions. Each solution contained 2 µl of 10× PCR buffer, 2 pmol primers, 200 µM dNTPs (dATP, dCTP, dGTP and dTTP), 0.5 µg/µl BSA, 1:30 000 SYBR® Green I (Roche, Germany), 1 U of DNA polymerase, Supertherm Taq (LPI, UK) or YEA Taq (Yeasterm Biotech Co. Ltd, Taiwan), 2 µl of DNA template and 2 µl of gold colloid solution. The total volume of the solution used in the conventional PCR machine was 40 µl, which included 4 µl of gold colloid solution and maintained the same concentrations of each reagent except SYBR® Green I and BSA. The experiment used the fragments, including three different lengths of purified DNA, cDNA of bladder cancer cell line and cDNA of lung cancer tissue, as shown in Table 1. The purified DNA fragments were used within the concentration of 102–109 copies/µl to confirm the efficiency of gold colloid in real-time PCR and in conventional PCR. The BNIP3 cDNA was amplified from bladder cancer cells, UB37 as well as from colorectal tissue.
Table 1.
The DNA fragment and Primer of the experiment
| Length (bp) | Primer (5′–3′) | Gene name | GC % | |
|---|---|---|---|---|
| EGFP-I | 173 | EGFP-N-S 5′-TGCAGTGCTTCAGCCGCTAC-3′ | pEGFP-N1, Clontech | 60 |
| EGFP-N-AS 5′-CAGCTCGATGCGGTTCACCA-3′ | ||||
| PT4K2B | 752 | SP6 5′-CATACGATTTAGGTGACACTATAG-3′ | phosphatidylinositol 4-kinase type-II beta | |
| T7 5′-TAATACGACTCACTATAGGGAGA-3′ | ||||
| MS1R | 1236 | Lafmid-L: 5′-AGGAAACAGCTATGACCATGATTACGC-3′ | Macrophage stimulating 1 receptor | |
| Lafmid-R: 5′-GGTTTTCCCAGTCACGACGTTGTAA-3′ | ||||
| BNIP3 | 238 | 5′-CGCAGCTGAAGCACATCC-3′ | Bcl-2 and Nineteen Kda interacting protein-3 | 50.48 |
| 5′-AACGAACCAAGTTAGACTCCA-3′ |
The real-time PCR and conventional PCR systems
Two different types of PCR machines were used in this study, the LightCycler PCR machine (Roche Diagnostic Systems, Penzberg, Germany) and the GeneAmp PCR System 9600 (Application Biosystems, Foster, USA). The LightCycler™ uses air as a heat transfer medium contributing to the high-speed cycling capabilities, and the average cooling/heating rate was ∼20°C/s. A total volume of up to 25 µl in a glass capillary was used to carry out the amplification. The LightCycler PCR machine has three different detection modes for detecting the fluorescence intensity of the DNA binding dyes or DNA probe. Fluorescence was detected at 520, 640 and 710 nm with the aid of photo hybrids. In this study, the fluorescence intensity of SYBR Green I was detected at 520 nm mode. The ABI 9600 PCR system follows the electro-thermal theory, the thermal cycle system utilizes electroformed silver and gold plating, and the average heating/cooling rate was at 1.5–2°C/s. A total volume of up to 200 µl in a PS tube was used to carry out the amplification. The agarose gel electrophoresis was used to detect the final PCR products of the conventional PCR system, ABI 9600.
RESULTS
The role of gold colloid in the PCR
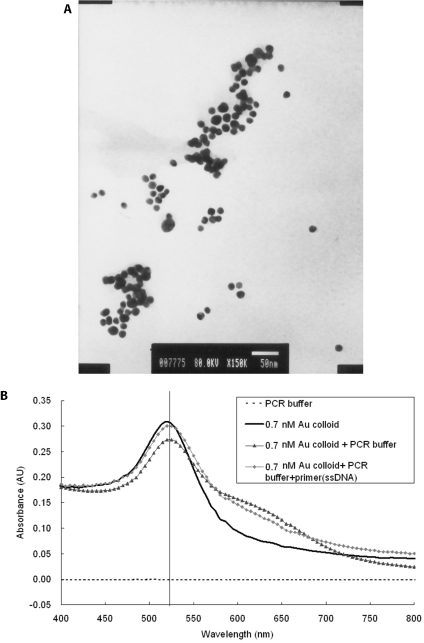
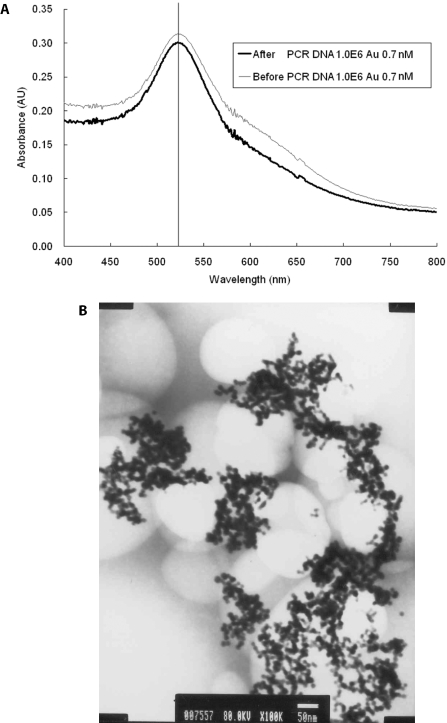
The gold nanoparticles used in this study had an average diameter of 12.7 ± 0.8 nm as measured by transmission electron microscopy (TEM) (Figure 1A). The UV-vis spectrum analysis demonstrated that the 13 nm diameter Au particles form a peak at the absorption wavelength of 520 nm. To maintain the stability of the nano-crystal, a capping layer is needed to protect the surface of the nanoparticle. A nanoparticle can be modified by adsorbing other organic molecules to make it biocompatible or to change its properties. The gold colloid uses sodium citrate as a capping layer and can be stored under sterile condition for several months. However, the PCR buffer contains 0.1 M of NaCl and MgCl2, which has been reported to cause the aggregation of Au colloid, unless a certain amount of single-stranded DNA (ssDNA) is added in the salt solution. The ssDNA will replace the citrate ions to bind to the Au colloid. Au nanoparticles were added in the PCR reagents to compare the PCR buffers with and without primers and the DNA fragments. The peaks of absorbance wavelength of Au colloid have little variety, as shown in Figure 1B, and the peak still exists after PCR (data not shown). This is due to the fact that the salt concentration in the PCR buffer was only three-tenth of that in the reference (21). In order to test the effect of citric acid, the buffer solution containing citric acid separated from gold nanoparticles solution was added in the PCR. Results showed that the citric acid will suppress the PCR (data not shown). The denaturation temperature of 95°C will not cause the aggregation of Au nanoparticles or its chemical properties to be changed, because sodium citrate cannot be reacted until the temperature reaches boiling temperature. A comparison of the OD value of the gold colloid, either before or after the PCR, shows that it still has the same peak at 520 nm of absorbance wavelength, as shown in Figure 2A. In Figure 2B, the image of TEM, shows the Au nanoparticles, which were added to the PCR reagent, and which did not change size after the PCR.
Figure 1.
Characterization of the prepared Au nanoparticle. (A) The electron microscopy image of 13 nm Au colloid. (B) Detection of Au nanoparticles by UV-vis spectrometer. In the visible region, a peak of 520 nm representing 13 nm Au nanoparticles is detected.
Figure 2.
The Au nanoparticles were not changed after real-time PCR. (A) Both of the PCR products before and after real-time PCR have absorbance in 520 nm by UV-vis spectrometer. (B) TEM image shows the Au nanoparticles were not changed (13 nm) after real-time PCR.
Based on the results of Au colloid UV-vis spectrum (Figure 1B), most of the primer and DNA proved not to bind to the gold colloid. Thus the ssDNA bound with Au colloid in salt solution cannot be the main explanation for the PCR enhancement. Contrary to the studies reported, the surface of gold colloids forms a highly ordered liquid layer, which leads to a higher thermal conductivity (16). This phenomenon can induce a fast heat transfer in DNAs that approach or bind to Au colloid and so enhance the local PCR. However, further investigation into the mechanism of gold nanoparticles affecting PCRs is required.
The exceptional efficiency in heat dispersion of Au nanoparticles in shortened PCR cycles
The heat transfer efficiency of Au nanoparticles in an aqueous solution or in oil has been reported (17). However, the physical properties of Au nanoparticles for enhancing the chemical reaction have rarely been explored. The effective concentration of gold colloid which influences the PCR efficiency was experimentally determined to be 0.7 nM. Exceeding 10 nM of 13 nm Au nanoparticles would disturb the reaction strongly. The Au nanoparticles do not enhance the yield amplification until a concentration of 0.01 nM has been reached (data not shown). When the characteristics of the DNA amplification curve were analyzed to determine the possible PCR factors affected by the Au colloid, the following factors were considered: the threshold cycle (CT), the fractional cycle number at which the fluorescence passes the fixed threshold, which is used for quantization of the PCR products. The current method of determining amplification efficiency is serial dilutions of the starting template. The input nucleic acid concentrations were varied over several orders of magnitude, and the series were usually diluted 5–10 times with sterilized water. In this study, the dilution series template molecules were amplified. The CT values were plotted against the log of the known starting concentration values, and from the slope of the regression line, the amplification efficiency was obtained. The PCR efficiency, E, can be calculated from the equation of the LightCycler instrument protocol.
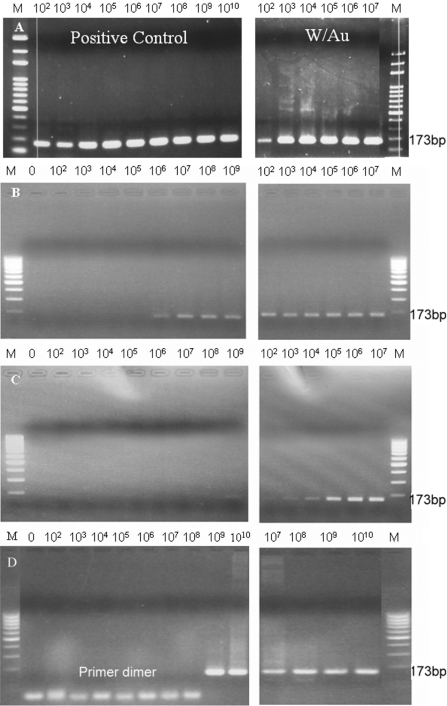
E is the efficiency of the reaction ranging from 1 to 2. Therefore, the CT value can be viewed as the PCR amplification efficiency in the same concentration of input nucleic acid of the testing kits. During PCR amplification, SYBR Green I was incorporated into the PCR product, which, in this experiment, was detected by emission fluorescence. We demonstrated that the heat conductivity of Au nanoparticles plays the most important role in increasing the efficiency of the PCR thermal reaction by dramatically shortening the time required. The low-cost test kit, SuperTherm Taq, was used in the real-time PCR system to amplify 173 bp EGFP-I DNA fragment. The PCR time with optimum amplification efficiency was chosen as the reference PCR time, which was denaturing at 95°C for 30 s, annealing at 58°C for 30 s, extending at 72°C for 60 s. The optimum amplification efficiency of the test kit without Au nanoparticles was 1.84 and the efficiency of the reagent with added Au nanoparticles was 1.72 (data not shown). The gold colloids seemed to disturb the reaction slightly. Figure 3 shows that the level of necessary initial DNA concentrations needed for obtaining visible bands after the PCR cycle times decreased. When the reaction time was shortened to one half of the reference reaction time, the amplification efficiency of the reagent with the Au nanoparticles (Eff = 1.59) became higher than that without the Au nanoparticles (Eff = 1.28), as shown in Figure 3B. The difference became larger as the reaction time was shortened. We reduced the per cycle reaction time to one-third and one-sixth of the reference condition, respectively. Figure 3D shows that in the positive control without Au nanoparticles, templates could only be detected accompanied with the formation of primer dimers at a concentration of >109. However, in the presence of gold nanoparticles, an intensive band was detected at 107 concentrations, and even more interesting, this was accomplished within only one-sixth of the reference reaction time (Figure 3D). The kinetics of the DNA synthesis curve demonstrated the same results as the PCR amplification. Commercial Au nanoparticles (Sigma) were also tested, and the results showed that commercial gold nanoparticles have a similar enhancement ability as well (data not shown).
Figure 3.
The effect of Au nanoparticles on PCR efficiency. The positive control group was compared with the gold colloid reagent group, in which 0.7 nM of gold nanoparticles were added. (A) The positive control group and the gold colloid reagent group with the reference reaction time; (B) with the reaction time reduced to half of the reference PCR time; (C) with the reaction time reduced to two-thirds of the reference PCR time; and (D) with the reaction time reduced to one-sixth of the PCR time.
PCR yield amplification by adding Au nanoparticles in quicker and slower thermal cycling PCR systems
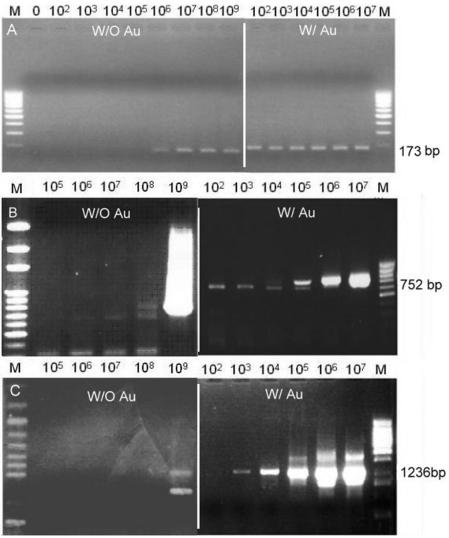
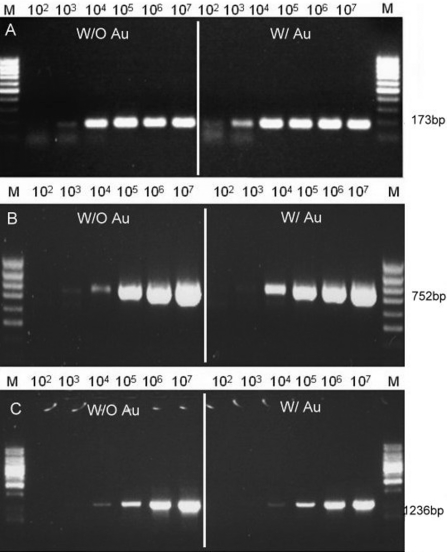
The results (Figures 4 and 5) illustrated that the rate of thermal cycling reinforced the level of enhancement of PCR by adding Au nanoparticles. In other words, the same amount of Au nanoparticles resulted in a different enhancement in quicker and slower PCR amplifications. Therefore, different experiments, in quicker thermal cycling using LightCycler and in slower thermal cycling using ABI 9600, were compared with different lengths of purification DNA fragments, EGFP-I, PT4K2B and MS1R (Table 1). Both instruments, LightCycler and ABI9600, were set in the same equilibration time of PCR temperature steps. The difference was that the heating/cooling rate of LightCycler is 10-fold faster than that of ABI 9600. When using the LightCyler with the parameters as per Table 2, the PCR yield amplification using Au nanoparticles were at least 104 times higher compared with the conditions without Au nanoparticles, as shown in Figure 4. However, using the ABI9600 PCR thermal cycler with gold colloid showed a lower enhancement ability than with the LightCycler. The addition of Au nanoparticles in ABI9600 only increased the yield amplification 5- to 10-fold compared with that without the addition of Au nanoparticles, as shown in Figure 5. With the addition of Au nanoparticles, the EGFP-I, 173 bp, was increased 10-fold, and the PT4K2B, 752 bp, was ∼5-fold. The amplification of the MS1R fragment (1236 bp) was similar to that without Au nanoparticles. With regards to heat transfer, the slower thermal cycling did not take advantage of the rapid heat exchange owing to the addition of Au nanoparticles. However, under similar amplification efficiencies, the required cycling time of the LightCycler was shortened dramatically. Au nanoparticles play an important role in the time required for heat to equilibrium, which is a very important factor for PCR thermal cycling. This conclusion was evidenced in the experiments when using the quicker thermal cycling PCR, LightCycler.
Figure 4.
The enhancement effect of Au nanoparticles in amplifying different sizes of DNA using the quicker thermal cycling PCR, LightCycler real-time PCR system. (A) EGFP-I 173 bp, (B) PT4K2B 752 bp, and (C) MSIR 1236 bp.
Figure 5.
The enhancement effect of Au nanoparticles in amplifying different sizes of DNA using the slower thermal cycling PCR, ABI 9600 conventional PCR system. (A) EGFP-I 173 bp, (B) PT4K2B 752 bp, and (C) MSIR 1236 bp.
Table 2.
The PCR system parameter setting
| DNA fragment | Denaturation | Annealing | Extension |
|---|---|---|---|
| Real-time PCR (LightCycler, Roche) detection point 83°C, 1 s | |||
| EGFP-I (173 bp) | 95°C, 5 s | 58°C, 5 s | 72°C, 10 s |
| PT4K2B(752 bp) | 95°C, 5 s | 55°C, 5 s | 72°C, 30 s |
| MS1R (1236 bp) | 95°C, 15 s | 53°C, 15 s | 72°C, 40 s |
| BNIP3 (238 bp) | 95°C, 10 s | 60°C, 5 s | 72°C, 12 s |
| BNIP3 tissue | 95°C, 10 s | 60°C, 5 s | 72°C, 10 s |
| Conventional PCR (ABI 9600; Applied Biosystem Launched) | |||
| EGFP-I (173 bp) | 95°C, 5 s | 58°C, 5 s | 72°C, 10 s |
| PT4K2B (752 bp) | 95°C, 5 s | 55°C, 5 s | 72°C, 30 s |
| MS1R (1236 bp) | 95°C, 15 s | 53°C, 15 s | 72°C, 40 s |
Determining the efficiency of adding Au nanoparticles using the real-time PCR
Conventional methods use agarose gels to detect the PCR amplification at the final phase or end point of the PCR. The agarose gel can only determine the concentration difference in the order of 5- or 10-fold. However, the real-time PCR can detect a difference as small as a 2-fold change. For analyzing the amplification effect of adding Au nanoparticles in real time, real-time PCRs were used in the main experiments. Only 106 copies of the EGFP-I DNA template are shown here. The group with 0.7 nM gold nanoparticles was compared with the positive group for CT kinetics. The CT of the Au nanoparticles reagent is 11 cycles greater than that of the positive control, which is 34.97. The target DNA was amplified much more efficiently compared with the reference reaction time. The efficiency is higher when the slope is smoother. The slope of the case with Au nanoparticles is −4.918 while the slope of the positive control is smaller (−8.566). This result indicates that the gold colloid increases the efficiency of the PCR and subsequently increases the yield of the PCR amplification.
Different DNA polymerases and DNA samples from tissue and cell
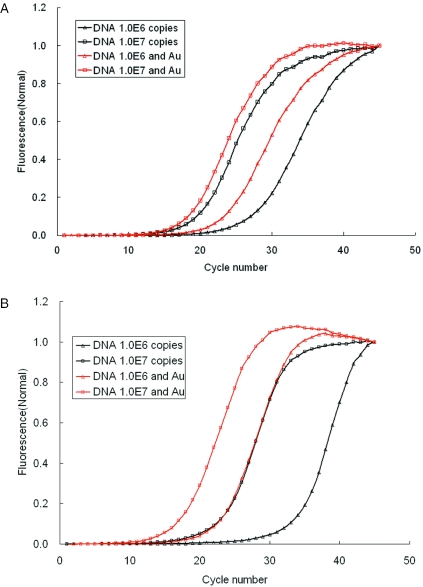
The PCR needs the DNA polymerase to amplify the DNA fragments. However, the DNA polymerase still exhibited many functional differences for complex samples and reagents. In this study we have demonstrated that the gold colloid can work with the SuperTherm DNA polymerase and YEA DNA polymerase under short time PCR conditions. The 106–107 copies of the 173 bp EGFP-I DNA template was detected by CT of the amplification curve in real-time PCR. In Figure 6A, YEA DNA polymerase was used to react with 1.0 × 107 copies of DNA, the CT values with and without Au nanoparticles are 19.1 and 20.5, respectively. For the PCR with 1.0 × 106 copies of DNA, the CT values with and without Au nanoparticles are 21.9 and 26.3, respectively. With Au nanoparticles the efficiency of the PCR was increased by 1.4–4.4 cycles. In contrast, in Figure 6B when using SuperTherm DNA polymerase to react with 1.0 × 107 copies of DNA, the CT values with and without Au nanoparticles were 18.3 and 24.3, respectively. In the PCRs with 1.0 × 106 copies of DNA, the CT values with and without Au nanoparticles were 23.9 and 35.0, respectively. With Au nanoparticles the efficiency of the PCR was increased by 6–12 cycles. Therefore, the enhancement of the PCR efficiency using Au nanoparticles depends on the DNA polymerases used. The results demonstrate that the gold colloid could also work with different Taq enzymes.
Figure 6.
The enhancement of Au nanoparticles in the efficiency of PCR depends on the DNA polymerases used. (A) Using YEA DNA polymerase to react with 1.0 × 107 and 1.0 × 106 copies of DNA. (B) Using SuperTherm DNA polymerase to react with 1.0 × 107 and 1.0 × 106 copies of DNA.
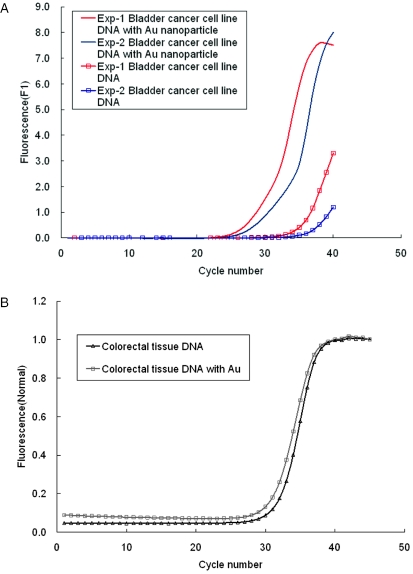
The aspect of gold colloid being able to enhance the PCR amplification can be applied directly to the detection of gene expression. The DNA template can be taken from the cells without purification. Figure 7A shows the PCR amplification of the BNIP3 gene in a bladder cancer cell line. In experiment 1, the CT values with and without Au nanoparticles are 28.6 and 38.5, respectively. In experiment 2, the CT values with and without Au nanoparticles were 27.2 and 37.3, respectively. With Au nanoparticles the average efficiency of PCR was increased by 10 cycles. In contrast, in Figure 7B, the PCR amplification of BNIP3 gene in colorectal tissue, the CT values with and without Au nanoparticles were 31.1 and 31.6, respectively. With Au nanoparticles the efficiency of PCR was increased by 0.5 cycle only. In summary, the Au nanoparticles enhanced the efficiency of the PCR using tumor cell lines and tumor samples. However, the enhancing effect of gold nanoparticles on PCR was not significant in colorectal tissue. It is possible that the contents in colorectal tissues such as ions, protease, RNase, DNase and enzyme inhibitors, suppress Taq polymerase activity in the presence of Au colloids, resulting in a reduction of the enhancing effect of Au colloids on PCR. It suggests that specific optimization of PCR conditions for diverse tissue samples are required.
Figure 7.
The Au nanoparticles enhance the efficiency of the PCR using tumor cell lines and tumor samples. (A) PCR amplification of BNIP3 gene in bladder cancer cell line. (B) PCR amplification of BNIP3 gene in colorectal tissue.
CONCLUSION
The 13 nm Au nanoparticles did indeed dramatically improve the PCR. With shortened PCR time in testing different sizes of the DNA fragments, the amplification yield of PCR reagent with Au nanoparticles was increased by at least 104- to 106-fold compared with the reagent without Au nanoparticles. Also, Au nanoparticles substantially enhanced the PCR efficiency in the quicker thermal cycler (higher heating/cooling rate). Based on these experimental results, it seems that the main reason that Au nanoparticles enhance the PCR yield amplification is related to the excellent heat dispersion of gold colloids. By applying this novel method, the amplification activity of low-cost DNA Taq enzymes with gold colloids was found to be increased in both conventional and real-time PCR systems. In conclusion, Au nanoparticles increase the reaction efficiency as well as generate a better DNA synthesis curve during real-time PCR analysis. It can be said that Au nanoparticles are extremely useful for improving the PCR.
Acknowledgments
The authors would like to thank the Center for Micro/Nano Technology Research, National Cheng Kung University, Tainan, Taiwan, People's Republic of China for access to their equipment and for their technical support. The funding from the Ministry of Education and the National Science Council of Taiwan, Republic of China under contract no. (NSC 93-2320-B-006-015) is gratefully acknowledged. Funding to pay the Open Access publication charges for this article was provided by National Science Council of Taiwan.
Conflict of interest statement. None declared.
REFERENCES
- 1.Brownie J., Shawcross S., Theaker J., Whitcombe D., Ferrie R., Newton C., Little S. The elimination of primer–dimer accumulation in PCR. Nucleic Acids Res. 1997;25:3235–3241. doi: 10.1093/nar/25.16.3235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Whitcombe D., Theaker J., Guy S.P., Brown T., Little S. Detection of PCR products using self-probing amplicons and fluorescence. Nat. Biotechnol. 1999;17:804–808. doi: 10.1038/11751. [DOI] [PubMed] [Google Scholar]
- 3.Freeman W.M., Walker S.J., Vrana K.E. Quantitative RT–PCR: pitfalls and potential. Biotechniques. 1999;26:112–125. doi: 10.2144/99261rv01. [DOI] [PubMed] [Google Scholar]
- 4.Smits C.B., Van Maanen C., Glas R.D., De Gee A.L., Dijkstrab T., Van Oirschot J.T., Rijsewijk F.A. Comparison of three polymerase chain reaction methods for routine detection of bovine herpesvirus 1 DNA in fresh bull semen. J. Virol. Methods. 2000;85:65–73. doi: 10.1016/s0166-0934(99)00153-6. [DOI] [PubMed] [Google Scholar]
- 5.Mullis K.B., Faloona F.A. Specific synthesis of Dna in vitro via a polymerase-catalyzed chain reaction. Methods Enzymol. 1987;155:335–350. doi: 10.1016/0076-6879(87)55023-6. [DOI] [PubMed] [Google Scholar]
- 6.Husmann M., Dragneva Y., Romahn E., Jehnichen P. Nuclear receptors modulate the interaction of Sp1 and GC-rich DNA via ternary complex formation. Biochem. J. 2000;352:763–772. [PMC free article] [PubMed] [Google Scholar]
- 7.Charles M.R. Quantifying gene expression. Mol. Biol. 2002;4:93–100. [PubMed] [Google Scholar]
- 8.Das S., Mohapatra S.C., Hsu J.T. Studies on primer–dimer formation in polymerase chain reaction (PCR) Biotechnol. Techniq. 1999;13:643–646. [Google Scholar]
- 9.Berg J., Nagl V., Muhlbauer G., Stekel H.A. A single-tube two-round polymerase chain reaction using the LightCycler™ instrument. J. Clin. Virol. 2001;20:71–75. doi: 10.1016/s1386-6532(00)00157-8. [DOI] [PubMed] [Google Scholar]
- 10.Ibrahim M.S., Lofts R.S., Jahrling P.B., Henchal E.A., Weedn V.W., Northrup M.A., Belgrader P. Real-time microchip PCR for detecting single-base differences in viral and human DNA. Anal. Chem. 1998;70:2013–20179. doi: 10.1021/ac971091u. [DOI] [PubMed] [Google Scholar]
- 11.Lagally E.T., Simpson P.C., Mathies R.A. Monolithic integrated microfluidic DNA amplification and capillary electrophoresis analysis system. Sensors Actuators B. 2000;63:138–146. [Google Scholar]
- 12.Chen F., Binder B., Hodson R.E. Flow cytometric detection of specific gene expression in prokaryotic cells using in situ RT-PCR. FEMS Microbiol. 2000;184:291–295. doi: 10.1111/j.1574-6968.2000.tb09029.x. [DOI] [PubMed] [Google Scholar]
- 13.Morenoa Y., Hernándeza M., Ferrúsa M.A., Alonsob J.L., Botellaa S., Montesa R., Hernándeza J. Direct detection of thermotolerant campylobacters in chicken products by PCR and in situ hybridization. Res. Microbiol. 2001;152:577–582. doi: 10.1016/s0923-2508(01)01232-3. [DOI] [PubMed] [Google Scholar]
- 14.Lin Y.C., Huang M.Y., Young K.C., Chang T.T., Wu C.T. A rapid micro-PCR system for hepatitis C virus amplification. Sensors Actuators B Chem. 2000;71:2–8. [Google Scholar]
- 15.Cheng J., Kricka L.J., editors. Biochip Technology. Singapore: Harwood Academic Publishers; p. 176. [Google Scholar]
- 16.Keblinski P., Phillpot S.R., Choi S.U.S., Eastmen J.A. Mechanism of heat flow in suspension of nano-sized particles (nanofluids) Int. J. Heat Mass Transfer. 2002;45:855–863. [Google Scholar]
- 17.Hu M., Hartland G.V. Heat dissipation for Au particles in aqueous solution: relaxation time versus size. J. Phys. Chem. B. 2002;106:7029–7033. [Google Scholar]
- 18.Zhang J.Z. Ultrafast studies of electron dynamics in semiconductor and metal colloidal nanoparticles: effects of size and surface. Acc. Chem. Res. 1997;30:423–429. [Google Scholar]
- 19.Link S., Burda C., Wang Z.L., El-Sayed M.A. Electron dynamics in gold and gold-silver alloy nanoparticles: the influence of a nonequilibrium electron distribution and the size dependence of the electron-phonon relaxation. J. Chem. Phys. 1999;111:1255–1264. [Google Scholar]
- 20.Hayat M.A. Colloidal Gold, Principle, Methods and Applications. Vol. 1. Academic press; 1989. pp. 13–31. Chap 2. [Google Scholar]
- 21.Li H., Rothberg L. Colorimetric detection of DNA sequences based on electrostatic interactions with unmodified gold nanoparticles. Proc. Natl Acad. Sci. USA. 2004;101:14036–14039. doi: 10.1073/pnas.0406115101. [DOI] [PMC free article] [PubMed] [Google Scholar]